Abstract
DNA viruses adopt various strategies to modulate the cellular environment for efficient genome replication and virion production. Previously, we demonstrated that the BGLF4 kinase of Epstein-Barr virus (EBV) induces premature chromosome condensation through the activation of condensin and topoisomerase IIα (C. P. Lee, J. Y. Chen, J. T. Wang, K. Kimura, A. Takemoto, C. C. Lu, and M. R. Chen, J. Virol. 81:5166-5180, 2007). In this study, we show that BGLF4 interacts with lamin A/C and phosphorylates lamin A protein in vitro. Using a green fluorescent protein (GFP)-lamin A system, we found that Ser-22, Ser-390, and Ser-392 of lamin A are important for the BGLF4-induced disassembly of the nuclear lamina and the EBV reactivation-mediated redistribution of nuclear lamin. Virion production and protein levels of two EBV primary envelope proteins, BFRF1 and BFLF2, were reduced significantly by the expression of GFP-lamin A(5A), which has five Ser residues replaced by Ala at amino acids 22, 390, 392, 652, and 657 of lamin A. Our data indicate that BGLF4 kinase phosphorylates lamin A/C to promote the reorganization of the nuclear lamina, which then may facilitate the interaction of BFRF1 and BFLF2s and subsequent virion maturation. UL kinases of alpha- and betaherpesviruses also induce the disassembly of the nuclear lamina through similar sites on lamin A/C, suggesting a conserved mechanism for the nuclear egress of herpesviruses.
Most DNA viruses replicate and assemble their genomes into nucleocapsids in the nuclei of infected cells. To facilitate efficient replication, viruses regulate the nuclear environment by affecting cellular chromatin and nuclear lamina (30, 35, 40). The nuclear lamina is a thin electron-dense meshwork lining the nucleoplasmic face of the inner nuclear membrane (INM) (14, 20) and provides structural support for the major components of the nuclear envelope (36, 39). The lamina also functions as a transverse scaffold for INM proteins (e.g., emerin and lamin B receptor), chromatin proteins (histone H2A/H2B dimers), and cytoskeleton-interacting proteins (nesprin1/2) (7, 52).
The nuclear lamina comprises a series of type V intermediate filaments composed of lamin types A, B1, B2, and C. Types A and C are products of RNA splicing variants of the lmnA transcripts, whereas types B1 and B2 are derived from two other genes, lmnB1 and B2 (15, 22). The INM-associated lamin B layer provides the fundamental structure of the lamina and is essential for the nuclear shape, whereas the lamin A/C layer adjacent to the nucleoplasm has more specialized functions and contributes to nuclear stiffness (7, 23, 52). Similarly to other intermediate filaments, lamins contain globular head and tail domains flanked by a central rod domain (15). The rod domains of two lamin molecules can intertwine to form dimers, whereas regions flanking the head/rod and rod/tail domains potentially interact with other lamin dimers to form longer filaments (45). Physiologically, the nuclear lamina is reorganized dynamically throughout the cell cycle via a mechanism regulated by phosphorylation. Phosphorylation by mitotic Cdc2 kinase at Ser-22, Ser-390, and Ser-392 residues on lamin A/C, or by protein kinase C (PKC) during apoptosis, leads to the depolymerization of lamin (disassembly of the nuclear lamina), which may lead to their release from the INM (11, 21, 44).
The intact meshwork of the nuclear lamina also presents a barrier to most DNA viruses. Upon infection, the viral genome may be imported by the importin β-mediated docking of viral nucleocapsid-nuclear pore complexes (NPCs) and the cytosol-dependent injection of DNA into the nuclei (19). After viral DNA replication and packaging, the ∼120-nm herpesviral nucleocapsids cannot be released into the cytoplasm through the nuclear pore, which has a diameter of 38 nm, or the nuclear lamina, which has an average crossover spacing of 52 nm (1, 2). Because assembled viral nucleocapsids must move toward and contact the INM for later budding at the cleft between the outer nuclear membrane (ONM) and the INM (primary envelopment), the structural destabilization of the nuclear lamina is required in the final stages of virus replication (reviewed in reference 37).
Indeed, infection with alpha- and betaherpesviruses results in changes to the nuclear lamina, including the reorganization and conformational changes of lamin A/C (5, 40, 45) and the redistribution of lamin B to perinuclear regions (43). Viral protein kinases have been suggested to be responsible for the changes in nuclear lamina during lytic infection. So far, two protein kinases, UL13 and Us3, were identified in alphaherpesviruses, while only UL homologs were found in beta- and gammaherpesviruses. In human herpes simplex virus type 1 (HSV-1), the changes of nuclear lamina during virus infection are attributable mainly to the phosphorylation function of Us3 kinase. The Us3 kinase induces the phosphorylation of two viral primary envelope proteins, UL34 and UL31, and cellular lamin A/C and emerin proteins are suggested to facilitate viral nucleocapsid egress (reviewed in references 37 and 40). As revealed by a deletion mutant, UL13 kinase is required for the proper subcellular localization of UL34 and UL31 (25). Because UL13 phosphorylated Us3 in vitro, the authors proposed that UL13-mediated Us3 phosphorylation is important for viral envelopment. However, the possibility of UL13 targeting nuclear lamina directly was not determined. In human cytomegalovirus (HCMV), cellular protein p32 was indicated to recruit UL97 to nuclear lamina and facilitate virion production (35). This raises the possibility that the reorganization of lamina in alphaherpesvirus-infected cells was governed mainly by its unique Us3 kinase, whereas the regulation in beta- and gammaherpesvirus was performed only by their UL kinases.
Epstein-Barr virus (EBV) is a ubiquitous human gammaherpesvirus. During lytic infection, EBV expresses BGLF4 protein kinase, which belongs to the conserved family of herpesviral UL kinases. BGLF4 can phosphorylate the viral DNA polymerase accessory factor BMRF1, EBNA-2, EBNA-LP, and BZLF as well as cellular EF-1δ and MCM4 at Ser/Thr-Pro motifs, indicating that BGLF4 is a proline-directed protein kinase (3, 9, 26, 27, 29, 58). Recently, BGLF4 was shown to initiate a DNA damage response and induce the phosphorylation of H2AX (50). The expression of BGLF4 also recruits and activates the nucleotide excision repair protein XPC to foster viral replication (32).
In our previous study, the expression of BGLF4 kinase alone was shown to induce chromosome condensation and the disassembly of the nuclear lamina (30). BGLF4 interacts with condensin complexes and induces condensin phosphorylation at Cdc2 consensus motifs. BGLF4 also stimulates the decatenation activity of topoisomerase II (Topo II), while it was not clear whether BGLF4 activates condensin and Topo II through direct phosphorylation. Here, we set out to determine how EBV BGLF4 kinase regulates the dynamics of the nuclear lamina, as well as chromosomal architecture, and to explore the mechanism used by BGLF4 to regulate the egress of nucleocapsids.
MATERIALS AND METHODS
Establishment of 293TREx_Flag_ERTA cells.
The tetracycline-regulated expression (T-REx) HEK293 cell line was purchased from Invitrogen (Carlsbad, CA) and maintained in accordance with the manufacturer's instructions. The expression plasmid pLenti4-Flag-ERTA was constructed by cloning coding sequences of N-terminal FLAG-tagged EBV BRLF1 (ERTA; NC_007605.1:C91078 to C92895) into pLenti4-TO-V5-DEST (Invitrogen), with minor modifications of the manufacturer's instructions. The expression of ERTA is controlled by the CMV/TetO2 promoter (Invitrogen) and is detectable using anti-FLAG M2 antibody (Sigma-Aldrich). Lentivirions harboring pLenti4-Flag-ERTA were produced using the ViraPower system (Invitrogen) and were used to infect T-REx HEK293 cells. Forty-eight hours after infection, the cells were selected with 400 μg of zeocin (Invitrogen)/ml for 2 weeks. Zeocin-resistant clones were subjected to doxycycline inducibility tests of ERTA expression by Western blot analysis. Finally, three independent positive clones with similar growth rates and expression levels were pooled (designated 293TREx_Flag_ERTA) and maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% tetracycline-free serum, 5 μg of blasticidin S HCl (Invitrogen)/ml, and 200 μg zeocin/ml for future study.
Establishment of EREV8 cells.
293TREx_Flag_ERTA cells were infected with recombinant Akata EBV by cocultivation with EBV-producing B cells as described previously (8). The infected cells then were selected by treatment with G418 (1.2 mg/ml; Invitrogen) and cloned by limiting dilution. The inducibility of EBV lytic gene expression by doxycycline treatment (50 ng/ml for 24 h) was examined using an immunofluorescence assay. One EBV-infected cell clone, designated 293-EREV8, showed the most significant induction of EBV reactivation upon doxycycline treatment and was used in this study.
Cell culture, virus induction, and transfection.
HeLa cells were derived from human cervical epithelial cells. EBV-negative NPC-TW01 was established from a Taiwanese nasopharyngeal carcinoma (31), and NA is recombinant Akata EBV-converted NPC-TW01 (8). All cells were cultured in DMEM (HyClone) and supplemented with 10% normal or tetracycline-depleted fetal calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C with 5% CO2. For EBV induction, NA cells were incubated with 40 ng/ml 12-O-tetradecanoylphorbol-13/acetate-3 mM sodium butyrate (TPA/SB). EREV8 cells were incubated with 100 ng/ml doxycycline (Sigma-Aldrich) to induce Rta expression and subsequent virus lytic replication. Plasmid or small interfering RNA (siRNA) was transfected using Lipofectamine 2000 in OptiMEM medium (Invitrogen). The BGLF4-targeted siRNAs, siBGLF4-1 (5′-CCCUCUAUGUAAAGCUGCCGGAGAA-3′) and siBGLF4-2 (5′-UGGGUAGGCUGGUCCUGACUGAUUA-3′), and a control siRNA (siCtrl; 5′-CCCGUAUAAAUGUCGGGCCACUGAA-3′) with comparable GC content were purchased from Invitrogen.
Plasmid construction.
pYPW17 and pYPW20 are plasmids expressing wild-type BGLF4 kinase and the K102I kinase-dead mutant, respectively (53). Flag-tagged HSV-1 UL13 (pTAG-UL13), HCMV UL97 (pTAG-UL97), and murine herpesvirus 68 (MHV68) ORF36 (pTAG-36) were generated independently by cloning BamHI-KpnI viral kinase gene fragments into pTAG-attR-C1 (Invitrogen) and were kindly provided by Ren Sun (UCLA). Flag-tagged Kaposi's sarcoma-associated herpesvirus (KSHV) ORF36 was generated by cloning the Flag-KSHV ORF36 gene fragment into the RsrII site of pLenti4-V5 (Invitrogen). For glutathione S-transferase (GST) fusion hCAP-G protein expression and purification, human condensin gene fragments hCAP-G(N-375), hCAP-G(345-764), and hCAP-G(745-C) with BamHI and SacI sites were generated by PCR using pBR322-hCAP-G (kindly provided by Keiji Kimura) as the template and fragment-specific primer pairs (data not shown). The amplicon was digested and cloned into the BamHI and XhoI sites of pCPL13, a modification of the pGEX-4T1 vector (GE Healthcare). For GST-Topo IIα(1178-C), a DNA fragment with SalI and NotI sites was generated by PCR using YEpWob6 (55) as the template and was cloned into pCPL13. For GST-lamin A fusion protein expression and purification, plasmids expressing GST-lamin A Head (amino acids [aa] 1 to 129), Rod 1 (aa 117 to 239), Rod 2 (aa 216 to 384), Tail 1 (aa 369 to 519), and Tail 2 (aa 490 to 660) proteins were designed and constructed as described previously (45). A green fluorescent protein (GFP)-tagged human lamin A expression plasmid (pEGFPhLA-WT, the wild type) was kindly provided by M. Izumi and D. M. Gilbert (24). All lamin A mutants, including GFP-lamin A(S22A), GFP-lamin A(S390A), GFP-lamin A(S392A), GFP-lamin A(S652A), GFP-lamin A(S657A), GFP-lamin A(S22/390A), GFP-lamin A(S22/392A), GFP-lamin A(S22/652A), GFP-lamin A(S22/657A), GFP-lamin A(S390/392A), GFP-lamin A(S652/657A), GFP-lamin A(4A) (which is S22/392/652/657A), and GFP-lamin A(5A) (which is S22/390/392/652/657A), were generated by a single-primer-based site-directed mutagenesis strategy (33 and data not shown).
Indirect immunofluorescence.
Slide-cultured cells were transfected with plasmids expressing BGLF4, K102I, vector pSG5, or GFP-lamin A mutants for 24 h. For BGLF4 and endogenous lamin A/C staining, the slides were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) (145 mM NaCl, 1.56 mM Na2HPO4, 1 mM KH2PO4, pH 7.2) at room temperature (RT) for 20 min, washed with PBS, and permeabilized with 0.1% Triton X-100 at RT for 5 min. The slides then were incubated with anti-BGLF4 2224 (53), rabbit anti-BGLF4 serum, anti-hCAP-E, Topo IIα (Santa Cruz), anti-lamin A/C (Santa Cruz), anti-Flag (Sigma-Aldrich), or anti-BMRF1 88A9 (51) at 37°C for 1.5 h. After extensive washes with PBS, slides were incubated with rhodamine-conjugated anti-mouse immunoglobulin G (IgG), fluorescein isothiocyanate-conjugated anti-rabbit IgG, or aminomethylcoumarin-conjugated anti-mouse IgG (Jackson) at 37°C for 1 h. Cellular DNA was stained with Hoechst 33258 at RT for 1.5 min. The staining patterns were observed under fluorescence (Axioskop 40 FL; Ziess) or confocal microscopy (Leica). Approximately 150 to 300 cells were counted for each test, and all of the experiments were repeated at least twice.
Coimmunoprecipitation assay.
HeLa cells (1 × 107) were transfected with plasmids expressing BGLF4, K102I, or vector pSG5. At 24 h posttransfection, cells were harvested and disrupted in NP-40 lysis buffer (1% NP-40, 50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM EDTA, and 1 mM Na3VO4). Cell lysates were centrifuged for 10 min at 16,000 × g to remove the insoluble fraction. Before immunoprecipitation, the lysate was precleared with 125 μl of 20% protein A-Sepharose beads (Pharmacia) for 1 h at 4°C. To immunoprecipitate condensin complexes, Topo IIα or lamin A/C lysate prepared in NP-40 buffer was incubated with hCAP-E and hCAP-G antibodies (1.5 μg each), Topo IIα antibody, or lamin A/C antibody (2 μg each) at 4°C for 1.5 h. Protein A-Sepharose beads (125 μl at 20%) then were added to pull down the immunocomplexes with rotation for 2 h at 4°C. The immunocomplexes then were washed extensively with NP-40 lysis buffer or cold PBS, disrupted in sodium dodecyl sulfate (SDS) sample buffer, and displayed in SDS-polyacrylamide gel electrophoresis (PAGE) using 8% (for condensin and Topo IIα) or 10% polyacrylamide (for lamin A/C) for immunoblotting detection as previously described (30).
Expression of BGLF4 and K102I proteins in recombinant baculovirus systems.
For recombinant BGLF4 and K102I baculoviruses, pFastBac-HTB-BGLF4 (pHYH4) and pFastBac-HTB-K102I (pHYH5) first were generated by cloning BamHI-BGLF4 or K102I-EcoRI fragments from pYPW17 or pYPW20 (53) into pFastBac-HTB vector (Invitrogen). pHYH4 and pHYH5 then were transformed into DH10Bac-competent cells that contain Bacmid (baculovirus shuttle vector) and a helper vector for BGLF4 or K102I gene recombination. After blue/white selection, Bacmid DNA harboring recombinant BGLF4 or K102I sequence (white colonies) was extracted and the sequence confirmed. The first generation of recombinant virus was produced by transfecting Bacmid DNA into insect Sf9 cells using TransFast (Promega). The recombinant viruses then were amplified according to the manufacturer's instructions (Invitrogen). For the expression of recombinant protein, Sf9 cells (1 × 107) were infected with baculovirus Bac-HTB-BGLF4 or Bac-HTB-K102I at RT for 48 to 84 h. The virus-infected cells were lysed in immunoprecipitation (IP) buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris, pH 7.4, 1 mM EGTA, 0.2 mM Na3VO4, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 0.5% NP-40, and 50 mM NaF) containing a protease inhibitor cocktail (Roche). After centrifugation (16,000 × g for 30 min at 4°C), supernatants with highly expressed protein were collected, aliquoted, and stored at −80°C.
Expression and purification of bacterially expressed GST fusion proteins.
To express GST, GST-tagged hCAP-G, Topo IIα(1178-C), and lamin A protein fragments, plasmids were transformed into Escherichia coli BL21(DE3). The bacteria then were cultured in M9TB medium (0.1% NH4Cl, 0.3% KH2PO4, 0.6% Na2HPO4, 1% tryptone, 0.5% NaCl, 1 mM MgSO4, and 0.4% glucose) until the optical density at 600 nm reached 0.6 to 0.8, and protein expression was induced with isopropylthiogalactopyranoside (IPTG; 0.5 mM for hCAP-G and 1 mM for Topo IIα or lamin A; Sigma-Aldrich) at 25°C for 2.5 h. Bacteria were pelleted and lysed in cold PBS containing 1% Triton X-100 and protease inhibitors (Roche). Lysozyme was added to a final concentration of 1 mg/ml, incubated on ice for 60 min, and frozen completely at −80°C overnight. After being quickly thawed, the lysates were sonicated as starting material and centrifuged at 16,000 × g at 4°C for 15 min. The pellet serves as the insoluble fraction. The supernatant was incubated with glutathione-agarose (50%; Sigma-Aldrich) at 4°C for 1 h. The beads then were pelleted and washed extensively with cold PBS with 1% Triton X-100, and the protein was eluted by elution buffer (50 mM Tris-HCl, pH 8.0, and 10 mM reduced glutathione). The protein expression was analyzed on 10% SDS-PAGE and detected by Coomassie blue staining or homemade anti-GST antibody.
Immunoprecipitation kinase assay.
BGLF4 kinase or kinase-dead K102I prepared from Sf9 cell lysate (100 μg) was immunoprecipitated using anti-BGLF4 2224 antibody as described previously. After collection by a brief centrifugation and extensive washing with IP buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris, pH 7.4, 1 mM EGTA, 0.2 mM Na3VO4, 0.2 mM PMSF, 0.5% NP-40, and 50 mM NaF) containing a protease inhibitor cocktail (Roche), immunocomplexes were washed once with kinase buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.5 mM dithiothreitol, 0.2 mM Na3VO4, and 0.1 mM NaF). For the kinase reaction, immunoprecipitates on protein A beads (10 μl) were incubated with kinase buffer containing 25 μM ATP, 5 μCi [γ-32P]ATP (Amersham), and 1 μg of histone H1 (Calbiochem) or 0.5 μg of purified GST fusion protein at 30°C for 30 min. After proteins were resolved by 10% SDS-PAGE, gels were dried, stained with Coomassie blue for the detection of substrate input, and subjected to autoradiography or transferred onto Hybond-C Extra membranes (Amersham) and detected with anti-BGLF4 2616 or Cdc2 antibody.
Isolation of secreted EBV particles, DNA extraction, and PCR assay.
EBV-positive EREV8 cells were mock transfected or transfected with plasmids expressing GFP-lamin A(WT), GFP-lamin A(S22/390A), or GFP-lamin A(5A). At 12 h posttransfection, cells were incubated with 100 ng/ml doxycycline (Sigma-Aldrich) for 72 h. The culture supernatants were collected, subjected to centrifugation at 10,000 × g for 30 min at 4°C, and filtered through a 0.45-μm nylon membrane to remove cell debris. To pellet viral particles, the resultant supernatant was centrifuged at 30,000 × g at 4°C for 90 min. The harvested viral particles then were incubated with DNase I to eliminate the contamination of cellular DNA as previously described (10). The viral particles and cells on the plates were lysed and prepared for PCR or immunoblot analysis. For EBV DNA detection, a 285-bp BamHI W fragment was amplified as previously described (53).
RESULTS
EBV BGLF4 interacts with and phosphorylates condensin and Topo IIα.
Previously, we showed that EBV BGLF4 kinase induces chromosome condensation through the phosphorylation of the condensin hCAP-G subunit at the MPM-2 epitopes and enhances the decatenation activity of Topo IIα (30). To identify a possible intracellular interaction between BGLF4 kinase and condensin or Topo IIα, immunofluorescence and IP assays were performed in BGLF4-transfected HeLa cells. In contrast to BGLF4-negative cells, which displayed predominantly cytoplasmic staining of condensin core subunit hCAP-E, condensin accumulated in the nuclei of BGLF4-expressing cells and colocalized with BGLF4 (Fig. 1A). In addition, the incubation of lysates with DNase I before or after IP or with 50 μg/ml ethidium bromide did not affect significantly the ability of anti-hCAP-G and -E antibodies to coimmunoprecipitate BGLF4 (data not shown), suggesting that the interaction between BGLF4 and condensin does not require the presence of DNA. In addition to condensin, we found that BGLF4 colocalizes with Topo IIα as punctate foci in transiently transfected HeLa cells and NA cells with EBV lytic infection (data not shown). A portion of BGLF4, but not K102I, could be coimmunoprecipitated with Topo IIα (data not shown).
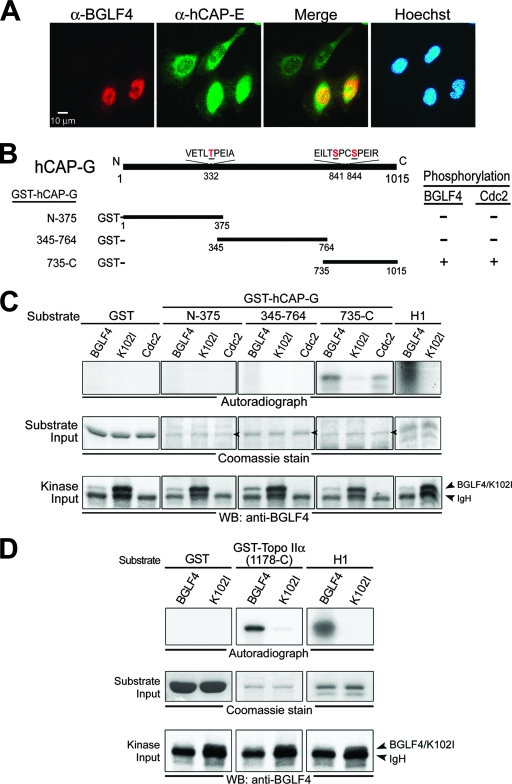
FIG. 1.
BGLF4 phosphorylates the condensin hCAP-G subunit and Topo IIα in vitro. (A) HeLa cells were transfected with a BGLF4-expressing plasmid. At 24 h posttransfection, cells were fixed with 4% paraformaldehyde and stained for BGLF4 (red), condensin hCAP-E (green), and DNA (blue) as described in Materials and Methods. BGLF4 induces hCAP-E nuclear accumulation and colocalizes with hCAP-E (yellow in the merge image) in the nucleus of transfected cells. (B) Schematic representation of human condensin hCAP-G protein. Three plasmids expressing GST fusion condensin protein, GST-hCAP-G(N-375), GST-hCAP-G(345-764) and GST-hCAP-G(735-C), were constructed and expressed in E. coli BL21(DE3) (data not shown). Putative BGLF4 and Cdc2 target sites Thr-332, Ser-841, and Ser-844 on condensin and the abilities of BGLF4 and Cdc2 to phosphorylate the condensin hCAP-G subunit are indicated. (C) Lysates from nocodazole-treated HeLa cells or insect Sf9 cells infected with BGLF4 or K102I recombinant baculovirus were immunoprecipitated with anti-Cdc2 or anti-BGLF4 2224 antibody. The immunoprecipitates then were incubated with kinase buffer containing [γ-32P]ATP in the presence of purified GST, GST-hCAP-G(N-375), GST-hCAP-G(345-764), GST-hCAP-G(735-C), or histone H1 at 30°C for 30 min. After proteins were resolved by 10% SDS-PAGE, gels were stained with Coomassie blue, dried, and subjected to autoradiography or transferred onto a nitrocellulose membrane and probed with anti-BGLF4 2616 antibody to detect the kinase input. The phosphorylation of histone H1 by BGLF4 served as a positive control for the kinase reaction. (D) The IP kinase assay was performed using recombinant baculovirus-infected Sf9 lysates and purified GST, GST-Topo IIα(1178-C), or histone H1 protein as the substrate. WB, Western blot.
The supercoiling and knotting activities of condensin and Topo IIα are activated by cellular Cdc2 kinase-induced phosphorylation (28, 56). To explore whether BGLF4 can phosphorylate condensin and Topo IIα directly, recombinant BGLF4 kinase and the kinase-dead mutant K102I were expressed in a recombinant baculovirus system. BGLF4 or K102I was immunoprecipitated from baculovirus-infected insect cell lysates, washed extensively, and incubated with pH 7.4 kinase buffer containing [γ-32P]ATP and histone H1 substrate. Significant phosphorylation signals on histone H1 were detected in the BGLF4, but not the K102I, kinase reaction, indicating that the assay conditions are suitable for the evaluation of the ability of BGLF4 to phosphorylate its target proteins (data not shown). Subsequently, bacterially expressed GST fusion protein fragments hCAP-G(N-375), hCAP-G(345-764), and hCAP-G(735-C) were used as substrates in a series of IP kinase assays (data not shown). For comparison, endogenous Cdc2 kinase was immunoprecipitated for a parallel IP kinase assay. Both BGLF4 and Cdc2 phosphorylate GST-hCAP-G(735-C) but not hCAP-G(N-375), hCAP-G(345-764), or GST protein in vitro (Fig. 1C). To explore the ability of BGLF4 to phosphorylate Topo IIα, GST-Topo IIα(1178-C) was expressed and purified as a substrate for the BGLF4 kinase assay (data not shown). We found that BGLF4, as well as Cdc2 kinase, phosphorylates GST-Topo IIα(1178-C) protein in vitro (Fig. 1D and 3C). Altogether, these results suggest that, similarly to Cdc2, BGLF4 can bind directly to and phosphorylate condensin and Topo IIα, leading to chromosome condensation.
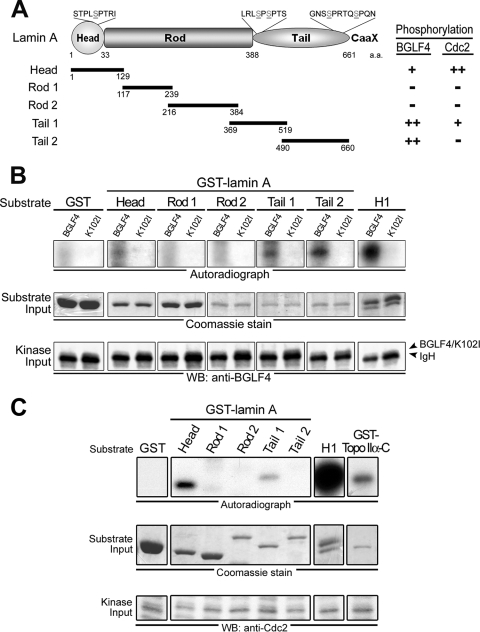
FIG. 3.
BGLF4 phosphorylates lamin A in vitro. (A) Structurally, human lamin A protein is composed of amino-terminal head, central rod, and carboxyl-terminal tail domains. GST-fused distinct fragments of lamin A (Head, aa 1 to 129; Rod 1, aa 117 to 239; Rod 2, aa 216 to 384; Tail 1, 369 to 519; Tail 2, aa 490 to 660) were expressed in E. coli and purified (data not shown). Putative BGLF4 and Cdc2 target sites on GFP-lamin A, including Ser-22, Ser-390, Ser-392, Ser-652, and Ser-657, are indicated. (B) BGLF4 or K102I proteins from insect Sf9 cells were immunoprecipitated with anti-BGLF4 2224 antibody. The IP kinase assay was performed by using purified GST, five GST-lamin A protein fragments, or histone H1 as the substrate. The results of the kinase reaction were analyzed as described in the legend to Fig. 1C. (C) Cellular Cdc2 phosphorylates lamin A and Topo IIα in vitro. HeLa cells were treated with nocodazole (100 ng/ml) for 20 h, harvested using the mechanical shake-off method, and lysed in IP buffer as described in Materials and Methods. Subsequently, IP kinase assays were performed using antibody against Cdc2 to precipitate endogenous Cdc2 kinase and purified GST-lamin A protein fragments or GST-Topo IIα(1178-C) as the kinase assay substrate. Histone H1 phosphorylation by BGLF4 served as a positive control in the kinase reaction. Inputs of kinase and the substrate were detected by anti-Cdc2 antibody or Coomassie blue staining. Cdc2 phosphorylates lamin A at Head and Tail 1 domains and phosphorylates Topo IIα at the carboxyl-terminal region. WB, Western blot.
EBV BGLF4 kinase interacts with lamin A/C and induces disassembly of the nuclear lamina.
Previously, we showed that BGLF4 kinase induces not only chromosome condensation but also the redistribution of nuclear lamin (30). To understand better the mechanism of the redistribution of the nuclear membrane in the presence of BGLF4, immunofluorescence staining was used to detect type A and C lamins in HeLa cells expressing BGLF4. As indicated in Fig. 2A, cells expressing BGLF4 show disrupted nuclear lamina with diffused, intranuclear aggregates of lamin A/C accompanied by condensed chromosomes, whereas K102I- or vector-transfected cells show an intact nuclear rim and a homogenous lamin staining pattern in the nuclei. Of note, BGLF4 colocalized with lamin A/C, especially at the inner edge of the nuclei (Fig. 2A). In addition, we found that a small proportion of BGLF4, but not K102I, was coimmunoprecipitated with lamin A/C (Fig. 2B). Taken together, these observations suggest that BGLF4 is able to interact with lamin proteins, which may subsequently affect the dynamics of the nuclear lamina.
FIG. 2.
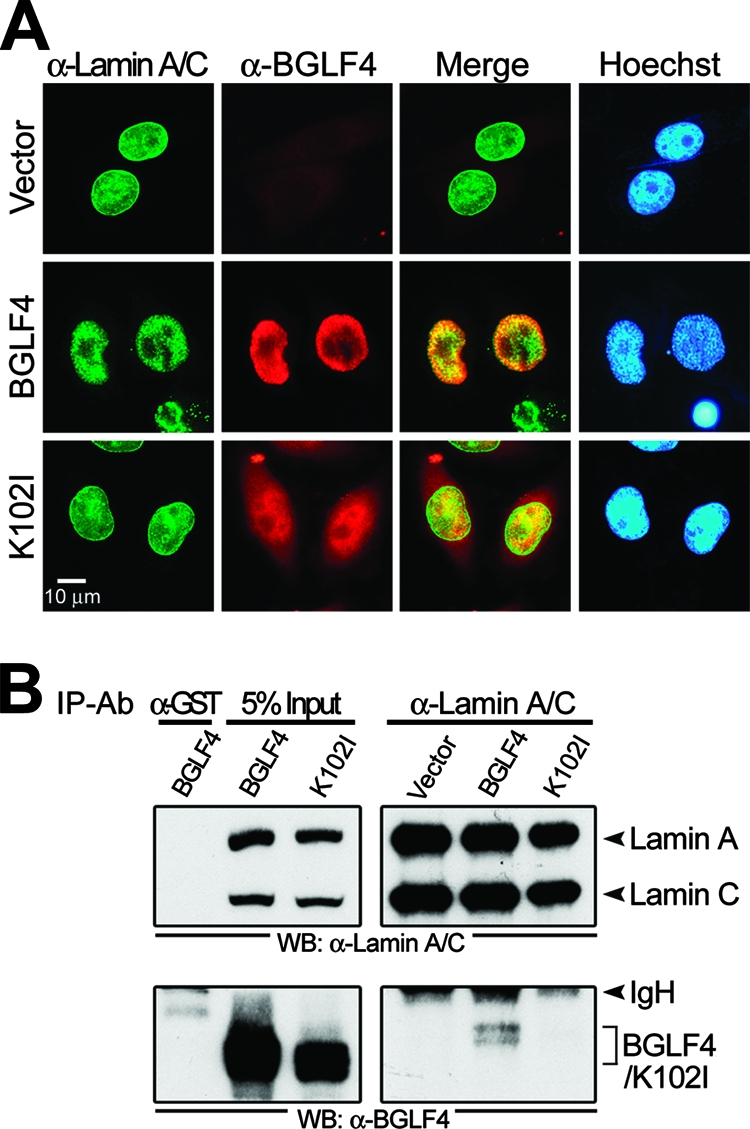
EBV BGLF4 kinase interacts with lamin A/C and induces the disassembly of the nuclear lamina accompanied by chromosome condensation. (A) HeLa cells were transfected with plasmid expressing BGLF4, K102I, or a control vector. At 24 h posttransfection, cells were fixed with 4% paraformaldehyde and stained for BGLF4 (red), lamin A/C (green), and DNA (blue) as described in Materials and Methods. BGLF4-expressing cells showed disrupted nuclear lamina with intranuclear aggregates of lamin A/C accompanied by condensed chromosomes. BGLF4 colocalized with lamin A/C in the nucleus (yellow in the merge images). (B) Lysates from HeLa cells with BGLF4 or K102I expression were immunoprecipitated with antibody against lamin A/C (α-lamin A/C) or GST (α-GST). The immunocomplexes then were resolved by 10% SDS-PAGE and immunoblotted with antibodies against lamin A/C (upper) or BGLF4 (lower). IgH, immunoglobulin heavy chain. Ab, antibody.
EBV BGLF4 kinase phosphorylates lamin A protein at multiple sites.
Because it plays a critical role in the correct localization of other nuclear envelope components, lamin A was monitored to investigate how BGLF4 regulates the integrity of the nuclear lamina. A series of recombinant GST-lamin A protein fragments was expressed in E. coli and purified as substrates for kinase assays (data not shown). For comparison, we first demonstrated that Cdc2 phosphorylated the amino-terminal Head fragment (aa 1 to 129) strongly and the carboxyl-terminal Tail 1 (aa 369 to 519) weakly (Fig. 3C), whereas BGLF4 phosphorylated the GST-lamin A Head region weakly and Tail 1 and Tail 2 (aa 490 to 660) regions strongly (Fig. 3A and B). It seems that BGLF4 and Cdc2 phosphorylate similar lamin A sites but with different efficiencies (Fig. 3A). BGLF4 phosphorylates additional sites within the Tail region of lamin A protein, implying that it has a broader kinase specificity than thought previously (27). Because the phosphorylation status of the lamin protein determines its solubility (21, 54), these results also suggest that BGLF4 induces the disassembly of the nuclear lamina through the direct phosphorylation of lamin A.
Ser-22, Ser-390, and Ser-392 of lamin A are important for BGLF4-induced disassembly of the nuclear lamina.
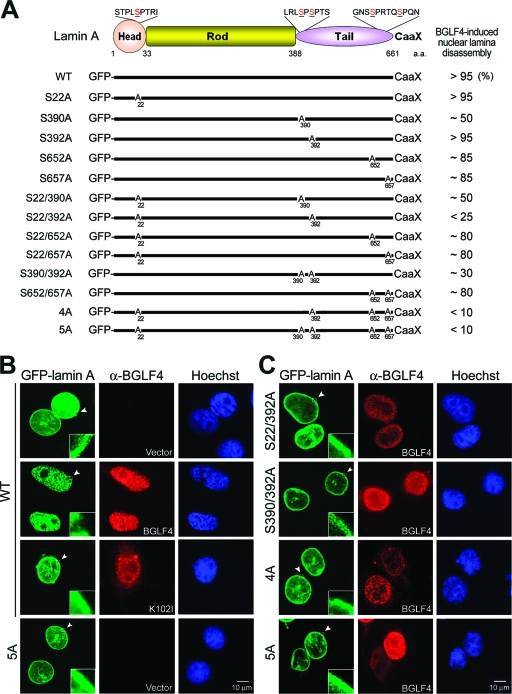
To investigate the regulatory function of BGLF4 on nuclear lamina, a chimeric GFP-lamin A, which was reported to be incorporated into the nuclear lamina with endogenous lamin A, B, and C (6, 21), was used in transient transfection. GFP-tagged wild-type lamin A localized at the nuclear rim, similarly to endogenous nuclear lamin, or associated with intranuclear tubule-like structures (Fig. 4B and data not shown). The intranuclear pattern of lamin A was reported previously to provide structural support for the nucleus (6). BGLF4, but not K102I, causes GFP-lamin A to be eliminated from the nuclear rim and dispersed into the nuclear space among condensed chromosomes, resembling disassembled nuclear lamina (Fig. 4B). These observations not only confirmed the effects of BGLF4 on endogenous lamina but also suggested that the GFP-tagged lamin A system is suitable for the study of nuclear lamina dynamics.
FIG. 4.
Confocal analysis of wild-type or mutant GFP-lamin A expression patterns in cells expressing BGLF4. (A) Schematic representation of GFP-tagged lamin A mutants. Putative BGLF4 target sites on GFP-lamin A, including Ser-22, Ser-390, Ser-392, Ser-652, and Ser-657, were mutated to Ala as indicated. The percentage of BGLF4-expressing cells with disassembled nuclear lamina was counted in 300 cells with double-positive signals of GFP and BGLF4 (data not shown) and are summarized at the right. (B) HeLa cells were cotransfected with GFP-lamin A wild type (WT), BGLF4, or K102I-expressing plasmid. At 24 h posttransfection, cells were fixed and stained for BGLF4 and DNA as described in Materials and Methods and were observed by confocal microscopy. Cells with BGLF4 expression show that GFP-lamin A released from the nuclear rim and formed intranuclear aggregates. (C) HeLa cells were cotransfected with BGLF4 and GFP-lamin A(S22/392A)-, GFP-lamin A(S390/392A)-, GFP-lamin A(4A)-, or GFP-lamin A(5A)-expressing plasmid. BGLF4 induces the disassembly of the nuclear lamina in cells with GFP-lamin A(S22/392A), GFP-lamin A(S390/392A), GFP-lamin A(4A), or GFP-lamin A(5A), estimated as 25, 30, 10, and 10% of the cells, respectively. Arrowheads indicate the regions enlarged.
Because Ser-22, Ser-390, and Ser-392 of lamin A are critical for Cdc2-mediated nuclear lamina disassembly, 13 mutants containing the putative Cdc2 and BGLF4 target sites replaced by alanine residues at various sites in GFP-Lamin A were coexpressed with BGLF4 in HeLa cells (Fig. 4 and data not shown). In the absence of BGLF4, the patterns of GFP-tagged wild-type or mutant lamin A localized at the nuclear rim were similar to that of the endogenous nuclear lamina (data not shown). In contrast, the extent of the disassembly of the nuclear lamina in cells expressing wild-type and lamin A mutants showed dramatic variation when coexpressed with BGLF4 kinase. As shown by the confocal images in Fig. 4C, BGLF4 induces the disassembly of the nuclear lamina in more than 95% of cells expressing wild-type GFP-lamin A [GFP-lamin A(WT)], GFP-lamin A(S22A), or GFP-lamin A(S392A) and in about 85% of cells expressing GFP-lamin A(S652A) or GFP-lamin A(S657A), whereas only 50% of cells expressing GFP-lamin A(S390A) showed the disassembly of the nuclear lamina. Interestingly, with S22A and an additional mutation, only 25% of cells expressing GFP-lamin A(S22/392A) were sensitive to the BGLF4-induced disassembly of the nuclear lamina, whereas GFP-lamin A(S22/390A), GFP-lamin A(S22/652A), and GFP-lamin A(S22/657A) behaved in a manner similar to that of the single mutants GFP-lamin A(S390A), GFP-lamin A(S652A), and GFP-lamin A(S657A). Data thus far indicate that Ser-22, Ser-390, and Ser-392 of lamin A mediated both the Cdc2- and BGLF4-induced disassembly of the nuclear lamina. Because the carboxyl end of lamin A (aa 490 to 660) was phosphorylated by BGLF4 in vitro, the possible contribution of this region was investigated further. BGLF4 induces the disassembly of the nuclear lamina in less than 10% of cells expressing GFP-lamin A(4A) or GFP-lamin A(5A) (Fig. 4C and data not shown). The nuclear lamina architecture in cells expressing GFP-lamin A(4A) or GFP-lamin A(5A) is more compact than that in cells with GFP-lamin A(S22/392A) or GFP-lamin A(S390/392A) expression according to confocal microscopy (Fig. 4C), suggesting that in addition to Ser-22, Ser-390, and Ser-392, both Ser-652 and Ser-657 in the Tail region of lamin A play a synergistic role in the BGLF4-mediated disassembly of the nuclear lamina.
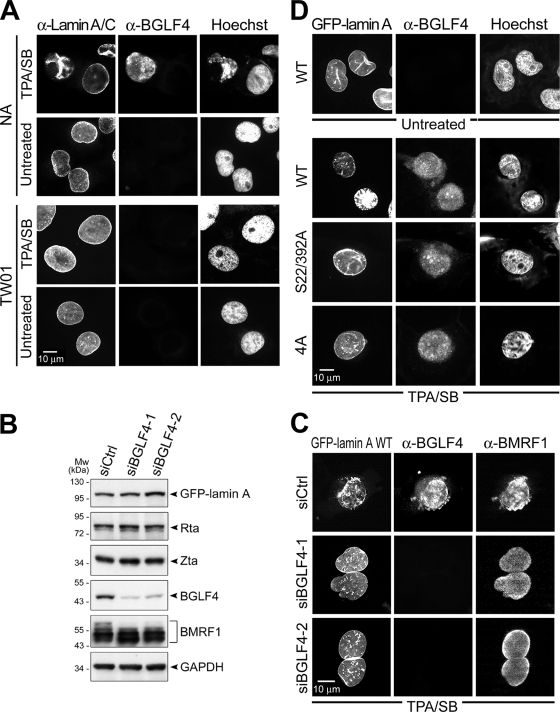
BGLF4 kinase is important for EBV reactivation-mediated redistribution of nuclear lamin.
We show by immunostaining that lamin A/C distribution was altered significantly and internalized in TPA/SB-induced EBV-positive NA cells with BGLF4 expression but not in untreated NA or TPA/SB-treated parental EBV-negative NPC-TW01 cells (Fig. 5A). Chromatin condensation also was observed only in cells with BGLF4 expression. To further demonstrate that BGLF4 plays a major role in nuclear lamin distribution during EBV reactivation, an siRNA strategy was used to knock down the expression of BGLF4 in TPA/SB-induced EBV-positive NA cells (Fig. 5B). At 48 h postinduction, BGLF4 was detected in 90% of cells with BMRF1 expression in control siRNA-transfected cells. Redistributed GFP-lamin was observed in 92% of cells with both BGLF4 and BMRF1 expression. In cells transfected with siBGLF4-1 or siBGLF4-2, less than 10% of cells with redistributed lamin were observed in BMRF1-positive, BGLF4-negative NA cells (Fig. 5C), indicating that the expression of BGLF4 is critical for the EBV reactivation-mediated redistribution of nuclear lamin.
FIG. 5.
Ser-22 and Ser-392 of lamin A are important for the redistribution of nuclear lamin induced by EBV reactivation. (A) EBV-positive NA or EBV-negative TW01 cells were left untreated or were treated with TPA/SB for EBV reactivation. At 48 h postinduction, cells were fixed and stained for lamin A/C to indicate nuclear lamina, BGLF4, and cellular DNA as described in Materials and Methods. Redistributed nuclear lamin and condensed cellular chromatin were observed in reactivated NA cells with BGLF4 expression but not in TPA/SB-treated TW01 cells. (B) NA cells were transfected with GFP-lamin A(WT) together with siBGLF4-1, siBGLF4-2, or a control siRNA (siCtrl). At 12 h posttransfection, cells were treated with TPA/SB for EBV reactivation. At 48 h posttreatment, cells were harvested for immunoblot analysis. The blots were probed with anti-BMRF1 48180 (Capricorn), anti-GFP JL-8, anti-Rta 467, anti-Zta 4F10, or anti-BGLF4 2616 in immunoblotting. GAPDH served as a loading control. (C) Slide-cultured NA cells under treatment similar to that for panel B were fixed and stained for BGLF4 (rabbit antiserum) and BMRF1 (88A9). (D) NA cells were transfected with GFP-lamin A(WT)-, GFP-lamin A(S22/392A)-, or GFP-lamin A(4A)-expressing plasmids. At 4 h posttransfection, cells were left untreated or were treated with TPA/SB to induce EBV reactivation. At 48 h posttreatment, cells were fixed and stained for BGLF4 to indicate EBV reactivation. EBV reactivation induces the disassembly of the nuclear rim and internalization in cells expressing GFP-lamin A but not in cells expressing the GFP-lamin A(S22/392A) or GFP-lamin A(4A) mutant. α-Lamin A/C, anti-lamin A/C antibody; α-BGLF4, anti-BGLF4 antibody; α-BMRF1, anti-BMRF1 antibody.
We were curious as to whether Ser-22 and Ser-392 of lamin A also are important for the EBV reactivation-mediated redistribution of nuclear lamin. GFP-lamin A(S22/392A) and GFP-lamin A(4A) mutants were transfected into NA cells to reflect nuclear lamina integrity. As observed previously, a disrupted lamina structure accompanied by intranuclear aggregates correlates with BGLF4 expression in EBV-reactivated cells (Fig. 5A). EBV-reactivated cells show significantly condensed chromosomes (Fig. 5D), while no significant alteration of the nuclear lamina was observed in cells with GFP-lamin A(S22/392A) or GFP-lamin A(4A) expression. These results indicate that BGLF4 target sites on lamin A also are important for the EBV reactivation-mediated redistribution of nuclear lamin.
Disassembly of the nuclear lamina is important for the protein expression levels of EBV BFRF1/BFLF2 and virion production.
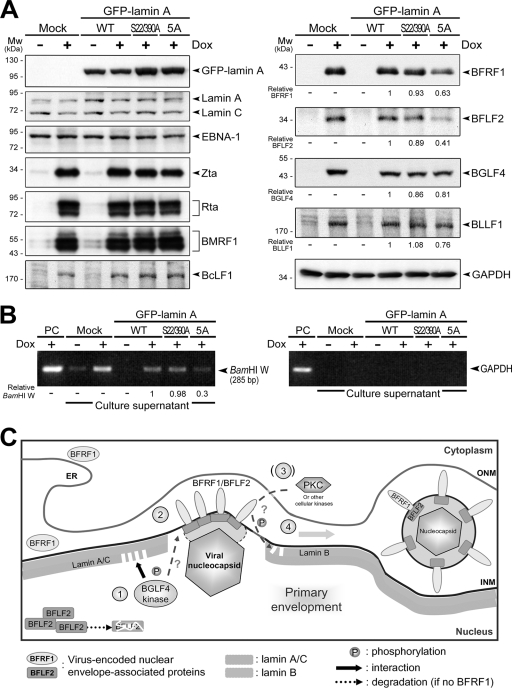
To illustrate the biological effects of the disassembly of the nuclear lamina during EBV replication, GFP-lamin A, GFP-lamin A(S22/390A), or GFP-lamin A(5A) was introduced into EREV8 cells, which harbor the Akata EBV genome and a tetracycline-inducible Rta. EBV reactivation was induced with doxycycline for 72 h. The cells were harvested to analyze the expression profiles of viral proteins by immunoblotting, and the virions in the culture supernatant were detected by PCR of EBV DNA. As shown in Fig. 6A, the addition of doxycycline induces the expression of the EBV immediate-early proteins Rta and Zta, early proteins BMRF1, BFRF1, BFLF2, and BGLF4, and late products BcLF1 (VCA) and BLLF1 (gp350) in EREV8 cells. The expression of GFP-lamin A(WT), GFP-lamin A(S22/390A), or GFP-lamin A(5A) in EREV8 cells has no effect on the expression of endogenous lamin A/C, viral latent protein EBNA-1, and lytic products Zta, BMRF1, and BcLF1 (Fig. 6A, left). However, the expression of GFP-lamin A(5A) reduces the protein levels of BFLF2 and BFRF1 to about 40 and 60% in doxycycline-treated EREV8 cells, while the expression of BGLF4 and BLLF1 also were slightly reduced (Fig. 6A, right). Because the expression of EBV BFRF1 and its interaction with BFLF2 protein play important roles for viral nucleocapsid egress (13, 18), our results suggest that the BGLF4-mediated redistribution of lamin is critical in the egress of EBV nucleocapsids. To this end, we analyzed the relative amounts of secreted virions in culture supernatants, using PCR to detect a region of BamHI W on the viral genome. The expression of GFP-lamin A(5A), but not GFP-lamin A(S22/390A), significantly reduces the amount of EBV DNA detected in the supernatants (about 30% of cells expressing wild-type lamin A) (Fig. 6B). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) DNA was not detected, which excludes the possibility of DNA contamination from cell debris in the supernatant. Taken together, these results indicate that the phosphorylation-mediated disassembly of the nuclear lamina is a crucial step for virion release. A hypothetical model for the contribution of BGLF4 to promoting the primary envelopment of the nucleocapsid is illustrated in Fig. 6C. The BGLF4-mediated disassembly of the nuclear lamina may facilitate the interaction of BFRF1, which is expressed in the perinuclear space between the ONM and INM, and BFLF2, which is a relatively labile intranuclear protein. The interaction of BFRF1 and BFLF2 can form patched complexes on the INM and finally promote the primary envelopment of the nucleocapsid.
FIG. 6.
Expression of lamin A with mutations at BGLF4 target sites reduces EBV BFRF1/BFLF2 protein expression and virion production. (A) EBV-positive tetracycline-inducible EREV8 cells were mock transfected or were transfected with plasmids expressing GFP-lamin A(WT), GFP-lamin A(S22/390A), or GFP-lamin A(5A). At 12 h posttransfection, cells were incubated with 100 ng/ml doxycycline (Dox) to induce the expression of Rta and subsequent EBV reactivation. The cells were harvested and subjected to immunoblotting analysis at 72 h postinduction. The protein expression was detected by anti-GFP JL-8, anti-lamin A/C 636, NPC-47 serum (for EBNA-1), anti-Zta 4F10, anti-Rta 467, anti-BMRF1 88A9, anti-BcLF1 L2, anti-BFRF1 E10, anti-BFLF2 C1, anti-BGLF4 2616, or anti-BLLF1 201 in immunoblotting. GAPDH served as a loading control. Mw, molecular mass. (B) The culture supernatants were centrifuged to harvest secreted EBV virions. The isolated viral particles were lysed and subjected to PCR analysis targeting the EBV DNA BamHI W fragment. The lack of GAPDH DNA rules out the possibility of cellular DNA contamination. PC, lysate from EREV8 cells that served as a PCR positive control. (C) A hypothetical model of the nuclear egression of EBV nucleocapsids. After EBV replication and genome encapsidation, the viral nucleocapsid moves close to the inner nuclear lamina. Simultaneously, lamina-targeted BGLF4 first induces the redistribution of lamin A/C adjacent to the nucleoplasm through direct phosphorylation (step 1) and consequently stabilizes BFLF2 protein by enhancing the interaction between nuclear envelope-associated BFRF1 and intranuclear BFLF2 (step 2). BGLF4 may further phosphorylate BFRF1 and BFLF2 for their correct localization in the nuclear membrane. Subsequently, the BFRF1/BFLF2 complexes may recruit a cellular kinase(s) such as PKC to facilitate the reorganization of and interaction with the lamina (step 3). Finally, EBV nucleocapsids pass through the barrier of the nuclear lamina and proceed to primary envelopment (step 4). ONM, outer nuclear membrane. ER, endoplasmic reticulum.
Human herpesviral UL protein kinases induce redistribution of nuclear lamin through a similar mechanism.
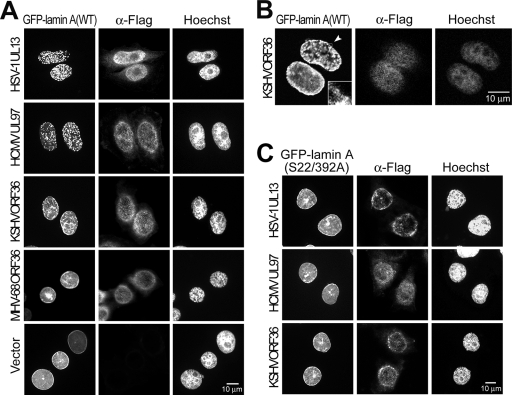
We showed that BGLF4 expression alone induces the dramatic reorganization of nuclear lamina. We noticed that HSV-1(ΔUL13) displayed a defect similar to that of HSV-1(ΔUs3) in primary envelopment in an earlier study (25). We examined the abilities of other herpesviral UL kinases to induce the disassembly of the nuclear lamina and whether HSV-1 UL13 can regulate nuclear lamina in the absence of Us3. Therefore, UL kinases of various herpesviruses and GFP-lamin A were transiently coexpressed in HeLa cells. In >95% of cells expressing UL97, the GFP-nuclear lamina was disassembled (Fig. 7). Notably, a similar GFP signal change was observed in >85% of cells expressing UL13, whereas KSHV ORF36 induced only partial nuclear rim alteration and lamin internalization without the complete disassembly of the nuclear lamina (Fig. 7B). Interestingly, although MHV68 ORF36 can induce chromosome condensation efficiently, it did not induce the disassembly of the nuclear lamina. Together with our previous observation of the induction of chromosome condensation by UL kinases (30), the abilities of conserved herpesviral protein kinases to induce the disassembly of the nuclear lamina and chromosome condensation are summarized in Table 1.
FIG. 7.
Human herpesviral UL kinases induce the disassembly of nuclear lamina. (A) HeLa cells were transfected with GFP-lamin A together with Flag-tagged HSV-1 UL13, HCMV UL97, KSHV ORF36, or MHV68 ORF36 expressing plasmid or vector control. At 24 h posttransfection, cells were fixed and stained for Flag-tagged protein and DNA as described in Materials and Methods. Disassembled nuclear rim and aggregated GFP-lamin protein was observed in cells expressing HSV-1 UL13 and HCMV UL97 cells but not in cells expressing MHV68 ORF36 or the control vector. (B) Confocal images of nuclear lamina in cells with KSHV ORF36 expression. KSHV ORF36 shows an ability to induce the partial disassembly of the nuclear lamina. The arrowhead indicates the region enlarged. (C) Ser-22 and Ser-392 of lamin A are important for human herpesviral UL kinases-mediated nuclear lamin redistribution. HeLa cells expressing GFP-lamin A(S22/392A) and Flag-tagged HSV-1 UL13-, HCMV UL97-, or KSHV ORF36-expressing plasmids were fixed and stained for Flag-tagged protein and DNA. An intact nuclear rim was observed in cells with both herpesviral kinases and GFP-lamin A(S22/392A) expression. α-Flag, anti-Flag antibody.
TABLE 1.
Ability of herpesviral kinases to induce chromosome condensation and lamina disassemblya
| Herpesvirus | Viral kinase | Chromosome condensation | Nuclear lamina disassembly |
|---|---|---|---|
| Alphaherpesvirus | |||
| HSV-1 | UL13 | +/− | ++ |
| Betaherpesvirus | |||
| HCMV | UL97 | − | ++ |
| Gammaherpesvirus | |||
| EBV | BGLF4 | ++ | ++ |
| KSHV | ORF36 | + | + |
| MHV68 | ORF36 | ++ | − |
Symbols: +∼++, 70 to 100% of kinase-positive cells express this phenomenon; +/−, 30 to 70% of kinase-positive cells express this phenomenon; −, <30% of kinase-positive cells express this phenomenon.
We then examined the role of Ser-22 and Ser-392 of lamin A in the disassembly of the nuclear lamina mediated by human herpesviral UL kinases. Compared to cells with GFP-lamin A expression (Fig. 7A), GFP-lamin A(S22/392A)-expressing cells are less sensitive to the HSV-1 UL13-, HCMV UL97-, or KSHV ORF36-induced disassembly of the nuclear lamina (Fig. 7C). This observation indicates that Ser-22 and Ser-392 of lamin A play crucial roles in the human herpesviral UL kinase-mediated disassembly of the nuclear lamina and also suggests that human herpesviral UL kinases have the ability to regulate the dynamics of nuclear lamina through a mechanism similar to that of Cdc2.
DISCUSSION
The nuclear lamina constitutes an intact meshwork for the maintenance of the nuclear structure. The nuclear lamina network also presents a natural barrier against most DNA viruses through which nucleocapsids must egress after viral genome replication for virion assembly and maturation. Previously, we demonstrated that EBV BGLF4 kinase induces chromosome condensation through the activation of condensin and Topo II (30). Here, we reveal the mechanisms whereby BGLG4 kinase phosphorylates cellular proteins involved in the disassembly of the nuclear lamina as well as chromosome condensation.
First, we showed that BGLF4 colocalizes and interacts with condensin independently of the presence of DNA (Fig. 1A and data not shown). Furthermore, BGLF4 colocalizes and interacts with Topo IIα in a kinase-dependent manner (data not shown), suggesting that the active form of BGLF4 has the correct conformation for the interaction. In addition, because BGLF4 phosphorylates the recombinant condensin hCAP-G subunit and Topo IIα proteins in vitro (Fig. 1C and D), it is possible that BGLF4 regulates chromosome condensation through direct phosphorylation. Both BGLF4 and Cdc2 phosphorylate the same region of the condensin hCAP-G subunit and Topo IIα, implying that BGLF4 has a regulatory mechanism similar to that of Cdc2 in inducing chromosome condensation.
Using an immunofluorescence assay, we found that condensin complexes accumulate within the nuclei of cells expressing BGLF4 (Fig. 1A). Because type I condensin complexes show strong chromosome targeting after the breakdown of the nuclear envelope (42), we propose that the BGLF4-mediated disassembly of the nuclear lamina partially contributes to the nuclear accumulation of condensin. In addition, BGLF4 colocalizes and interacts with lamin A/C in a kinase-dependent manner (Fig. 2). BGLF4 phosphorylates lamin A protein in the Head and Tail regions (aa 1 to 129 and 388 to 660) (Fig. 3B), suggesting that it has the ability to regulate nuclear lamina dynamics through direct phosphorylation. Notably, BGLF4, but not Cdc2, phosphorylates the Tail II region (aa 490 to 660) of lamin A. Phosphorylation within the Tail II region (Ser-652) of lamin A has been detected in screening proteins phosphorylated in vivo in HeLa cells, but the biological significance is unclear so far (4).
During mitosis, the phosphorylation of lamin A or C by Cdc2 at Ser-22, Ser-390, and Ser-392 is critical for the disassembly of the nuclear lamina (12, 21, 44, 54). Here, we showed that BGLF4 also acts to modify similar residues on lamin A/C (Fig. 4C and data not shown). Interestingly, we also observed that BGLF4 targets a broader range of substrates than Cdc2 kinase (Fig. 3A). Previously, the BGLF4-targeting motifs have been identified as QHVS133PMRQ in cellular EF-1δ and GRAT19PAQT and VRGT110PVRQ in MCM4 (the phosphorylation sites are underlined) (27, 29). BGLF4 also targets viral EBNA-LP on RSPS35PTRG, EBNA-2 on PTPS243PPRM, and BMRF1 on PSPS337PPPP and PPRT344PTWE motifs (26, 57, 58). Taking these findings together with BGLF4 targeting sites on condensin, Topo IIα, and lamin A phosphorylation resolved in this study, we propose that the optimal phosphorylation consensus sequence of BGLF4 kinase is [R/K/H/L/S/T]-X-S/T-P. S/T is the target site for the phosphorylation of either serine or threonine, and X may be absent or any amino acid (preferably proline). The mapping of possible phosphorylation sites on lamin A (Fig. 3) indicates that BGLF4 also shares the same phosphorylation sequence S/T-P-X-[R/K] with cyclin-dependent kinases (CDKs) (34, 49), suggesting that BGLF4 has the ability to regulate signaling pathways mediated by other CDKs.
How herpesvirus egresses through the nuclear membrane after genome replication is an age-old question. Two virus-encoded nuclear membrane-associated proteins, UL34 and UL31 in alphaherpesvirus HSV-1, pUL50 and pUL53 in HCMV, and BFRF1 and BFLF2 in EBV, are critical for efficient nucleocapsid egress. In HSV-1-infected cells, UL34 and UL31 interact with lamin A/C and potentially recruit nuclear cellular PKCs to nuclear lamina, which may promote the egress of nucleocapsids (5, 43, 45). The correct nuclear localization of UL34 and UL31 is determined by virus-encoded Us3 and UL13 kinases (25, 46-48), while Us3 can phosphorylate lamin A/C in vitro (40). Similarly, HCMV pUL50 and pUL53 can interact with the lamin A/C and lamin B receptors and recruit cellular PKC to the nuclear lamina (38). EBV BFRF1 and BFLF2 also have been shown to interact with the nuclear lamin and are important for efficient primary viral envelopment and egress (13, 18).
Here, we demonstrated that EBV reactivation induces changes in the nuclear lamina (Fig. 5A). The BGLF4-targeted sites on lamin A also are determinants of the EBV reactivation-induced nuclear redistribution of lamin and virion production (Fig. 5D and 6B). The outcome of expressing lamin A with mutated BGLF4-targeting sites (Fig. 6A and B) was similar to that of the BGLF4 knockdown experiment (16), which displays a 14-fold reduction of virion release and the approximate extinction of BFLF2 in BGLF4 knockdown 293/EBV+ cells, while no significant change of the intracellular viral DNA copy number was observed. Because BFLF2 can be stabilized through interaction with BFRF1 in the cells with reactivated EBV (18), we suggest that the disappearance of BFLF2 observed in the BGLF4 knockdown experiment partially results from its inefficient interaction with BFRF1, which consequently caused the nuclear retention of nucleocapsids. Additionally, because EBV capsids acquire an envelope highly enriched for BLLF1 (gp350) at the trans-Golgi or plasma membrane after nuclear egress (17), inefficient nucleocapsid reenvelopment may lead to the slightly reduced expression level of BLLF1.
A model for nuclear egress (Fig. 6C) is proposed to bring together our observations and the behavior of EBV homologs in other herpesviruses. After DNA replication and genome encapsidation, viral nucleocapsids move close to the inner nuclear lamina, which is composed mainly of lamin A/C proteins. Simultaneously, BGLF4 kinase induces the redistribution of lamin A/C adjacent to the nucleoplasm through direct phosphorylation (Fig. 3 and 4) and consequently enhances the efficacy of interaction between nuclear envelope-associated BFRF1 and intranuclear BFLF2, enabling the BFLF2 protein to escape proteasome-mediated degradation. Because one BGLF4 putative phosphorylation site in BFRF1 and four in BFLF2 were found, it also is possible that BGLF4 phosphorylates BFRF1/BFLF2 to further enable them to form complexes and localize correctly in the nuclear membrane and to complete the subsequent disassembly of local nuclear lamina. Finally, EBV nucleocapsids can breach the barrier of the nuclear lamina and proceed to primary envelopment.
In this model, the molecular mechanism through which BFRF1 prevents the degradation of BFLF2 remains unclear. In addition, because PKC is recruited to the nuclear rim during HSV-1 and murine CMV replication, which has been suggested to increase lamin B phosphorylation (41, 43), we do not exclude the possible contribution of cellular kinases such as PKC during the egress of EBV nucleocapsids. The recruited PKC potentially coordinates with BGLF4 to increase lamin B protein phosphorylation and leads to nucleocapsid egress. Although some steps in this model need further investigation, this study provides novel evidence linking herpesviral UL kinase, the structural changes of the nuclear lamina, and the egress of viral nucleocapsids.
Using a GFP tagging system to explore lamin dynamics, we found that the expression of human herpesviral UL kinases, including HSV-1 UL13, HCMV UL97, EBV BGLF4, and KSHV ORF36, can induce the disassembly of the nuclear lamina (Fig. 7A and B; Table 1). These UL viral kinases seem to regulate the lamina through a similar pathway (Fig. 7C). Nevertheless, EBV BGLF4 seems to be the most powerful of the human herpes UL kinases in terms of inducing nuclear changes (Table 1). The observation that HSV-1 UL13 also can induce nuclear lamina reorganization provides a clue for its direct contribution to nucleocapsid egress in addition to its phosphorylation of Us3 kinase, which was suggested previously (25). The dynamics of nuclear lamina is physiologically controlled by CDKs through phosphorylation at S/T-P-X-[R/K] motifs or PKCs at [K/R]-(X)1∼2-S/T or S/T-X-[K/R] motifs, where X is any amino acid. However, the consensus sequence of the Us3 kinase target sequence is (R)>2-X-S/T-Y-Y. X is absent or is any amino acid, preferably Arg, Ala, Val, Pro, or Ser. Y is preferably an acidic residue except for proline or is an absent amino acid (40). It does not match the well-characterized motifs contributing to the regulation of nuclear lamina. Although Us3 phosphorylates lamin A in vitro (40), it cannot be excluded that Us kinase may regulate the nuclear lamina through a cellular kinase-mediated pathway or, alternatively, a distinct pathway is involved in Us kinase-mediated nuclear lamina change.
Overall, we demonstrate that BGLF4 is a major player that creates a beneficial nuclear environment for EBV replication. The BGLF4-mediated disassembly of the nuclear lamina is important for virion production. Because lamins can bind the nesprin family of cytoskeleton linker proteins, nuclear pore complex, and emerin, connecting them to intermediate filaments, microtubules, and the actin cytoskeleton (reviewed in references 7 and 52), it will be interesting to study whether the BGLF4-induced rearrangement of actin and microtubules observed in our previous study (30) is dependent on the nuclear lamina structure. Our proposed model of how herpesviral UL kinases modulate multiple nuclear events to help viral genome replication and conquer natural lamina barriers for nuclear egress also provides a perspective for future studies of other DNA viruses.
Acknowledgments
We thank David M. Baines at Cornell University for GST-tagged lamin A constructs and Joel D. Gilbert at the State University of New York Upstate Medical University for GFP-lamin A-expressing plasmids. We also thank Alberto Faggioni and Roberta Gonnella at Università La Sapienza, Rome, Italy, for BFRF1 and BFLF2 antibodies. We are grateful to Tim J. Harrison of University College London for the critical reading and modification of the manuscript.
This study was supported by grants NHRI-EX96-9609BI and NHRI-EX97-9609BI from National Health Research Institutes and grants NSC95-2320-B002-087-MY3 and NSC97-2811-B002-015 from the National Science Council, Taiwan.
Footnotes
Published ahead of print on 24 September 2008.
REFERENCES
- 1.Aebi, U., J. Cohn, L. Buhle, and L. Gerace. 1986. The nuclear lamina is a meshwork of intermediate-type filaments. Nature 323560-564. [DOI] [PubMed] [Google Scholar]
- 2.Alber, F., S. Dokudovskaya, L. M. Veenhoff, W. Zhang, J. Kipper, D. Devos, A. Suprapto, O. Karni-Schmidt, R. Williams, B. T. Chait, A. Sali, and M. P. Rout. 2007. The molecular architecture of the nuclear pore complex. Nature 450695-701. [DOI] [PubMed] [Google Scholar]
- 3.Asai, R., A. Kato, K. Kato, M. Kanamori-Koyama, K. Sugimoto, T. Sairenji, Y. Nishiyama, and Y. Kawaguchi. 2006. Epstein-Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J. Virol. 805125-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beausoleil, S. A., M. Jedrychowski, D. Schwartz, J. E. Elias, J. Villen, J. Li, M. A. Cohn, L. C. Cantley, and S. P. Gygi. 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA 10112130-12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjerke, S. L., and R. J. Roller. 2006. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347261-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broers, J. L., B. M. Machiels, G. J. van Eys, H. J. Kuijpers, E. M. Manders, R. van Driel, and F. C. Ramaekers. 1999. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J. Cell Sci. 1123463-3475. [DOI] [PubMed] [Google Scholar]
- 7.Broers, J. L., F. C. Ramaekers, G. Bonne, R. B. Yaou, and C. J. Hutchison. 2006. Nuclear lamins: laminopathies and their role in premature ageing. Physiol. Rev. 86967-1008. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y., C. H. Tung, Y. T. Huang, J. Lu, J. Y. Chen, and C. H. Tsai. 1999. Requirement for cell-to-cell contact in Epstein-Barr virus infection of nasopharyngeal carcinoma cells and keratinocytes. J. Virol. 738857-8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, M. R., S. J. Chang, H. Huang, and J. Y. Chen. 2000. A protein kinase activity associated with Epstein-Barr virus BGLF4 phosphorylates the viral early antigen EA-D in vitro. J. Virol. 743093-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chua, H. H., H. H. Lee, S. S. Chang, C. C. Lu, T. H. Yeh, T. Y. Hsu, T. H. Cheng, J. T. Cheng, M. R. Chen, and C. H. Tsai. 2007. Role of the TSG101 gene in Epstein-Barr virus late gene transcription. J. Virol. 812459-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross, T., G. Griffiths, E. Deacon, R. Sallis, M. Gough, D. Watters, and J. M. Lord. 2000. PKC-delta is an apoptotic lamin kinase. Oncogene 192331-2337. [DOI] [PubMed] [Google Scholar]
- 12.Eggert, M., N. Radomski, D. Tripier, P. Traub, and E. Jost. 1991. Identification of phosphorylation sites on murine nuclear lamin C by RP-HPLC and microsequencing. FEBS Lett. 292205-209. [DOI] [PubMed] [Google Scholar]
- 13.Farina, A., R. Feederle, S. Raffa, R. Gonnella, R. Santarelli, L. Frati, A. Angeloni, M. R. Torrisi, A. Faggioni, and H. J. Delecluse. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 793703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fawcett, D. W. 1966. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am. J. Anat. 119129-145. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, D. Z., N. Chaudhary, and G. Blobel. 1986. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl. Acad. Sci. USA 836450-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershburg, E., S. Raffa, M. R. Torrisi, and J. S. Pagano. 2007. Epstein-Barr virus-encoded protein kinase (BGLF4) is involved in production of infectious virus. J. Virol. 815407-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong, M., and E. Kieff. 1990. Intracellular trafficking of two major Epstein-Barr virus glycoproteins, gp350/220 and gp110. J. Virol. 641507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonnella, R., A. Farina, R. Santarelli, S. Raffa, R. Feederle, R. Bei, M. Granato, A. Modesti, L. Frati, H. J. Delecluse, M. R. Torrisi, A. Angeloni, and A. Faggioni. 2005. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J. Virol. 793713-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greber, U. F., and A. Fassati. 2003. Nuclear import of viral DNA genomes. Traffic 4136-143. [DOI] [PubMed] [Google Scholar]
- 20.Gruenbaum, Y., A. Margalit, R. D. Goldman, D. K. Shumaker, and K. L. Wilson. 2005. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 621-31. [DOI] [PubMed] [Google Scholar]
- 21.Heald, R., and F. McKeon. 1990. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell 61579-589. [DOI] [PubMed] [Google Scholar]
- 22.Höger, T. H., K. Zatloukal, I. Waizenegger, and G. Krohne. 1990. Characterization of a second highly conserved B-type lamin present in cells previously thought to contain only a single B-type lamin. Chromosoma 10067-69. [DOI] [PubMed] [Google Scholar]
- 23.Hutchison, C. J., M. Alvarez-Reyes, and O. A. Vaughan. 2001. Lamins in disease: why do ubiquitously expressed nuclear envelope proteins give rise to tissue-specific disease phenotypes? J. Cell Sci. 1149-19. [DOI] [PubMed] [Google Scholar]
- 24.Izumi, M., O. A. Vaughan, C. J. Hutchison, and D. M. Gilbert. 2000. Head and/or CaaX domain deletions of lamin proteins disrupt preformed lamin A and C but not lamin B structure in mammalian cells. Mol. Biol. Cell 114323-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato, A., M. Yamamoto, T. Ohno, M. Tanaka, T. Sata, Y. Nishiyama, and Y. Kawaguchi. 2006. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 801476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato, K., A. Yokoyama, Y. Tohya, H. Akashi, Y. Nishiyama, and Y. Kawaguchi. 2003. Identification of protein kinases responsible for phosphorylation of Epstein-Barr virus nuclear antigen leader protein at serine-35, which regulates its coactivator function. J. Gen. Virol. 843381-3392. [DOI] [PubMed] [Google Scholar]
- 27.Kawaguchi, Y., K. Kato, M. Tanaka, M. Kanamori, Y. Nishiyama, and Y. Yamanashi. 2003. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J. Virol. 772359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura, K., O. Cuvier, and T. Hirano. 2001. Chromosome condensation by a human condensin complex in Xenopus egg extracts. J. Biol. Chem. 2765417-5420. [DOI] [PubMed] [Google Scholar]
- 29.Kudoh, A., T. Daikoku, Y. Ishimi, Y. Kawaguchi, N. Shirata, S. Iwahori, H. Isomura, and T. Tsurumi. 2006. Phosphorylation of MCM4 at sites inactivating DNA helicase activity of the MCM4-MCM6-MCM7 complex during Epstein-Barr virus productive replication. J. Virol. 8010064-10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, C. P., J. Y. Chen, J. T. Wang, K. Kimura, A. Takemoto, C. C. Lu, and M. R. Chen. 2007. Epstein-Barr virus BGLF4 kinase induces premature chromosome condensation through activation of condensin and topoisomerase II. J. Virol. 815166-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, C. T., C. I. Wong, W. Y. Chan, K. W. Tzung, J. K. Ho, M. M. Hsu, and S. M. Chuang. 1990. Establishment and characterization of two nasopharyngeal carcinoma cell lines. Lab. Investig. 62713-724. [PubMed] [Google Scholar]
- 32.Lu, C. C., Y. C. Chen, J. T. Wang, P. W. Yang, and M. R. Chen. 2007. Xeroderma pigmentosum C is involved in Epstein Barr virus DNA replication. J. Gen. Virol. 883234-3243. [DOI] [PubMed] [Google Scholar]
- 33.Makarova, O., E. Kamberov, and B. Margolis. 2000. Generation of deletion and point mutations with one primer in a single cloning step. BioTechniques 29970-972. [DOI] [PubMed] [Google Scholar]
- 34.Marin, O., F. Meggio, G. Draetta, and L. A. Pinna. 1992. The consensus sequences for cdc2 kinase and for casein kinase-2 are mutually incompatible. A study with peptides derived from the beta-subunit of casein kinase-2. FEBS Lett. 301111-114. [DOI] [PubMed] [Google Scholar]
- 35.Marschall, M., A. Marzi, P. aus dem Siepen, R. Jochmann, M. Kalmer, S. Auerochs, P. Lischka, M. Leis, and T. Stamminger. 2005. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 28033357-33367. [DOI] [PubMed] [Google Scholar]
- 36.McKeon, F. D., M. W. Kirschner, and D. Caput. 1986. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature 319463-468. [DOI] [PubMed] [Google Scholar]
- 37.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9423-429. [DOI] [PubMed] [Google Scholar]
- 38.Milbradt, J., S. Auerochs, and M. Marschall. 2007. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 882642-2650. [DOI] [PubMed] [Google Scholar]
- 39.Moir, R. D., T. P. Spann, and R. D. Goldman. 1995. The dynamic properties and possible functions of nuclear lamins. Int. Rev. Cytol. 162B141-182. [DOI] [PubMed] [Google Scholar]
- 40.Mou, F., T. Forest, and J. D. Baines. 2007. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 816459-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297854-857. [DOI] [PubMed] [Google Scholar]
- 42.Ono, T., Y. Fang, D. L. Spector, and T. Hirano. 2004. Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell 153296-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park, R., and J. D. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peter, M., J. Nakagawa, M. Doree, J. C. Labbe, and E. A. Nigg. 1990. In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell 61591-602. [DOI] [PubMed] [Google Scholar]
- 45.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 785564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 758803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 768939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryckman, B. J., and R. J. Roller. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Songyang, Z., S. Blechner, N. Hoagland, M. F. Hoekstra, H. Piwnica-Worms, and L. C. Cantley. 1994. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 4973-982. [DOI] [PubMed] [Google Scholar]
- 50.Tarakanova, V., V. Leung-Pineda, S. Hwang, C. Yang, K. Matatall, M. Basson, R. Sun, H. Piwnica-Worms, B. Sleckman, and H. Virgin IV. 2007. Gamma-herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe 1275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai, C. H., M. V. Williams, and R. Glaser. 1991. Characterization of two monoclonal antibodies to Epstein-Barr virus diffuse early antigen which react to two different epitopes and have different biological function. J. Virol. Methods 3347-52. [DOI] [PubMed] [Google Scholar]
- 52.Vlcek, S., and R. Foisner. 2007. Lamins and lamin-associated proteins in aging and disease. Curr. Opin. Cell Biol. 19298-304. [DOI] [PubMed] [Google Scholar]
- 53.Wang, J. T., P. W. Yang, C. P. Lee, C. H. Han, C. H. Tsai, and M. R. Chen. 2005. Detection of Epstein-Barr virus BGLF4 protein kinase in virus replication compartments and virus particles. J. Gen. Virol. 863215-3225. [DOI] [PubMed] [Google Scholar]
- 54.Ward, G. E., and M. W. Kirschner. 1990. Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell 61561-577. [DOI] [PubMed] [Google Scholar]
- 55.Wasserman, R. A., C. A. Austin, L. M. Fisher, and J. C. Wang. 1993. Use of yeast in the study of anticancer drugs targeting DNA topoisomerases: expression of a functional recombinant human DNA topoisomerase II alpha in yeast. Cancer Res. 533591-3596. [PubMed] [Google Scholar]
- 56.Wells, N. J., and I. D. Hickson. 1995. Human topoisomerase II alpha is phosphorylated in a cell-cycle phase-dependent manner by a proline-directed kinase. Eur. J. Biochem. 231491-497. [DOI] [PubMed] [Google Scholar]
- 57.Yang, P. W., S. S. Chang, C. H. Tsai, Y. H. Chao, and M. R. Chen. 2008. Effect of phosphorylation on the transactivation activity of Epstein-Barr virus BMRF1, a major target of the viral BGLF4 kinase. J. Gen. Virol. 89884-895. [DOI] [PubMed] [Google Scholar]
- 58.Yue, W., E. Gershburg, and J. S. Pagano. 2005. Hyperphosphorylation of EBNA2 by Epstein-Barr virus protein kinase suppresses transactivation of the LMP1 promoter. J. Virol. 795880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]