Abstract
We screened a panel of R5X4 and X4 human immunodeficiency virus type 1 (HIV-1) strains for their sensitivities to AMD3100, a small-molecule CXCR4 antagonist that blocks HIV-1 infection via this coreceptor. While no longer under clinical development, AMD3100 is a useful tool with which to probe interactions between the viral envelope (Env) protein and CXCR4 and to identify pathways by which HIV-1 may become resistant to this class of antiviral agents. While infection by most virus strains was completely blocked by AMD3100, we identified several R5X4 and X4 isolates that exhibited plateau effects: as the AMD3100 concentration was increased, virus infection and membrane fusion diminished to variable degrees. Once saturating concentrations of AMD3100 were achieved, further inhibition was not observed, indicating a noncompetitive mode of viral resistance to the drug. The magnitude of the plateau varied depending on the virus isolate, as well as the cell type used, with considerable variation observed when primary human T cells from different human donors were used. Structure-function studies indicated that the V1/V2 region of the R5X4 HIV-1 isolate DH12 was necessary for AMD3100 resistance and could confer this property on two heterologous Env proteins. We conclude that some R5X4 and X4 HIV-1 isolates can utilize the AMD3100-bound conformation of CXCR4, with the efficiency being influenced by both viral and host factors. Baseline resistance to this CXCR4 antagonist could influence the clinical use of such compounds.
The entry of human immunodeficiency virus type 1 (HIV-1) into cells can be prevented by a variety of small-molecule inhibitors that target the viral envelope (Env) protein or the coreceptors to which it binds (reviewed in references 3, 12, and 46). Entry inhibitors have been used as molecular tools to characterize how sequential interactions between Env, CD4, and a coreceptor lead to the conformational changes in Env that result in membrane fusion and virus infection (39, 43). They have also been used successfully in the clinic (12). A particularly useful application of coreceptor antagonists is to identify the efficiency with which a virus uses the chemokine receptor CCR5 or CXCR4 to infect primary cells. While many HIV-1 strains can use either CCR5 or CXCR4 (R5X4 viruses) to infect cell lines, the efficiency with which a given virus uses each coreceptor for infection can vary widely and does not always predict the mechanism of entry into human T cells or macrophages (18). Thus, some R5X4 viruses use only CXCR4 to infect certain primary cells, others use only CCR5, and some viruses use both coreceptors to infect multiple cell types (16, 17, 26, 36, 59-61). The use of specific and potent coreceptor antagonists can be utilized to prevent entry via one coreceptor, revealing the efficiency with which the alternative coreceptor can support virus infection. Viral resistance to these drugs may elucidate mechanisms of interaction between the coreceptor and the Env protein.
Coreceptor antagonists have also been used in the clinic to treat HIV-infected individuals, with one CCR5 antagonist (maraviroc) having been licensed for use in 2007. There are several variables that affect the potency of these agents, including the impressive genetic variability of Env (15). In general, the potencies with which entry inhibitors fully suppress infection of primary virus strains vary to a greater extent than do those of antiviral agents that target more conserved viral proteins, such as reverse transcriptase, integrase, and protease (20, 31). In addition, host cell factors also influence the efficiencies with which entry inhibitors prevent virus infection of primary cells obtained from different individuals (27, 35, 37). One such host factor that influences entry inhibitor potency is coreceptor expression levels, which can vary considerably among individuals (33, 49, 56, 58). In general, higher levels of coreceptor expression accelerate fusion kinetics, necessitating higher levels of fusion inhibitors (such as enfuvirtide [ENF]) and coreceptor antagonists to fully suppress infection (27, 37, 43, 48).
In this study, we examined factors that influence the potency of the CXCR4 antagonist AMD3100 (reviewed in reference 6). AMD3100 is an antagonist of CXCR4 that inhibits the entry of a variety of X4-tropic strains (7, 10, 31, 51, 52). Although no longer being pursued for clinical use as an anti-HIV therapy (23), AMD3100 is a useful molecular tool with which to study interactions between HIV-1 and CXCR4, to examine the extent to which HIV-1 strains vary in their sensitivities to CXCR4 antagonists, and to ask whether differential CXCR4 domain use by HIV-1 Env impacts virus tropism. With these questions in mind, we examined a panel of R5X4- and X4-tropic virus strains for their sensitivities to AMD3100 and found three strains that continued to use CXCR4 for entry even in the face of saturating AMD3100 concentrations. These viruses exhibited a “plateau effect” in which membrane fusion and infection levels were reduced and then remained constant once saturating concentrations of AMD3100 were achieved. This pattern of resistance was AMD3100/CXCR4 specific, as these viruses could be fully inhibited by other classes of entry inhibitors. Resistance mapped to the V1/V2 region of Env and could be transferred to heterologous Env backgrounds by introducing the V1/V2 loop from an AMD3100-resistant virus. Our results indicate that, at baseline, some HIV-1 strains can utilize the drug-bound form of CXCR4. This finding illustrates how differential use of CXCR4 by HIV-1 strains can have practical implications, including resistance to otherwise potent antiviral agents.
MATERIALS AND METHODS
Reagents.
The CXCR4 inhibitor AMD3100 was obtained through the AIDS Research and Reference Reagent Program. The CCR5 inhibitor CMPD167 was obtained from Merck. The CCR5 inhibitor aplaviroc was provided by GlaxoSmithKline. The CXCR4-specific monoclonal antibody (MAb) 12G5 was a generous gift from James Hoxie (University of Pennsylvania) (11). The phycoerythrin-conjugated MAb 12G5 and the immunoglobulin G2a isotype control were purchased from BD Pharmingen.
Viruses and plasmids.
The following HIV-1 strains were obtained through the Penn Center for AIDS Research Virus and Molecular Core: DH12, NL4-3, 89.6, SPL-3, and TYBE. The following HIV-1 strains were obtained through the AIDS Research and Reference Reagent Program (with specific contributors in parentheses after the virus name): 93UG065 (Joint United Nations Program on HIV/AIDS), BK132/GS, and BZ167/GS (Nelson Michael). HIV-1 R3A (38) was a gift from Lishan Su (University of North Carolina, Chapel Hill). The AD8/DH12 Env chimeras (5) and truncated DH12 Env (28) were generous gifts from Michael Cho (Case Western Reserve University).
Entry PCR assay.
U87.CD4.CXCR4 and U87.CD4 cells (obtained through the AIDS Research and Reference Reagent Program from H. K. Deng and D. R. Littman) were infected with DNase-treated (50 U/ml DNase incubated at room temperature for 20 min) replication-competent virus for 3 days. The cells were then washed with phosphate-buffered saline (PBS) two times and lysed with cell lysis buffer (1 M KCl, 1% NP-40, 1 M Tris, pH 8.0, 15 mg/ml proteinase K, and water). The lysates were then incubated at 60°C for 2 h and 98°C for 15 min. For each reaction, 1 μl of DNA lysate was transferred to a MicroAmp fast optical 96-well reaction plate with a barcode (Applied Biosystems) containing 12.5 μl TaqMan Universal PCR Master Mix (Applied Biosystems), 9 μl of sterile water (Fisher Scientific), and 1.5 μl of the primer/probe sets recognizing an early HIV replication product (the first strong stop or long terminal repeat [LTR]) or the cellular GAPDH (glyceraldehyde 3-phosphate dehydrogenase) gene (61). Real-Time PCR was then performed using the Applied Biosystems 7500 Fast Real-Time PCR System. Final values were calculated for each sample by taking the value determined for the LTR and dividing it by the respective GAPDH value. The linear range of the assay was determined by the value of the LTR/GAPDH ratio, being equal to or less than 1. To determine the amount of virus (the titer was determined by the amount of p24 antigen) needed to be within the linear range of the assay, titration experiments were performed. Based on these results, the amounts of virus (ng of p24) used for each strain were as follows: DH12, 0.3 ng; R3A, 1.7 ng; NL4-3, 5 ng; 89.6, 3.6 ng; SPL-3, 4.4 ng; TYBE, 1.8 ng; BK132/GS, 3 ng; BZ167/GS, 7.3 ng; and 93UG065, 3.9 ng. In entry PCR inhibition assays, cells were incubated with drug for 1 h prior to virus addition (the addition of virus brought the drug concentrations to the concentrations reported).
Viral pseudotypes.
Viral pseudotypes were made by transfecting 293T cells with the env plasmid of interest and the NL4-3 env-deficient (through frameshift) genome with a luciferase reporter gene in place of nef. The cells were incubated for 2 to 3 days, and then the supernatants were harvested and filtered with a Corning 0.45-μm syringe filter. Single-use aliquots were then frozen and stored at −80°C. The titer of each pseudotype preparation was determined by measuring the amount of p24 antigen present. U87.CD4.CXCR4 or U87.CD4 cells were infected with the indicated viral pseudotypes by adding 10 ng of p24 of the pseudotype to the cells and centrifuging them at 1,200 rpm for 1.5 to 2 h at room temperature. The cells were then incubated for 3 days at 37°C. Then, the cells were washed with PBS and lysed with lysis buffer (0.1% Triton X-100 in PBS). Luciferase substrate was then added, and the luciferase activity was read (as a measure of infection).
For pseudotype assays done using primary human CD4+ T cells, the cells were obtained through the Penn Center for AIDS Research Immunology Core and stimulated with phytohemagglutinin for 3 days at 37°C. The cells were then centrifuged at 1,200 rpm for 5 min and then resuspended in RPMI medium with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and interleukin 2 at 20 U/ml. The cells were transferred to a 96-well plate at a density of 200,000 cells/well. Thirty-five nanograms of p24 DH12gp140 and 30 ng p24 R3A viral pseudotypes were then added to the wells, and the plates were incubated for 4 days at 37°C, after which the cells were transferred to Eppendorf tubes and centrifuged at 1,200 rpm for 5 min to pellet the cells. The supernatant was removed, and lysis buffer was added. The cells were then transferred to a 96-well white-bottom plate, luciferase substrate was added, and enzyme activity was assayed as a measure of infection. For pseudotype inhibition assays, the cells (both CD4+ T cells and U87 cells) were incubated with a 2× concentration of drug for 1 h at 37°C. The addition of the pseudotypes brought the drug to the final concentration reported in the assays.
Cell-cell fusion assay.
The cell-cell fusion assay has been described in detail previously (50). Briefly, quail fibroblasts (QT6) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 1% l-glutamine, and 1% penicillin/streptomycin. Target cells were transfected with plasmids expressing CD4, CXCR4, or CCR5 under the control of the CMV promoter and a luciferase plasmid under the control of a T7 promoter. The effector cells were transfected with the appropriate env expression plasmid and infected with a vaccinia virus encoding the T7 RNA polymerase (vTF1.1) (1). The two cell populations were then mixed together and incubated at 37°C for 7 to 8 h. The cells were then lysed with 0.1% Triton X-100 and transferred to a white-bottom plate. Fusion between the two cell groups was detected by reading the T7 polymerase-driven luciferase expression. The extent of fusion in the absence of AMD3100 was set to 100%, and all other fusion values were compared to it. In cell-cell fusion inhibition assays, the target cells were incubated with a 2× concentration of the appropriate drug for 1 h prior to the addition of the effector cells. Upon addition, the concentration of drug becomes 1× (the final concentration is reported) and is maintained throughout the rest of the experiment.
FACS analysis.
U87.CD4.CXCR4 cells were incubated with AMD3100 for 1 hour at 37°C. The cells were then washed with fluorescence-activated cell sorter (FACS) buffer (5 mM EDTA, 2% FBS, and 0.05% Na azide in PBS) with the appropriate concentration of AMD3100 present. The cells were centrifuged (1,300 rpm for 5 min) and washed again. Next, the cells were stained in a 50-μl total volume of FACS buffer plus AMD3100 plus 10 μl 12G5-phycoerythrin, FACS buffer plus cells only, or FACS buffer plus immunoglobulin G2a isotype control MAb for 30 min at 4°C. The cells were washed two more times (again with AMD3100 present) and then resuspended in 300 μl FACS buffer. FACS analysis was then performed using the FACSCalibur from Becton Dickinson, and the data were analyzed using the program CellQuest (Becton Dickinson).
Chimera construction.
Chimeras between the Envs of DH12 and R3A were created using overlapping PCR. Briefly, primers were designed to amplify one specific region from each of the two Envs, with one primer having overlapping sequence of both pieces. The two pieces were then used as the template for the second PCR, in which one full-length env gene was made using both DH12 and R3A pieces. The PCR mixture contained 2.5 U Platinum Pfx DNA polymerase from Invitrogen, 10× Pfx buffer, 10× PCRx Enhancer, 10 mM deoxynucleotide triphosphates, 50 mM MgSO4, and water. The PCR cycling conditions were as follows: a 5-min hot start at 94°C and 20 cycles of 94°C for 30 s, 50°C for 30 s, and 68°C for 2.5 min, followed by a 7-min extension time. The PCR products were run on 1% agarose gels and excised using the Zymoclean kit from Zymo Research Laboratories. The purified chimeric DNA was then cloned into the mammalian expression vector pciPRE by utilizing the KpnI and XbaI restriction sites. All chimeras were screened by sequencing, and functional assessment was done through cell-cell fusion assays.
RESULTS
Identification of R5X4 and X4 virus strains partially resistant to inhibition by AMD3100.
Due to the genetic variability of the viral Env protein, primary HIV-1 strains can exhibit an impressive range of sensitivities to entry inhibitors (22, 31, 47). Recently, we identified a series of R5 Env proteins from an HIV-infected individual that exhibited only partial sensitivity to CCR5 inhibitors: viruses bearing these Env proteins could infect cells in a CCR5-dependent manner, albeit less efficiently, in the face of saturating concentrations of drug, suggesting an inherent ability of some R5 viruses to utilize the drug-bound conformation of CCR5 (J. C. Tilton, H. Amrine-Madsen, J. L. Miamidian, K. M. Kitrinos, J. Pfaff, J. F. Demarest, N. Ray, J. L. Jeffrey, C. C. LaBranche, and R. W. Doms, unpublished data). Such altered use of CCR5 could impact the clinical response to CCR5 inhibitors and might also affect viral tropism.
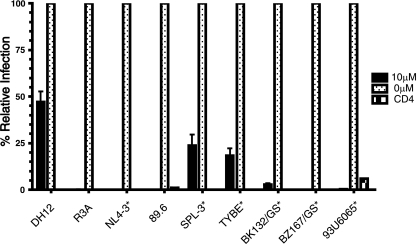
To determine if some X4 and R5X4 virus strains exhibit a resistance pattern similar to that of the small-molecule CXCR4 inhibitor AMD3100, we infected human astroglioma U87 cells expressing CD4 and CXCR4 with a panel of nine virus strains in the presence of a high concentration of AMD3100 (10 μM) that routinely blocks CXCR4-dependent virus infection (Fig. 1). A quantitative PCR assay was employed to assess the efficiency of virus entry by measuring the appearance of early reverse transcriptase products. We found that while 10 μM AMD3100 inhibited six virus strains by ≥95%, three virus strains were not fully inhibited, including the R5X4 strain DH12 (53) and the X4 strains SPL-3 (45) and TYBE (60). Entry of DH12 was reduced by only 45% in the presence of 10 μM AMD3100. None of the viruses entered U87 cells expressing CD4 alone, indicating that entry was indeed CXCR4 dependent. Importantly, both DH12 and R3A were inhibited by high concentrations of the reverse transcriptase inhibitor zidovudine and the fusion inhibitor ENF, as measured by p24 production, showing that complete viral inhibition could be obtained on U87.CD4.CXCR4 cells and primary CD4+ T cells (data not shown).
FIG. 1.
Susceptibilities of HIV-1 strains to inhibition by AMD3100. U87.CD4.CXCR4 cells were incubated with 10 μM AMD3100 or medium without inhibitor for 1 h at 37°C. Between 0.3 ng and 7.3 ng p24 replication-competent virus was then added (see Materials and Methods for specific virus amounts), and the extents of virus infection were determined 3 days later using an entry PCR assay employing primer/probes specific for the HIV-1 LTR or the cellular GAPDH gene as a control. The quantity of virus added was determined by titration experiments to find the amount, measured by p24 antigen, needed to obtain results in the linear range of the assay (defined by the ratio LTR/GAPDH ≤ 1). X4-tropic viruses are indicated with asterisks; all other viruses are R5X4. The results are expressed as a percentage of infection in the absence of AMD3100 for each virus, and the error bars represent the averages plus standard deviations of three independent experiments.
The Env protein is responsible for the resistant phenotype of DH12.
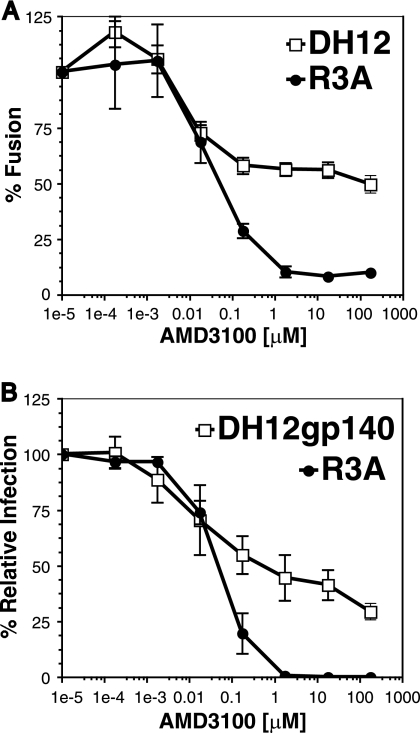
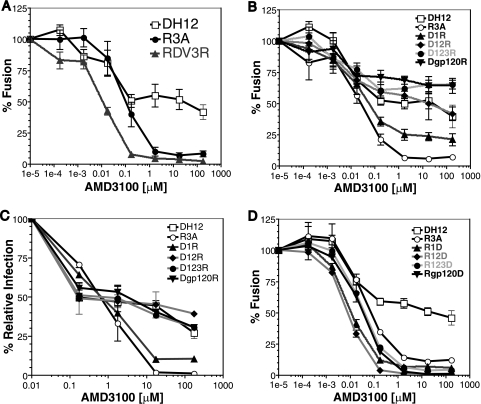
To determine if the DH12 Env protein was solely responsible for the AMD3100-resistant phenotype, cell-cell fusion and virus pseudotype assays were performed. For the cell-cell fusion assay, quail fibroblasts (QT6) were separated into two populations: targets expressing CD4, CXCR4, and the luciferase gene under the control of a T7 promoter and effectors expressing the Env of choice cloned into a plasmid (pcDNA3.1) containing a T7 promoter boosted by infection with a vaccinia virus that encodes T7 polymerase. The targets were then incubated with increasing concentrations of AMD3100 for 1 h at 37°C and then mixed and incubated at 37°C for 7 to 8 h. If fusion between the cells occurs, luciferase is produced and can be easily measured. We found that cell fusion mediated by the R3A Env was efficiently inhibited by AMD3100 (50% inhibitory concentration, ∼40 nM) while fusion mediated by the DH12 Env was inhibited by only 50% at similar drug concentrations (Fig. 2A). As the drug concentrations were increased, a plateau was observed, suggesting that the DH12 Env is able to use the drug-bound form of AMD3100, albeit less efficiently than the unbound receptor. This phenomenon has been seen in studies investigating CCR5 antagonists (44, 57).
FIG. 2.
The DH12 Env protein exhibits partial resistance to AMD3100. (A) Cell-cell fusion inhibition assays were performed with QT6 cells expressing human CD4 and CXCR4 and QT6 cells expressing either the DH12 or R3A Env protein. Receptor- and Env-expressing cells were mixed in the presence of the indicated concentrations of AMD3100, and the extents of cell-cell fusion were determined 7 to 8 h later. The extent of fusion in the absence of AMD3100 was set to 100%. (B) Infection assays using viral pseudotypes expressing either the DH12gp140 or R3A Env protein were performed on U87.CD4.CXCR4 cells in the presence or absence of AMD3100 at the indicated concentrations, and the extents of virus infection were determined 3 days later. The results are expressed as percentages of fusion/infection in the absence of AMD3100, and the error bars represent the averages ± standard errors of the mean of three independent experiments.
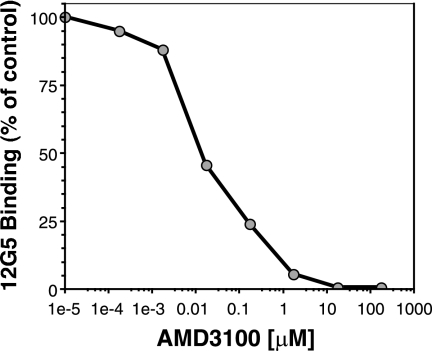
To examine the sensitivity of the DH12 Env protein to AMD3100 in the context of virus infection, pseudotypes were produced using the env genes of R3A and DH12 in the expression vector pciPRE and the NL4-3 core containing a frameshift in env and luciferase in place of its own nef gene. However, we found that the DH12 Env protein was inefficiently incorporated into these virus particles, resulting in very low levels of luciferase activity (data not shown). To overcome this, we utilized a DH12 env construct in which 140 amino acids were truncated from the cytoplasmic domain (named DH12gp140) (28). Western blot analysis showed that the truncated DH12 Env was incorporated into the pseudoviral particles approximately five times more efficiently than the wild-type DH12 Env (data not shown). We also found that DH12gp140 efficiently elicited cell-cell fusion and that saturating levels of AMD3100 inhibited fusion activity by only 50%, similar to the wild-type DH12 Env protein (data not shown). Pseudotype assays using DH12gp140 were performed by infecting U87 cells expressing CD4 and CXCR4 in the presence of different concentrations of drug (Fig. 2B). Aliquots of drug-incubated cells were also stained with a MAb to CXCR4 (12G5) to assess the degree of receptor occupancy, since AMD3100 prevents the binding of this antibody to CXCR4 (14). We found that 12G5 binding was completely inhibited by 17.5 μM AMD3100 (Fig. 3), at which point DH12 infection was inhibited only about 55% (Fig. 2B). When AMD3100 was used at 175 μM, a concentration at least 10-fold above saturating levels and still nontoxic to the cells, DH12 could still enter U87 cells expressing CD4 and CXCR4 (Fig. 2B). In contrast, R3A was fully inhibited by these concentrations of drug. These results suggest that DH12 can either utilize the AMD3100-bound conformation of CXCR4 in both cell fusion and virus infection assays or utilize low levels of receptor that fail to bind the drug or MAb 12G5, perhaps due to coreceptor heterogeneity.
FIG. 3.
Receptor occupancy as judged by FACS staining. U87.CD4.CXCR4 cells were incubated with the indicated concentrations of AMD3100 for 1 h at 37°C. Cells (200,000) from each drug condition were washed two times with FACS buffer containing the appropriate concentration of AMD3100. Next, the cells were stained in 50 μl total FACS buffer plus 10 μl of the CXCR4-specific MAb 12G5. The cells were then washed two more times, and FACS was performed on a FACSCalibur from Becton Dickinson. The mean fluorescence intensity of an isotype control was identical to that seen when AMD3100 concentrations were equal to or greater than 17.5 μM (data not shown).
DH12 is sensitive to inhibition by CCR5 and fusion inhibitors.
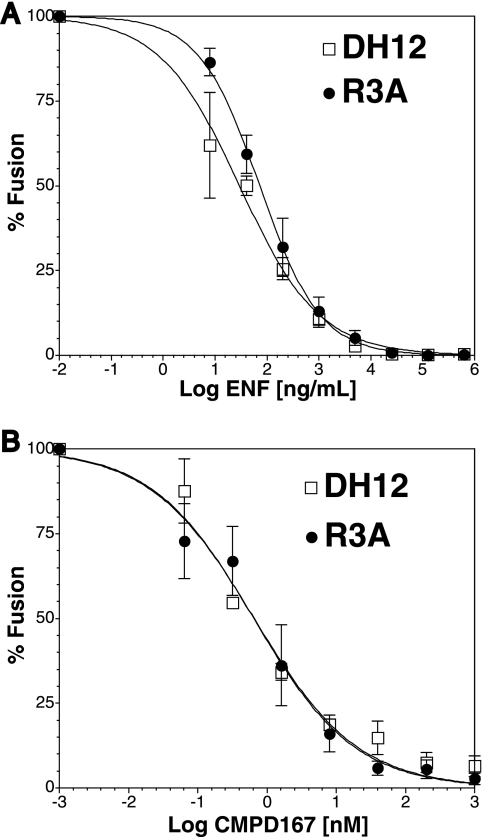
To determine if DH12 is broadly resistant to various entry inhibitors, cell-cell fusion assays using QT6 cells expressing CD4 and CCR5 in the presence of various concentrations of the CCR5 antagonist CMPD167 or using cells expressing CD4 and CXCR4 in the presence of the fusion inhibitor ENF were performed. We found that the membrane fusion activity of DH12 was fully sensitive to ENF (Fig. 4A) and to CMPD167 (Fig. 4B), with 50% inhibitory concentrations comparable to that of the R3A Env protein. Essentially identical results were obtained with virus pseudotypes; when cells expressing CD4 and CCR5 or CD4 and CXCR4 were used as targets, infection mediated by the DH12 Env protein was completely inhibited by ENF and by CCR5 antagonists (data not shown). Taken together, these results show that DH12 is not broadly resistant to entry inhibitors. Rather, it is specifically resistant to the CXCR4 inhibitor AMD3100.
FIG. 4.
DH12 and R3A are fully sensitive to other entry inhibitors. Cell-cell fusion inhibition assays were performed on QT6 cells expressing the DH12 or R3A Env protein with QT6 cells expressing CD4 and CXCR4 (A) or CD4 and CCR5 (B) and incubated with the indicated concentrations of the fusion inhibitor ENF (A) or the CCR5 inhibitor CMPD167 (B). The extent of fusion in the absence of drug was set to 100%. Least-squares regression lines were fitted using Prism 4.0a software (GraphPad, California). The error bars represent the averages ± standard errors of the mean of three independent experiments.
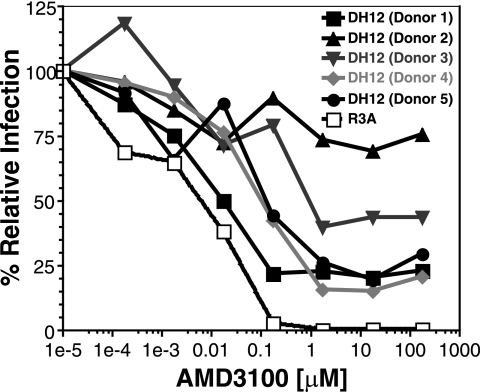
Variable sensitivity of DH12 Env to AMD3100 on primary cells.
Both virus and host factors can influence the sensitivity of HIV-1 to inhibition by coreceptor antagonists. To determine if the ability of DH12 to utilize CXCR4 in the face of saturating concentrations of AMD3100 was influenced by host cell factors, we performed pseudotype inhibition assays on primary human CD4+ T cells derived from five different donors. While DH12 primarily uses CXCR4 to infect human T cells, infection via CCR5 is also possible, albeit unlikely (59, 61). Thus, we included an inhibitory concentration of the CCR5 antagonist aplaviroc, to which DH12 is fully sensitive, in all experiments. Cells were incubated with various concentrations of AMD3100 for 1 h before the addition of either the R3A or DH12gp140 viral pseudotype. The cells were then assayed for luciferase activity 4 days after infection. We found that AMD3100 fully inhibited R3A viral-pseudotype infection of primary T cells from all five donors and did so at similar drug concentrations. Thus, a single representative inhibition curve is shown in Fig. 5. In contrast, infection mediated by the DH12gp140 viral pseudotypes was never fully inhibited by AMD3100. Rather, as AMD3100 concentrations were increased, variable degrees of inhibition were observed, depending on the donor. The concentrations at which partial inhibition was achieved varied by about 1 log unit, while the extents of maximal inhibition varied from approximately 20% (donor 4) to 75% (donor 1). Thus, the DH12 Env protein can continue to mediate CXCR4-dependent membrane fusion, even in the face of saturating concentrations of AMD3100, on both cell lines and primary T cells. The extent to which AMD3100 inhibits virus infection can vary considerably depending on the donor from whom the T cells are obtained.
FIG. 5.
DH12gp140 is resistant to AMD3100 inhibition on primary CD4+ T cells. Pseudotype inhibition assays were performed on primary CD4+ T cells, using the DH12 C-terminally truncated and the full-length R3A env genes in the NL4-3 background in the presence of increasing concentrations of AMD3100. The extent of infection in the absence of drug was set to 100%. Each line represents a different T-cell donor. R3A inhibition curves for all five donor CD4+ T cells were almost identical and hence are shown as a single representative inhibition curve.
The V1/V2 loop of DH12 is sufficient to impart AMD3100 resistance to heterologous Envs.
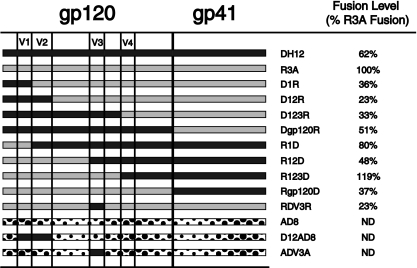
To identify the determinants within the DH12 Env protein responsible for the AMD3100-resistant phenotype, chimeras between the env genes of DH12 and R3A were constructed (Fig. 6). All chimeras were screened for functionality in a cell-cell fusion assay and were found to elicit fusion with efficiencies ranging from approximately one-third to twice the levels obtained with the R3A Env protein (Fig. 6). Because the V3 loop of Env plays a critical role in coreceptor interactions and can also play a role in resistance to coreceptor inhibitors (29, 57), we first introduced the V3 loop from DH12 into the R3A background. However, this chimera (RDV3R) was fully sensitive to AMD3100 (Fig. 7A). We then examined chimeras in which increasingly large N-terminal portions of DH12 gp120 were introduced into the R3A background (Fig. 7B). We found that the addition of the first 150 residues of DH12 (D1R), which includes the V1 region, resulted in an Env chimera that could not be fully inhibited by AMD3100, with approximately 20% membrane fusion activity remaining when saturating amounts of AMD3100 were used. A chimera containing the first 299 residues of DH12 (D12R), extending up to the last cysteine residue of V2, behaved like DH12 in the fusion assay, with AMD3100 inhibiting its fusion activity by approximately 50% at high concentrations. Env chimeras containing larger portions of the DH12 gp120 subunit, or even the entire subunit, likewise exhibited resistance to AMD3100. Essentially identical results were seen when U87.CD4.CXCR4 cells were infected with DH12/R3A Env chimera pseudotype viruses (Fig. 7C), demonstrating again that the V1/V2 loop of HIV-1 DH12 is necessary and sufficient to impart AMD3100 resistance to the R3A Env.
FIG. 6.
Panel of HIV-1 Env chimeras. Chimeras between DH12 and R3A env genes were made using overlapping PCR. The chimeric env genes were then cloned into the pciPRE mammalian expression vector using the KpnI and XbaI restriction sites. The chimeras were sequenced to confirm their identities, and cell-cell fusion assays were performed to test Env functionality. The DH12/AD8 chimeras were previously described by Cho et al. (5). The fusion levels obtained with each chimera in the absence of drug were assessed and compared to the consistently high fusion levels of R3A.
FIG. 7.
The V1/V2 loop of DH12 is necessary and sufficient to confer AMD3100 resistance in the R3A Env background. (A, B, and D) Cell-cell fusion inhibition assays were performed with QT6 cells expressing the chimeric Env proteins indicated using QT6 cells expressing human CD4 and CXCR4 as targets. Prior to cell mixing, the CD4- and CXCR4-expressing cells were incubated with the specified concentrations of AMD3100 for 1 h. Cell-cell fusion was determined via luciferase reading 7 to 8 h after cell mixing. The extent of fusion for each Env in the absence of AMD3100 was set to 100%. (C) Infection assays using viral pseudotypes expressing either the DH12gp140, R3A, or chimeric Env protein were performed on U87.CD4.CXCR4 cells in the presence or absence of AMD3100 at the indicated concentrations, and the extents of virus infection were determined 3 days later. The results are expressed as percentages of fusion/infection in the absence of AMD3100, and the error bars represent the averages ± standard errors of the mean of three independent experiments.
We also constructed a panel of reciprocal chimeras in which N-terminal portions of the DH12 gp120 protein were replaced with the corresponding regions from the R3A Env protein. We found that replacing the N-terminal portion of DH12 gp120 up to and including the V1 region completely abolished the AMD3100 resistance phenotype (Fig. 7C). Chimeras containing larger regions of the R3A Env protein continued to be fully sensitive to AMD3100. Thus, we conclude that the V1/V2 region of the DH12 Env protein is both necessary and sufficient for conferring resistance to AMD3100 on the R3A Env protein.
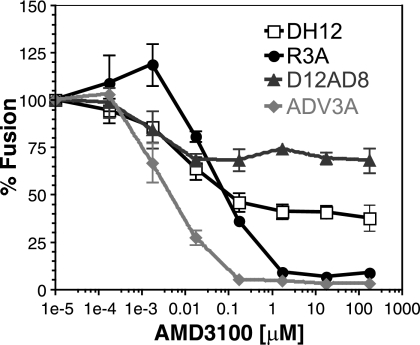
To test if the V1/V2 loop of DH12 alone could confer AMD3100 resistance on other HIV-1 strains, chimeras between DH12 and the R5-tropic HIV-1 AD8 were tested. Cho and colleagues have shown that both the DH12 V1/V2 loop and the V3 loop can confer CXCR4 usage when introduced into the AD8 Env protein, which otherwise uses only CCR5 (4). We first tested both chimeras in the cell-cell fusion assay to confirm the use of CXCR4 in addition to CCR5 for cell entry (data not shown). When the experiment was repeated in the presence of increasing concentrations of AMD3100, we found that the AD8 chimera containing the DH12 V1/V2 region exhibited resistance to AMD3100 while the AD8 chimera containing the DH12 V3 loop did not (Fig. 8). Thus, only the V1/V2 loop of DH12, but not the V3 region, was sufficient to confer AMD3100 resistance in a second, heterologous HIV-1 Env background.
FIG. 8.
The V1/V2 loop of DH12 can confer AMD3100 resistance on the AD8 HIV-1 strain. Cell-cell fusion inhibition assays were performed with QT6 cells expressing the indicated DH12/AD8 chimeric Env protein and QT6 cells expressing human CD4 and CXCR4. The cells were incubated with increasing concentrations of AMD3100 for 1 h prior to being mixed. The extent of fusion for each Env in the absence of AMD3100 was set to 100%. The error bars represent the averages ± standard errors of the mean of three independent experiments.
Other primary HIV-1 strains display similar AMD3100 resistance.
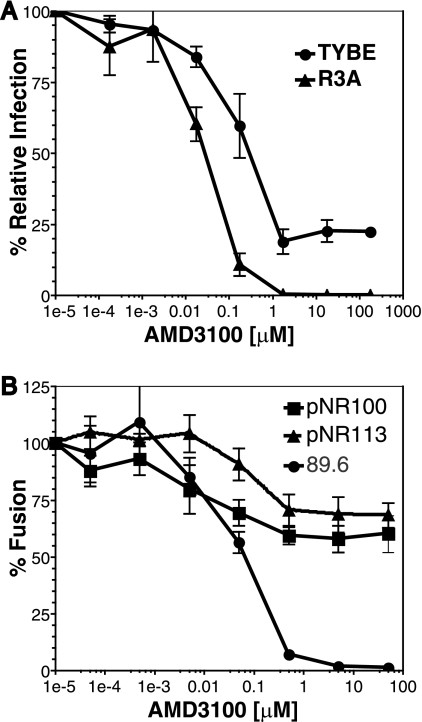
Two other HIV-1 strains, SPL-3 and TYBE, were identified in the preliminary resistance screen as being incompletely inhibited by AMD3100 (Fig. 1). In addition, we identified two X4-tropic Env clones (pNR100 and pNR113) from a patient who underwent and failed ENF therapy that also exhibited resistance to AMD3100 (47). Full AMD3100 inhibition curves were performed using TYBE in pseudotype assays and pNR100 and pNR113 in cell-cell fusion inhibition assays. Figure 9A and B show that all three HIV-1 Envs exhibited a plateau effect as AMD3100 concentrations were increased. Saturating concentrations of AMD3100 inhibited infection of TYBE pseudotypes by approximately 80% (Fig. 9A), while cell fusion mediated by the pNR100 and pNR113 Env proteins was inhibited by approximately 35% and 25%, respectively (Fig. 9B). These results show that the baseline resistance to AMD3100, manifested as incomplete suppression in the face of saturating concentrations of drug, is a characteristic exhibited by several primary HIV-1 strains.
FIG. 9.
Other primary HIV-1 strains exhibit resistance to AMD3100 inhibition. (A) Pseudotype inhibition assays were performed on U87.CD4.CXCR4 cells. Increasing concentrations of AMD3100 were incubated with the cells prior to TYBE pseudotype addition. (B) Cell-cell fusion assays were performed using QT6 cells expressing pNR100 and pNR113 Env proteins and QT6 cells expressing CD4 and CXCR4. Target cells were incubated with AMD3100 1 h prior to effector-target cell mixing. The extent of infection/fusion in the absence of AMD3100 was set to 100%.The error bars represent the averages ± standard errors of the mean of three independent experiments.
DISCUSSION
The failure of AMD3100 to completely inhibit infection by HIV-1 DH12 and several other clade B virus strains could not be ascribed to an artifact of any particular experimental system: incomplete inhibition in the form of a stable, residual amount of virus infection in the face of saturating concentrations of AMD3100 was observed with replication-competent virus, virus pseudotypes, and cell fusion assays and was seen with both cell lines and primary cell types. The fact that we identified several R5X4 and X4 virus strains that exhibited this form of baseline resistance to AMD3100 suggests that this phenotype may be relatively common. However, this pattern of resistance is quite different from what is observed with other classes of antiretroviral agents, such as reverse transcriptase (reviewed in reference 2) and protease inhibitors (40). Resistance to these drugs typically involves mutations that modify the drug binding site, resulting in higher levels of drug being needed to achieve the same antiviral effect. In the case of CXCR4 and CCR5 coreceptor antagonists, this pathway is not an option, as these drugs target host molecules. As a result, other mechanisms, such as the accrual of mutations in Env that enable a virus to utilize a different coreceptor or to utilize the drug-bound conformation of the targeted coreceptor, must be employed.
In the case of DH12 and the other AMD3100-resistant viruses studied here, we feel that the most likely mechanism to account for their resistance to AMD3100 is utilization of the drug-bound form of CXCR4. The reasons for this conclusion are several. First, the mode of resistance is clearly noncompetitive in nature. Once AMD3100 concentrations reached 10 μM, virus infection was not further inhibited even if drug concentrations were increased by an additional several log units. This finding is not consistent with virus outcompeting AMD3100 for CXCR4 binding. Second, the plateau concentration of AMD3100, beyond which further viral inhibition was not observed, was similar to the concentration needed to fully prevent binding of a monoclonal antibody to CXCR4, suggesting that all coreceptor molecules were drug bound. Third, the plateau effect was observed in multiple cell types, and was always dependent upon the presence of CXCR4, arguing that DH12 did not utilize a coreceptor other than CXCR4. Thus, we conclude that DH12, TYBE, SPL-3, pNR113, and pNR100 could use AMD3100-bound CXCR4 to infect cells, with the magnitude of the plateau reflecting the efficiency with which each virus could use the drug-bound form of CXCR4 relative to its drug-free conformation.
The ability of HIV strains to use the AMD3100-bound form of CXCR4 has not previously been described, to the best of our knowledge, though utilization of drug-bound CCR5 has been described for several virus strains derived by passage in the presence of increasing concentrations of CCR5 antagonists (29, 37, 44, 57) and by viruses into which V3 loop deletions have been introduced (30, 34, 42). Taken together, these studies indicate that mutations conferring resistance to CCR5 antagonists often, but not always, involve mutations in the V3 loop, and when introduced into other Env backgrounds, these mutations have not conferred the drug-resistant phenotype (37). In the case of DH12, we found that the V1/V2 region was necessary and sufficient to confer the ability to recognize the AMD3100-bound conformation of CXCR4 on the R3A and AD8 Env proteins. This differs markedly from a virus that was made resistant to AMD3100 through in vitro passaging (8). De Vreese et al. passaged the X4-tropic HIV-1 NL4-3 in the presence of increasing concentrations of AMD3100. The resulting drug-resistant virus had several mutations distributed throughout the gp120 subunit of Env, with the majority focused in or around the V3 loop. The properties of this virus differ in two important respects from those of the viruses described here. First, the in vitro-passaged virus described by De Vreese et al. could be fully inhibited by AMD3100, albeit at higher drug concentrations (9). Second, the determinants for this resistant phenotype likely involved the V3 loop. The V3 loop is clearly the major determinant of coreceptor choice (reviewed in references 19 and 21), while the V1/V2 region plays a subsidiary role (4, 24, 32, 54, 55). However, the DH12 Env, as well as its V1/V2 region, exhibits some unusual properties. First, DH12 was selected for study by Shibata and colleagues, as it was one of 23 HIV-1 isolates tested that could efficiently infect chimpanzee T cells (53). Second, the DH12 Env is highly fusogenic, and the virus is highly cytopathic (4, 5, 53). It uses both CCR5 and CXCR4 in an efficient manner. Third, either the V1/V2 region or the V3 loop from DH12 can confer the ability to use CXCR4 on the R5 virus strain AD8, while introduction of both of these regions improves the efficiency with which CXCR4 can be used (4, 5). It will be important to introduce the DH12 V1/V2 region into additional Env proteins to determine if the resistant phenotype can be conferred on other heterologous viruses.
How might a virus utilize the drug-bound form of CXCR4? A possible model is suggested by studies of how HIV-1 Env interacts with CCR5 and CXCR4 in general, as well as insights gleaned from a virus strain that can efficiently utilize drug-bound forms of CCR5 due to partial deletion of its V3 loop (30, 34, 42). There is good evidence that the distal portion of the V3 loop interacts with the extracellular loops of CCR5 or CXCR4, while the base of the V3 loop and bridging sheet bind to the amino-terminal domain of the coreceptor, with sulfated tyrosine residues playing a particularly important role in the case of CCR5 (13, 25, 41, 42). Removal of 15 residues from the center of the V3 loop from the R5X4 virus strain R3A renders the virus completely resistant to CCR5 antagonists through a mechanism that most likely involves efficient utilization of the drug-bound coreceptor (30). We have found that the resulting virus, termed TA1, is more sensitive than the parental virus to mutations in the N-terminal domain of CCR5, suggesting a model in which TA1 can utilize drug-bound CCR5 by interacting primarily with the N-terminal domain of the coreceptor, whose conformation may not be greatly affected by drug binding. It is possible that a similar mechanism might account for the ability of DH12 and other viruses to utilize AMD3100-bound CXCR4, though as yet we have no direct evidence for this hypothesis. If so, the V1/V2 region must play a significant role in this process. It is possible that the V1/V2 region interacts directly with some portion of CXCR4 and that the DH12 V1/V2 region does so particularly well, though structure-function studies have not provided strong evidence for such a model. An alternative explanation is that changes in the conformation or orientation of the V1/V2 region change the conformation or exposure of the base of the V3 loop, which is known to play an important role in binding the N-terminal domain of the coreceptor (25). However, it should be noted that elimination of the V1/V2 region in R5 Envs has not been associated with drug resistance (30). The panel of disparate, AMD3100-resistant X4 and R5X4 viruses described here will provide the molecular tools needed to more fully understand this unusual phenotype.
It is important to also consider the fact that when DH12 was used to infect human peripheral blood mononuclear cells in the presence of saturating amounts of AMD3100, the magnitude of the plateau varied depending on the donor from whom the cells were collected. Thus, host cell factors also contribute to the AMD3100-resistant phenotype, though the mechanism(s) responsible for this donor-dependent effect is not known. Finally, the virus strains studied here exhibited significant resistance to AMD3100 even though all were cloned from patients who had not received the drug. Such baseline resistance to this CXCR4 antagonist, if common, could affect future clinical use of this class of antiviral agents.
Acknowledgments
We thank Lamorris Loftin, Farida Shaheen, Lingshu Wang, and Yanjie Yi for excellent technical advice and reagents; Michael Cho for the DH12/AD8 chimeras; and Rick Bushman, Jim Hoxie, and Dennis Kolson for invaluable advice.
This work was supported by NIH grants T32 AI 07632 to J.E.H., F32 AI 068442 to N.R., 5R25 GM 071745-03 to L.-J.S, and AI R01 40880 to R.W.D. and by the University of Pennsylvania Center for AIDS Research (P30 AI 045008).
Footnotes
Published ahead of print on 17 September 2008.
REFERENCES
- 1.Alexander, W. A., B. Moss, and T. R. Fuerst. 1992. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J. Virol. 662934-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer, P. L., S. G. Sarafianos, P. K. Clark, E. Arnold, and S. H. Hughes. 2006. Why do HIV-1 and HIV-2 use different pathways to develop AZT resistance? PLoS Pathog. 2e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briz, V., E. Poveda, and V. Soriano. 2006. HIV entry inhibitors: mechanisms of action and resistance pathways. J. Antimicrob. Chemother. 57619-627. [DOI] [PubMed] [Google Scholar]
- 4.Cho, M. W., M. K. Lee, M. C. Carney, J. F. Berson, R. W. Doms, and M. A. Martin. 1998. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J. Virol. 722509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, M. W., R. Shibata, and M. A. Martin. 1996. Infection of chimpanzee peripheral blood mononuclear cells by human immunodeficiency virus type 1 requires cooperative interaction between multiple variable regions of gp120. J. Virol. 707318-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Clercq, E. 2003. The bicyclam AMD3100 story. Nat. Rev. Drug Discov. 2581-587. [DOI] [PubMed] [Google Scholar]
- 7.De Clercq, E., N. Yamamoto, R. Pauwels, J. Balzarini, M. Witvrouw, K. De Vreese, Z. Debyser, B. Rosenwirth, P. Peichl, R. Datema, et al. 1994. Highly potent and selective inhibition of human immunodeficiency virus by the bicyclam derivative JM3100. Antimicrob. Agents Chemother. 38668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vreese, K., V. Kofler-Mongold, C. Leutgeb, V. Weber, K. Vermeire, S. Schacht, J. Anne, E. de Clercq, R. Datema, and G. Werner. 1996. The molecular target of bicyclams, potent inhibitors of human immunodeficiency virus replication. J. Virol. 70689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Vreese, K., I. Van Nerum, K. Vermeire, J. Anne, and E. De Clercq. 1997. Sensitivity of human immunodeficiency virus to bicyclam derivatives is influenced by the three-dimensional structure of gp120. Antimicrob. Agents Chemother. 412616-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donzella, G. A., D. Schols, S. W. Lin, J. A. Este, K. A. Nagashima, P. J. Maddon, G. P. Allaway, T. P. Sakmar, G. Henson, E. De Clercq, and J. P. Moore. 1998. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 472-77. [DOI] [PubMed] [Google Scholar]
- 11.Endres, M. J., P. R. Clapham, M. Marsh, M. Ahuja, J. D. Turner, A. McKnight, J. F. Thomas, B. Stoebenau-Haggarty, S. Choe, P. J. Vance, T. N. Wells, C. A. Power, S. S. Sutterwala, R. W. Doms, N. R. Landau, and J. A. Hoxie. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87745-756. [DOI] [PubMed] [Google Scholar]
- 12.Este, J. A., and A. Telenti. 2007. HIV entry inhibitors. Lancet 37081-88. [DOI] [PubMed] [Google Scholar]
- 13.Farzan, M., T. Mirzabekov, P. Kolchinsky, R. Wyatt, M. Cayabyab, N. P. Gerard, C. Gerard, J. Sodroski, and H. Choe. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96667-676. [DOI] [PubMed] [Google Scholar]
- 14.Fricker, S. P., V. Anastassov, J. Cox, M. C. Darkes, O. Grujic, S. R. Idzan, J. Labrecque, G. Lau, R. M. Mosi, K. L. Nelson, L. Qin, Z. Santucci, and R. S. Wong. 2006. Characterization of the molecular pharmacology of AMD3100: a specific antagonist of the G-protein coupled chemokine receptor, CXCR4. Biochem. Pharmacol. 72588-596. [DOI] [PubMed] [Google Scholar]
- 15.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 2962354-2360. [DOI] [PubMed] [Google Scholar]
- 16.Ghaffari, G., D. L. Tuttle, D. Briggs, B. R. Burkhardt, D. Bhatt, W. A. Andiman, J. W. Sleasman, and M. M. Goodenow. 2005. Complex determinants in human immunodeficiency virus type 1 envelope gp120 mediate CXCR4-dependent infection of macrophages. J. Virol. 7913250-13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glushakova, S., Y. Yi, J. C. Grivel, A. Singh, D. Schols, E. De Clercq, R. G. Collman, and L. Margolis. 1999. Preferential coreceptor utilization and cytopathicity by dual-tropic HIV-1 in human lymphoid tissue ex vivo. J. Clin. Investig. 104R7-R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodenow, M. M., and R. G. Collman. 2006. HIV-1 coreceptor preference is distinct from target cell tropism: a dual-parameter nomenclature to define viral phenotypes. J. Leukoc. Biol. 80965-972. [DOI] [PubMed] [Google Scholar]
- 19.Gorry, P. R., R. L. Dunfee, M. E. Mefford, K. Kunstman, T. Morgan, J. P. Moore, J. R. Mascola, K. Agopian, G. H. Holm, A. Mehle, J. Taylor, M. Farzan, H. Wang, P. Ellery, S. J. Willey, P. R. Clapham, S. M. Wolinsky, S. M. Crowe, and D. Gabuzda. 2007. Changes in the V3 region of gp120 contribute to unusually broad coreceptor usage of an HIV-1 isolate from a CCR5 Delta32 heterozygote. Virology 362163-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrigan, P. R., J. S. Montaner, S. A. Wegner, W. Verbiest, V. Miller, R. Wood, and B. A. Larder. 2001. World-wide variation in HIV-1 phenotypic susceptibility in untreated individuals: biologically relevant values for resistance testing. AIDS 151671-1677. [DOI] [PubMed] [Google Scholar]
- 21.Hartley, O., P. J. Klasse, Q. J. Sattentau, and J. P. Moore. 2005. V3: HIV's switch-hitter. AIDS Res. Hum Retrovir. 21171-189. [DOI] [PubMed] [Google Scholar]
- 22.Heil, M. L., J. M. Decker, J. N. Sfakianos, G. M. Shaw, E. Hunter, and C. A. Derdeyn. 2004. Determinants of human immunodeficiency virus type 1 baseline susceptibility to the fusion inhibitors enfuvirtide and T-649 reside outside the peptide interaction site. J. Virol. 787582-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrix, C. W., A. C. Collier, M. M. Lederman, D. Schols, R. B. Pollard, S. Brown, J. B. Jackson, R. W. Coombs, M. J. Glesby, C. W. Flexner, G. J. Bridger, K. Badel, R. T. MacFarland, G. W. Henson, and G. Calandra. 2004. Safety, pharmacokinetics, and antiviral activity of AMD3100, a selective CXCR4 receptor inhibitor, in HIV-1 infection. J. Acquir. Immune Defic. Syndr. 371253-1262. [DOI] [PubMed] [Google Scholar]
- 24.Hoffman, T. L., E. B. Stephens, O. Narayan, and R. W. Doms. 1998. HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc. Natl. Acad. Sci. USA 9511360-11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, C. C., S. N. Lam, P. Acharya, M. Tang, S. H. Xiang, S. S. Hussan, R. L. Stanfield, J. Robinson, J. Sodroski, I. A. Wilson, R. Wyatt, C. A. Bewley, and P. D. Kwong. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 3171930-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igarashi, T., O. K. Donau, H. Imamichi, M. J. Dumaurier, R. Sadjadpour, R. J. Plishka, A. Buckler-White, C. Buckler, A. F. Suffredini, H. C. Lane, J. P. Moore, and M. A. Martin. 2003. Macrophage-tropic simian/human immunodeficiency virus chimeras use CXCR4, not CCR5, for infections of rhesus macaque peripheral blood mononuclear cells and alveolar macrophages. J. Virol. 7713042-13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ketas, T. J., S. E. Kuhmann, A. Palmer, J. Zurita, W. He, S. K. Ahuja, P. J. Klasse, and J. P. Moore. 2007. Cell surface expression of CCR5 and other host factors influence the inhibition of HIV-1 infection of human lymphocytes by CCR5 ligands. Virology 364281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, Y. B., M. K. Lee, D. P. Han, and M. W. Cho. 2001. Development of a safe and rapid neutralization assay using murine leukemia virus pseudotyped with HIV type 1 envelope glycoprotein lacking the cytoplasmic domain. AIDS Res. Hum. Retrovir. 171715-1724. [DOI] [PubMed] [Google Scholar]
- 29.Kuhmann, S. E., P. Pugach, K. J. Kunstman, J. Taylor, R. L. Stanfield, A. Snyder, J. M. Strizki, J. Riley, B. M. Baroudy, I. A. Wilson, B. T. Korber, S. M. Wolinsky, and J. P. Moore. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 782790-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laakso, M. M., F. H. Lee, B. Haggarty, C. Agrawal, K. M. Nolan, M. Biscone, J. Romano, A. P. Jordan, G. J. Leslie, E. G. Meissner, L. Su, J. A. Hoxie, and R. W. Doms. 2007. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 3e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labrosse, B., J. L. Labernardiere, E. Dam, V. Trouplin, K. Skrabal, F. Clavel, and F. Mammano. 2003. Baseline susceptibility of primary human immunodeficiency virus type 1 to entry inhibitors. J. Virol. 771610-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labrosse, B., C. Treboute, A. Brelot, and M. Alizon. 2001. Cooperation of the V1/V2 and V3 domains of human immunodeficiency virus type 1 gp120 for interaction with the CXCR4 receptor. J. Virol. 755457-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, B., M. Sharron, L. J. Montaner, D. Weissman, and R. W. Doms. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. USA 965215-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, G., A. Bertolotti-Ciarlet, B. Haggarty, J. Romano, K. M. Nolan, G. J. Leslie, A. P. Jordan, C. C. Huang, P. D. Kwong, R. W. Doms, and J. A. Hoxie. 2007. Replication-competent variants of human immunodeficiency virus type 2 lacking the V3 loop exhibit resistance to chemokine receptor antagonists. J. Virol. 819956-9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda, K., H. Nakata, Y. Koh, T. Miyakawa, H. Ogata, Y. Takaoka, S. Shibayama, K. Sagawa, D. Fukushima, J. Moravek, Y. Koyanagi, and H. Mitsuya. 2004. Spirodiketopiperazine-based CCR5 inhibitor which preserves CC-chemokine/CCR5 interactions and exerts potent activity against R5 human immunodeficiency virus type 1 in vitro. J. Virol. 788654-8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malkevitch, N., D. H. McDermott, Y. Yi, J. C. Grivel, D. Schols, E. De Clercq, P. M. Murphy, S. Glushakova, R. G. Collman, and L. Margolis. 2001. Coreceptor choice and T cell depletion by R5, X4, and R5X4 HIV-1 variants in CCR5-deficient (CCR5delta32) and normal human lymphoid tissue. Virology. 281239-247. [DOI] [PubMed] [Google Scholar]
- 37.Marozsan, A. J., S. E. Kuhmann, T. Morgan, C. Herrera, E. Rivera-Troche, S. Xu, B. M. Baroudy, J. Strizki, and J. P. Moore. 2005. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D). Virology 338182-199. [DOI] [PubMed] [Google Scholar]
- 38.Meissner, E. G., K. M. Duus, F. Gao, X. F. Yu, and L. Su. 2004. Characterization of a thymus-tropic HIV-1 isolate from a rapid progressor: role of the envelope. Virology 32874-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2760-766. [DOI] [PubMed] [Google Scholar]
- 41.Moore, J. P., S. G. Kitchen, P. Pugach, and J. A. Zack. 2004. The CCR5 and CXCR4 coreceptors—central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retrovir. 20111-126. [DOI] [PubMed] [Google Scholar]
- 42.Nolan, K. M., A. P. Jordan, and J. A. Hoxie. 2008. Effects of partial deletions within the human immunodeficiency virus type 1 V3 loop on coreceptor tropism and sensitivity to entry inhibitors. J. Virol. 82664-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Platt, E. J., J. P. Durnin, and D. Kabat. 2005. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J. Virol. 794347-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pugach, P., A. J. Marozsan, T. J. Ketas, E. L. Landes, J. P. Moore, and S. E. Kuhmann. 2007. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology 361212-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rana, S., G. Besson, D. G. Cook, J. Rucker, R. J. Smyth, Y. Yi, J. D. Turner, H. H. Guo, J. G. Du, S. C. Peiper, E. Lavi, M. Samson, F. Libert, C. Liesnard, G. Vassart, R. W. Doms, M. Parmentier, and R. G. Collman. 1997. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the delta ccr5 mutation. J. Virol. 713219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ray, N., and R. W. Doms. 2006. HIV-1 coreceptors and their inhibitors. Curr. Top. Microbiol. Immunol. 30397-120. [DOI] [PubMed] [Google Scholar]
- 47.Ray, N., J. E. Harrison, L. A. Blackburn, J. N. Martin, S. G. Deeks, and R. W. Doms. 2007. Clinical resistance to enfuvirtide does not affect susceptibility of human immunodeficiency virus type 1 to other classes of entry inhibitors. J. Virol. 813240-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 9916249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reynes, J., P. Portales, M. Segondy, V. Baillat, P. Andre, O. Avinens, M. C. Picot, J. Clot, J. F. Eliaou, and P. Corbeau. 2001. CD4 T cell surface CCR5 density as a host factor in HIV-1 disease progression. AIDS 151627-1634. [DOI] [PubMed] [Google Scholar]
- 50.Rucker, J., B. J. Doranz, A. L. Edinger, D. Long, J. F. Berson, and R. W. Doms. 1997. Cell-cell fusion assay to study role of chemokine receptors in human immunodeficiency virus type 1 entry. Methods Enzymol. 288118-133. [DOI] [PubMed] [Google Scholar]
- 51.Schols, D., J. A. Este, G. Henson, and E. De Clercq. 1997. Bicyclams, a class of potent anti-HIV agents, are targeted at the HIV coreceptor fusin/CXCR-4. Antivir. Res. 35147-156. [DOI] [PubMed] [Google Scholar]
- 52.Schols, D., S. Struyf, J. Van Damme, J. A. Este, G. Henson, and E. De Clercq. 1997. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J. Exp. Med. 1861383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shibata, R., M. D. Hoggan, C. Broscius, G. Englund, T. S. Theodore, A. Buckler-White, L. O. Arthur, Z. Israel, A. Schultz, H. C. Lane, et al. 1995. Isolation and characterization of a syncytium-inducing, macrophage/T-cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J. Virol. 694453-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shieh, J. T., J. Martin, G. Baltuch, M. H. Malim, and F. Gonzalez-Scarano. 2000. Determinants of syncytium formation in microglia by human immunodeficiency virus type 1: role of the V1/V2 domains. J. Virol. 74693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smyth, R. J., Y. Yi, A. Singh, and R. G. Collman. 1998. Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate. J. Virol. 724478-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384184-187. [DOI] [PubMed] [Google Scholar]
- 57.Westby, M., C. Smith-Burchnell, J. Mori, M. Lewis, M. Mosley, M. Stockdale, P. Dorr, G. Ciaramella, and M. Perros. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 812359-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, L., W. A. Paxton, N. Kassam, N. Ruffing, J. B. Rottman, N. Sullivan, H. Choe, J. Sodroski, W. Newman, R. A. Koup, and C. R. Mackay. 1997. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J. Exp. Med. 1851681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yi, Y., S. N. Isaacs, D. A. Williams, I. Frank, D. Schols, E. De Clercq, D. L. Kolson, and R. G. Collman. 1999. Role of CXCR4 in cell-cell fusion and infection of monocyte-derived macrophages by primary human immunodeficiency virus type 1 (HIV-1) strains: two distinct mechanisms of HIV-1 dual tropism. J. Virol. 737117-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yi, Y., C. Lee, Q. H. Liu, B. D. Freedman, and R. G. Collman. 2004. Chemokine receptor utilization and macrophage signaling by human immunodeficiency virus type 1 gp120: implications for neuropathogenesis. J. Neurovirol 10(Suppl. 1)91-96. [DOI] [PubMed] [Google Scholar]
- 61.Yi, Y., F. Shaheen, and R. G. Collman. 2005. Preferential use of CXCR4 by R5X4 human immunodeficiency virus type 1 isolates for infection of primary lymphocytes. J. Virol. 791480-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]