Abstract
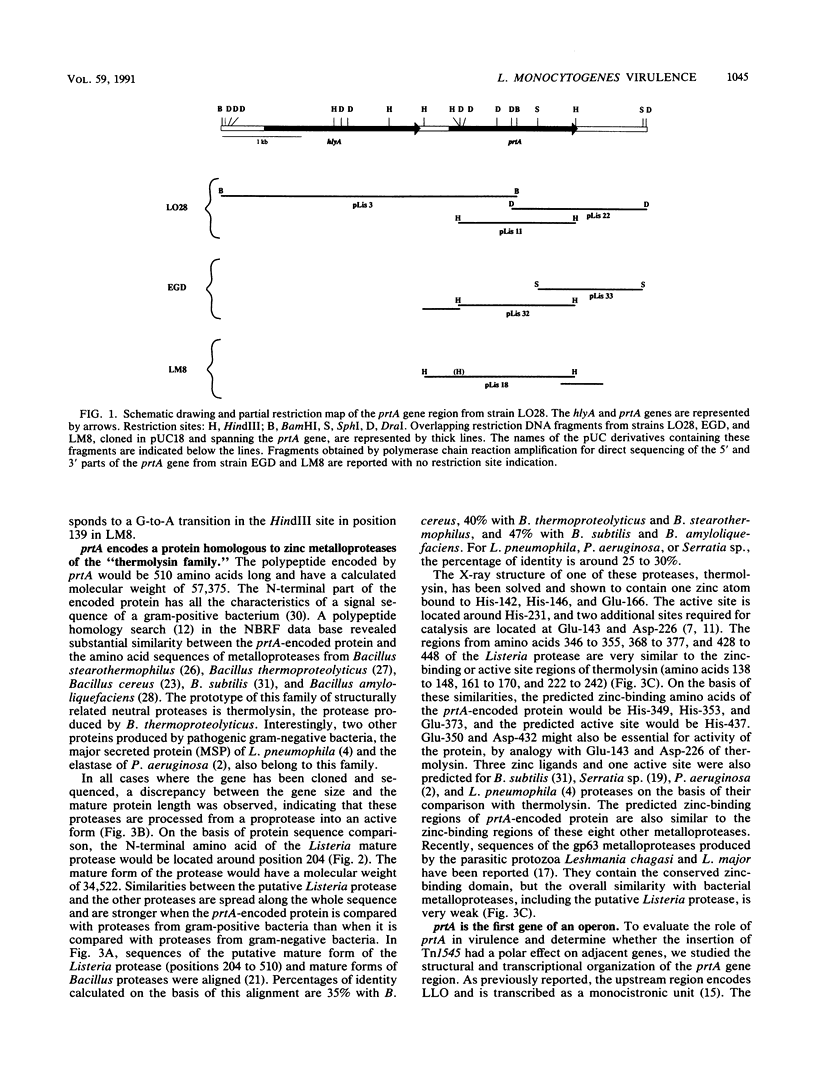
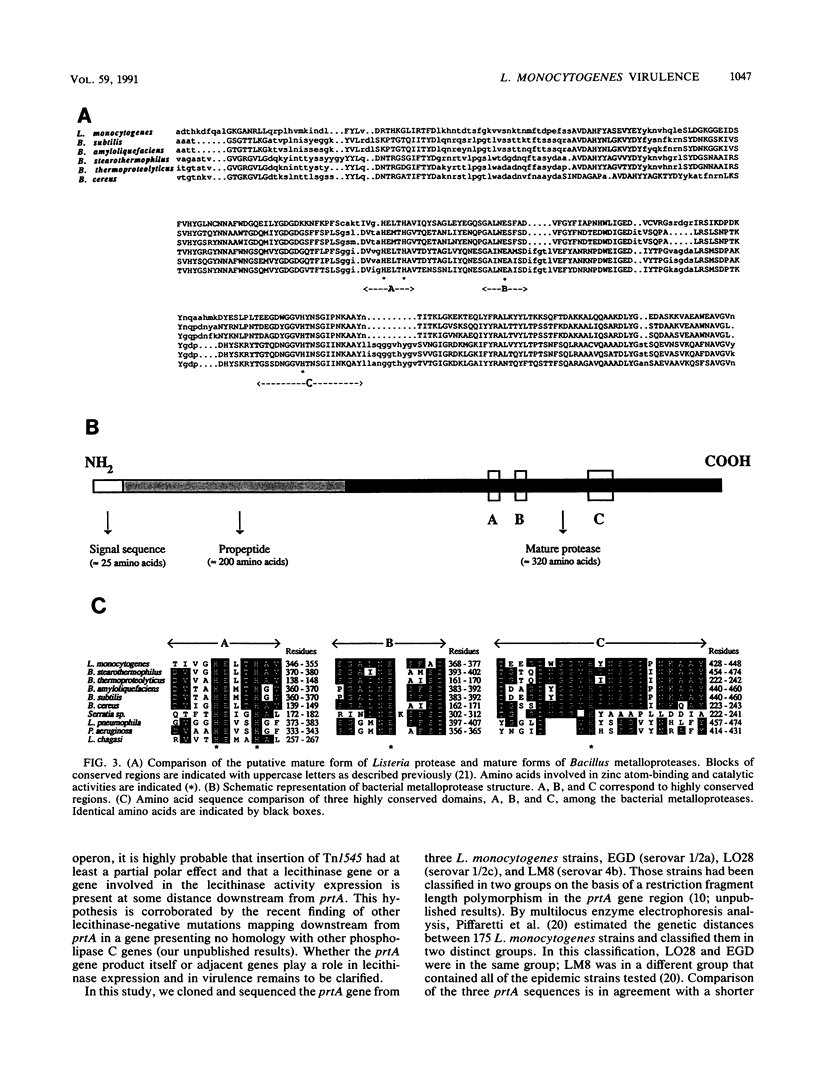
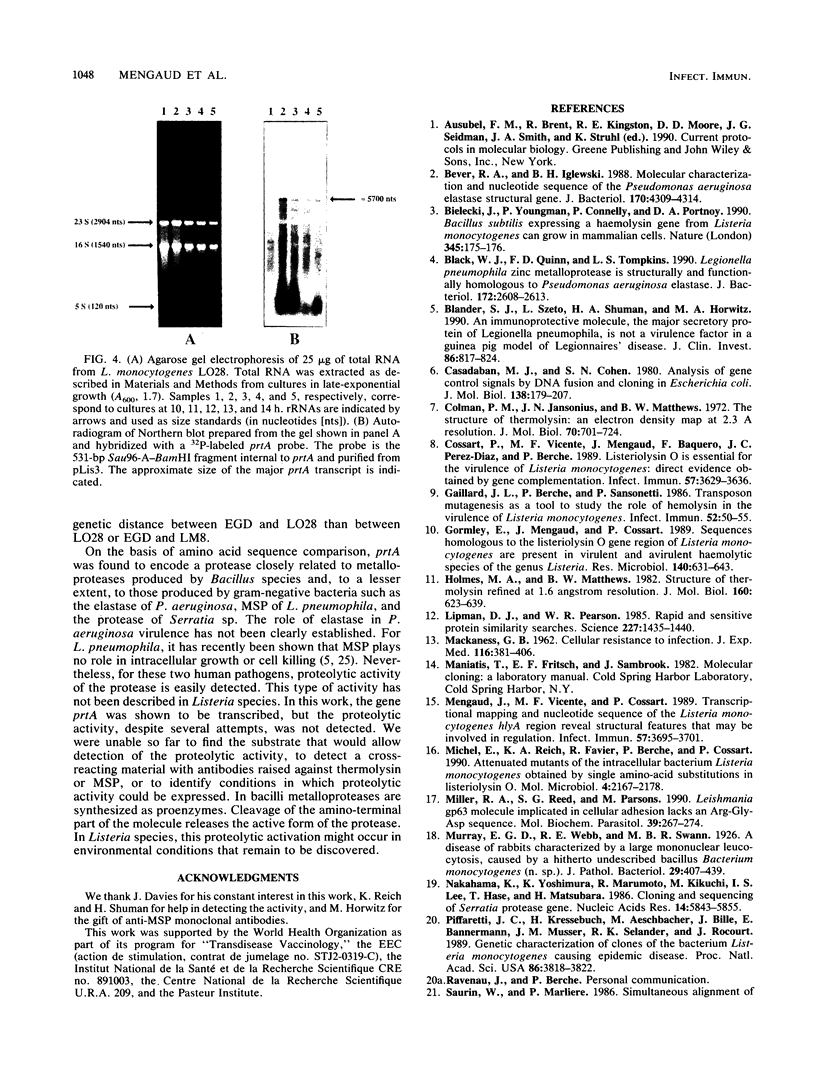
The region flanking the transposon in a Tn1545-induced lecithinase-negative mutant of Listeria monocytogenes EGD was cloned and sequenced. The transposon had inserted in ORF D, the open reading frame previously identified downstream from hlyA, the gene encoding listeriolysin O. The complete sequence of ORF D from strain EGD has been determined as well as that of two other strains: LO28, a clinical isolate; and LM8, an epidemic strain. ORF D is 1,533 bp long and encodes a protein highly homologous to metalloproteases of bacilli, Serratia sp., Legionella pneumophila, and Pseudomonas aeruginosa. It was renamed prtA. Northern RNA blot analysis indicated that prtA is the first gene of a 6-kb operon, suggesting that the lecithinase-negative phenotype of the mutant might be due to a polar effect of the transposon insertion.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bever R. A., Iglewski B. H. Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J Bacteriol. 1988 Sep;170(9):4309–4314. doi: 10.1128/jb.170.9.4309-4314.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielecki J., Youngman P., Connelly P., Portnoy D. A. Bacillus subtilis expressing a haemolysin gene from Listeria monocytogenes can grow in mammalian cells. Nature. 1990 May 10;345(6271):175–176. doi: 10.1038/345175a0. [DOI] [PubMed] [Google Scholar]
- Black W. J., Quinn F. D., Tompkins L. S. Legionella pneumophila zinc metalloprotease is structurally and functionally homologous to Pseudomonas aeruginosa elastase. J Bacteriol. 1990 May;172(5):2608–2613. doi: 10.1128/jb.172.5.2608-2613.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander S. J., Szeto L., Shuman H. A., Horwitz M. A. An immunoprotective molecule, the major secretory protein of Legionella pneumophila, is not a virulence factor in a guinea pig model of Legionnaires' disease. J Clin Invest. 1990 Sep;86(3):817–824. doi: 10.1172/JCI114779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Colman P. M., Jansonius J. N., Matthews B. W. The structure of thermolysin: an electron density map at 2-3 A resolution. J Mol Biol. 1972 Oct 14;70(3):701–724. doi: 10.1016/0022-2836(72)90569-4. [DOI] [PubMed] [Google Scholar]
- Cossart P., Vicente M. F., Mengaud J., Baquero F., Perez-Diaz J. C., Berche P. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect Immun. 1989 Nov;57(11):3629–3636. doi: 10.1128/iai.57.11.3629-3636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard J. L., Berche P., Sansonetti P. Transposon mutagenesis as a tool to study the role of hemolysin in the virulence of Listeria monocytogenes. Infect Immun. 1986 Apr;52(1):50–55. doi: 10.1128/iai.52.1.50-55.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormley E., Mengaud J., Cossart P. Sequences homologous to the listeriolysin O gene region of Listeria monocytogenes are present in virulent and avirulent haemolytic species of the genus Listeria. Res Microbiol. 1989 Nov-Dec;140(9):631–643. doi: 10.1016/0923-2508(89)90195-2. [DOI] [PubMed] [Google Scholar]
- Holmes M. A., Matthews B. W. Structure of thermolysin refined at 1.6 A resolution. J Mol Biol. 1982 Oct 5;160(4):623–639. doi: 10.1016/0022-2836(82)90319-9. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- MACKANESS G. B. Cellular resistance to infection. J Exp Med. 1962 Sep 1;116:381–406. doi: 10.1084/jem.116.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengaud J., Vicente M. F., Cossart P. Transcriptional mapping and nucleotide sequence of the Listeria monocytogenes hlyA region reveal structural features that may be involved in regulation. Infect Immun. 1989 Dec;57(12):3695–3701. doi: 10.1128/iai.57.12.3695-3701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel E., Reich K. A., Favier R., Berche P., Cossart P. Attenuated mutants of the intracellular bacterium Listeria monocytogenes obtained by single amino acid substitutions in listeriolysin O. Mol Microbiol. 1990 Dec;4(12):2167–2178. doi: 10.1111/j.1365-2958.1990.tb00578.x. [DOI] [PubMed] [Google Scholar]
- Miller R. A., Reed S. G., Parsons M. Leishmania gp63 molecule implicated in cellular adhesion lacks an Arg-Gly-Asp sequence. Mol Biochem Parasitol. 1990 Mar;39(2):267–274. doi: 10.1016/0166-6851(90)90065-t. [DOI] [PubMed] [Google Scholar]
- Nakahama K., Yoshimura K., Marumoto R., Kikuchi M., Lee I. S., Hase T., Matsubara H. Cloning and sequencing of Serratia protease gene. Nucleic Acids Res. 1986 Jul 25;14(14):5843–5855. doi: 10.1093/nar/14.14.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffaretti J. C., Kressebuch H., Aeschbacher M., Bille J., Bannerman E., Musser J. M., Selander R. K., Rocourt J. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc Natl Acad Sci U S A. 1989 May;86(10):3818–3822. doi: 10.1073/pnas.86.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin W., Marliere P. Comparaison de plusieurs séquences protéiques par reconnaissance de blocs conservés. C R Acad Sci III. 1986;303(13):541–546. [PubMed] [Google Scholar]
- Sidler W., Niederer E., Suter F., Zuber H. The primary structure of Bacillus cereus neutral proteinase and comparison with thermolysin and Bacillus subtilis neutral proteinase. Biol Chem Hoppe Seyler. 1986 Jul;367(7):643–657. doi: 10.1515/bchm3.1986.367.2.643. [DOI] [PubMed] [Google Scholar]
- Szeto L., Shuman H. A. The Legionella pneumophila major secretory protein, a protease, is not required for intracellular growth or cell killing. Infect Immun. 1990 Aug;58(8):2585–2592. doi: 10.1128/iai.58.8.2585-2592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Imanaka T., Aiba S. Nucleotide sequence and promoter region for the neutral protease gene from Bacillus stearothermophilus. J Bacteriol. 1985 Sep;163(3):824–831. doi: 10.1128/jb.163.3.824-831.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasantha N., Thompson L. D., Rhodes C., Banner C., Nagle J., Filpula D. Genes for alkaline protease and neutral protease from Bacillus amyloliquefaciens contain a large open reading frame between the regions coding for signal sequence and mature protein. J Bacteriol. 1984 Sep;159(3):811–819. doi: 10.1128/jb.159.3.811-819.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M. Y., Ferrari E., Henner D. J. Cloning of the neutral protease gene of Bacillus subtilis and the use of the cloned gene to create an in vitro-derived deletion mutation. J Bacteriol. 1984 Oct;160(1):15–21. doi: 10.1128/jb.160.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]