Abstract
The retroviral restriction factor, TRIM5α, blocks infection of a spectrum of retroviruses soon after virus entry into the cell. TRIM5α consists of RING, B-box 2, coiled-coil, and B30.2(SPRY) domains. The B-box 2 domain is essential for retrovirus restriction by TRIM5α, but its specific function is unknown. We show here that the B-box 2 domain mediates higher-order self-association of TRIM5αrh oligomers. This self-association increases the efficiency of TRIM5α binding to the retroviral capsid, thus potentiating restriction of retroviral infection. The contribution of the B-box 2 domain to cooperative TRIM5α association with the retroviral capsid explains the conditional nature of the restriction phenotype exhibited by some B-box 2 TRIM5α mutants; the potentiation of capsid binding that results from B-box 2-mediated self-association is essential for restriction when B30.2(SPRY) domain-mediated interactions with the retroviral capsid are weak. Thus, B-box 2-dependent higher-order self-association and B30.2(SPRY)-dependent capsid binding represent complementary mechanisms whereby sufficiently dense arrays of capsid-bound TRIM5α proteins can be achieved.
The host restriction factor TRIM5α blocks the infection of some retroviruses at an early, postentry step in a species-specific manner (38). For example, rhesus monkey TRIM5α (TRIM5αrh) potently blocks the infection of human immunodeficiency virus (HIV-1) and several other retroviruses; by contrast, human TRIM5α (TRIM5αhu) only modestly restricts HIV-1 infection, but potently blocks infection by N-tropic murine leukemia viruses (N-MLV) (13, 17, 28, 33, 35, 38, 45). Restricting TRIM5α proteins specifically recognize the viral cores and accelerate the uncoating process, which may interfere with the orderly disassembly of the viral capsid (11, 32, 39); Viral cDNA synthesis is typically interrupted, although experiments with proteasome inhibitors indicate that this is not necessary for restriction of infection (1, 39, 42).
TRIM5α is a member of the tripartite motif (TRIM) protein family and consists of RING, B-box 2, coiled-coil, and B30.2(SPRY) domains (26, 34). The role of individual TRIM5α domains in antiretroviral restriction has been investigated. The B30.2(SPRY) domain of TRIM5α determines viral specificity and restriction potency by mediating recognition of the retroviral capsid (21, 30, 40, 46). The coiled-coil domain is essential for TRIM5α oligomerization, which contributes to binding avidity for the viral capsid and thus restriction potency (16, 27, 30). The RING finger domain is the signature of a class of E3 ubiquitin ligases involved in proteasome-mediated protein degradation (26); TRIM5α has recently been shown to possess E3 ubiquitin ligase activity and can ubiquitinate itself (43). However, deletion or disruption of the TRIM5α RING domain only partially attenuates antiviral activity (14, 38, 40); moreover, TRIM5αrh efficiently restricts HIV-1 even at a nonpermissive temperature in cells expressing a temperature-sensitive ubiquitin-activating (E1) enzyme or in the presence of proteasome inhibitors (31, 39, 42). The B-box 2 domain is unique to members of the TRIM protein family; some TRIM proteins also possess an adjacent B-box 1 domain (34). The B-boxes play an important but poorly understood role in the function of TRIM proteins (26, 34). It has been shown that the B-box 2 domain of MID1 (TRIM18) adopts a ββα RING-like tertiary fold and coordinates two zinc atoms in a cross-brace pattern, similar to the RING and B-box 1 structures (24, 25). The tandem B-box 1 and B-box 2 domains in MID1 pack against each other in a stable association, reminiscent of the RING dimer interaction (41). The B-box 2 domains of several TRIM proteins have been implicated in protein-protein interactions, multimerization, and subcellular localization (2, 5, 22, 29, 49).
Some changes or deletions affecting the B-box 2 domain of TRIM5α eliminate its antiretroviral activity (7, 9, 14, 30, 39). The retroviral capsid can still be recognized by these mutant TRIM5α proteins; however, because B-box 2 changes often decrease TRIM5α turnover and result in high steady-state levels of expression (7, 8) and because the input levels of TRIM5α mutants have not always been adjusted in capsid-binding assays, it remains possible that quantitative differences in capsid binding result from B-box 2 disruption. Indeed, some B-box 2 mutants exhibit conditionally defective phenotypes linked with defects in capsid binding (20). Paired with a B30.2(SPRY) domain that specifies efficient capsid binding and potent restriction, these B-box 2 domain alterations are functionally tolerated; on the other hand, when paired with a B30.2(SPRY) domain that confers weaker capsid binding and/or restriction, these mutants exhibit little or no detectable antiviral activity (20). These conditionally defective TRIM5α B-box 2 mutants bind the targeted capsid less efficiently than the unmodified TRIM5α protein (20). Based on these observations, a functional interplay between the TRIM5α B-box 2 and B30.2(SPRY) domains has been proposed (20). Supporting this interpretation is the observation that TRIMCyp, an owl monkey restriction factor that consists of the TRIM5 RING, B-box 2, and coiled-coil domains fused to cyclophilin A (Cyp A), is much less sensitive to B-box 2 changes (6, 15, 20, 44). Thus, replacing the B30.2(SPRY) domain with a different HIV-1 capsid-binding moiety, Cyp A (23), significantly reduces the requirements for an intact B-box 2 domain (20).
In the present study, we test the hypothesis that the TRIM5α B-box 2 domain contributes to capsid binding by promoting higher-order interactions among TRIM5α oligomers. We utilize two TRIM5α B-box variants, R121E and ER/RE. TRIM5αrh R121E contains an alteration of an arginine residue that is exposed on the B-box 2 surface (7); this mutant, despite high levels of expression, is markedly attenuated for HIV-1-restricting activity (7). A second TRIM5αrh mutant, ER/RE, contains an additional change of glutamic acid 120 to arginine and exhibits a conditionally defective phenotype. The abilities of these two prototypic TRIM5αrh B-box 2 mutants to oligomerize, achieve higher-order self-association, and bind HIV-1 capsid complexes were examined.
MATERIALS AND METHODS
Creation of cells stably expressing TRIM5 proteins.
Mutations specifying the R121E and ER/RE changes in the B-box 2 domain were introduced into the pLPCX-TRIM5αrh expression plasmid (38) by QuikChange (Stratagene). The TRIM5αrh RBCC-L2 mutant, which lacks the B30.2(SPRY) domain (16), has the following C-terminal sequence: … MFRELTDA. The sequence of the TRIM5αrh ΔV1-HA mutant (6, 36) near the deletion is: … PQIMY*NFNYCT …, with the asterisk marking the site of the deletion in the V1 region of the B30.2(SPRY) domain. Recombinant viruses were produced in 293T cells (American Type Culture Collection) by cotransfecting the pLPCX plasmids (Clontech) expressing TRIM5 proteins with the pVPack-GP and pVPack-VSV-G packaging plasmids (Stratagene) (38). The pVPack-VSV-G plasmid encodes the vesicular stomatitis virus G envelope glycoprotein, which allows efficient entry into a wide range of vertebrate cells (47). The resulting virus particles were used to transduce 2 × 105 HeLa cells or Cf2Th cells in six-well plates. The transduced cells were then selected in 1 μg (HeLa) or 5 μg (Cf2Th) of puromycin (Sigma)/ml.
Infection with viruses expressing GFP.
Recombinant HIV-1 and N-MLV viruses expressing green fluorescent protein (GFP) were made as previously described (33, 38). A 20-μl portion of the preparation of HIV-1-GFP used in these experiments corresponds to ∼3,000 cpm reverse transcriptase units; a 100-μl portion of N-MLV-GFP corresponds to ∼400 cpm reverse transcriptase units. For infection, 3 × 104 cells were seeded in 24-well plates and incubated with the viruses for 60 h. Cells were then washed with phosphate-buffered saline (PBS), fixed with 3.7% formaldehyde, and subjected to fluorescence-activated cell sorting (FACS) analysis with a FACScan (Becton Dickinson).
Cross-linking of TRIM5 proteins.
Cell lysates prepared in 1% NP-40-PBS-protease inhibitor cocktail were incubated with various concentrations (final concentrations of 0, 1, 2, 4, or 8 mM) of glutaraldehyde (Sigma) at room temperature for 5 min, followed by adding excess glycine to quench the reaction (16, 27). The cross-linked lysates were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotted with horseradish peroxidase (HRP)-conjugated anti-HA antibody (Roche).
Half-life of TRIM5α mutants.
HeLa cells expressing wild-type TRIM5αrh or TRIM5αrh ER/RE were seeded in six-well plates 1 day prior to the experiment. When the cells reached 40 to 60% confluence, cycloheximide at 100 μg/ml (Sigma) was added to block protein synthesis. Treated cells were lysed at different time points, and cell lysates containing equal amounts of total protein were subjected to SDS-PAGE and Western blotting.
Subcellular localization using immunofluorescence confocal microscopy.
Cells stably expressing HA epitope-tagged TRIM5α variants were cultured on eight-well chamber slides. After 24 h, the cells were fixed with 4% formaldehyde for 30 min; permeabilized with 0.5% (vol/vol) Triton X-100 in PBS for 5 min; and blocked with 10% goat serum, 1% bovine serum albumin, and 0.2% Triton X-100. The cells were then incubated with primary rat anti-HA 3F10 antibody (1:100; Roche) and secondary goat anti-rat immunoglobulin G conjugated with Alexa-488 (1:200; Molecular Probes). The processed cells were analyzed by using a Zeiss LSM510 META confocal microscope.
Coimmunoprecipitation.
293T cells in six-well plates were transiently transfected with pLPCX vectors encoding HA- or FLAG-tagged TRIM5 variants. The FLAG epitope tags were placed at the N termini of the TRIM5 proteins, the HA epitope tags at the C termini. Two days later, cells were lysed in 1 ml of lysis buffer (1% NP-40-PBS-protease inhibitor cocktail). The lysates were cleared of insoluble materials and aggregates by centrifugation at 13,200 rpm for 1 h at 4°C. Cleared lysates containing different TRIM variants were then mixed and incubated with 20 μl (packed volume) of protein A-Sepharose beads (Pharmacia) for 3 h at 4°C to remove proteins that had bound nonspecifically to the beads. To achieve a comparable level of input TRIM5 protein, lysates of 293T cells transiently transfected with the empty pLPCX vector were at times used to dilute the lysates containing the TRIM5 variants. Samples of the precleared lysate mixture were taken at this point for analysis of the input proteins. The remaining lysates were incubated with 20 μl of fresh beads and 1 μl (ca. 5 to 6 μg) of anti-FLAG antibody (Sigma) overnight at 4°C on a rocker. The immunoprecipitates were washed three times with buffer I (300 mM NaCl, 50 mM Tris-HCl, 1% NP-40) at 4°C for 10 min each on a rocker and once with buffer II (150 mM NaCl, 10 mM Tris-HCl) for 10 min at 4°C. The beads were then treated with 2× SDS sample buffer (125 mM Tris-HCl, 3% SDS, 16.7% glycerol, 3% β-mercaptoethanol, 0.01% bromophenol blue) and boiled for 5 min to release the precipitated proteins. Supernatants were analyzed by SDS-PAGE and Western blotting with HRP-conjugated anti-HA antibody (1:500; Roche) or anti-FLAG antibody (1:500; Sigma).
HIV-1 CA-NC recruitment assay.
Purification of recombinant HIV-1 CA-NC protein produced in Escherichia coli was carried out as previously described (12). For a source of TRIM5 proteins, 293T cells transiently transfected with pLPCX plasmids expressing TRIM5 variants were lysed by freeze-thawing in hypotonic lysis buffer (10 mM Tris [pH 7.4], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol) containing protease inhibitors (Roche). The lysates from different TRIM variants were then mixed and incubated with the in vitro-assembled HIV-1 CA-NC complexes. The binding assay was carried out as previously described (18, 19, 21, 39).
RESULTS
Expression, oligomerization, and antiretroviral activity of TRIM5αrh B-box 2 mutants.
The phenotypes of two TRIM5αrh B-box 2 mutants, R121E and ER/RE, were examined in human (HeLa) and canine (Cf2Th) cells stably expressing these proteins. The steady-state levels of R121E expression were higher than those of ER/RE, which in turn were higher than those of wild-type TRIM5αrh (Fig. 1A). Both mutants assembled into oligomers as efficiently as the wild-type TRIM5αrh protein (Fig. 1B). Perhaps due to the contribution of the endogenous human TRIM5α in HeLa cells, R121E exhibited a mild anti-HIV-1 activity; in Cf2Th cells, no restriction of HIV-1 infection by R121E was observed (Fig. 1C). The ER/RE mutant exhibited potent anti-HIV-1 activity in HeLa cells, but the anti-HIV-1 activity of ER/RE was slightly weaker than that of the wild-type TRIM5αrh protein in Cf2Th cells. In both cell types, the ER/RE mutant restricted HIV-1 more potently than R121E, indicating that the E120R change in the ER/RE mutant partially reverts the R121E phenotype. Neither mutant TRIM5αrh protein inhibited N-MLV infection; in fact, HeLa cells expressing these mutants were slightly more susceptible to N-MLV infection than the control cells transduced with the empty LPCX vector, suggesting a dominant-negative effect of the mutants on the endogenous TRIM5αhu protein.
FIG. 1.
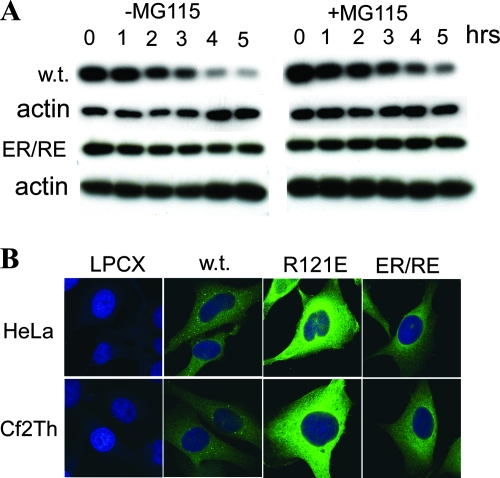
Expression and antiretroviral activity of TRIM5α variants. (A) The expression level of wild-type (w.t.) TRIM5αrh, R121E, and ER/RE with C-terminal HA tags was determined by Western blotting lysates from HeLa and Cf2Th cells stably expressing these proteins. Cell lysates with equal amount of total proteins were resolved by SDS-PAGE, and the Western blot was probed with an anti-HA antibody and an anti-β-actin antibody to control for loading. (B) Oligomerization of TRIM5α variants was examined. Cell lysates from 293T cells transiently expressing the indicated TRIM5α proteins were cross-linked with increasing concentrations of glutaraldehyde (0, 1, 2, 4, and 8 mM). The cross-linked products were resolved by SDS-PAGE and visualized by Western blotting with an anti-HA antibody. (C) The effects of the TRIM5α B-box 2 variants on retroviral infection were assessed. HeLa or Cf2Th cells stably expressing the wild-type TRIM5αrh protein (w.t.) and mutant TRIM proteins, or control cells transduced with the empty LPCX vector, were incubated with various amounts of HIV-1-GFP or N-MLV-GFP. Infected GFP-positive cells were counted by FACS. The experiments were repeated with comparable results.
The degree of defectiveness of the TRIM5αrh ER/RE mutant varied, depending upon the retrovirus being restricted. Such conditional phenotypes for B-box 2 mutants have been previously observed and were found to depend upon the affinity of the TRIM5α parent protein for the retroviral capsid (20). Because capsid-binding affinity is determined by the B30.2(SPRY) domain (21, 30, 40, 46), we tested the phenotype of the ER/RE B-box 2 changes in the context of the TRIM5α R(H286-493) protein. The R(H286-493) protein is identical to TRIM5αrh except that the B30.2(SPRY) domain is derived from human TRIM5α (40). As a result of this B30.2(SPRY) domain substitution, the binding affinity of the R(H286-493) protein for the HIV-1 capsid is reduced relative to that of TRIM5αrh(40). However, the human TRIM5α B30.2(SPRY) domain allows the R(H286-493) protein to restrict N-MLV infection even more potently than the wild-type TRIM5αrh protein (40). As expected (40), the R(H286-493) protein partially restricted HIV-1 infection but potently inhibited N-MLV infection (Fig. 1C, bottom row). The R(H286-493) ER/RE protein exhibited little anti-HIV-1 activity but significantly inhibited N-MLV infection. Thus, ER/RE exhibits a conditional restriction phenotype that is dependent on the restriction potency of the parental TRIM5α protein, a property determined by the B30.2(SPRY) domain.
Turnover and subcellular localization of TRIM5αrh B-box 2 mutants.
Rhesus TRIM5α is turned over rapidly by a process that is only minimally affected by proteasome inhibitors (8). The turnover of the TRIM5αrh B-box mutants was examined in the absence or presence of the proteasome inhibitor, MG115. Similar to R121E (7) and unlike the wild-type TRIM5αrh protein, the ER/RE mutant maintained high levels of protein expression during a 5-h period of cycloheximide treatment (Fig. 2A). Treatment of the cells with MG115 minimally changed the turnover of the wild-type and ER/RE TRIM5αrh proteins. Thus, the rapid degradation of TRIM5αrh is not required for the HIV-1-restricting ability of the ER/RE mutant observed in HeLa cells. Observations on a panel of other B-box 2 mutants (7) led to a similar conclusion: the rapid turnover of TRIM5α is not a prerequisite for efficient retrovirus-restricting ability.
FIG. 2.
Turnover and subcellular localization of TRIM5αrh B-box 2 variants. (A) HeLa cells expressing wild-type TRIM5αrh and TRIM5αrh ER/RE were treated with cycloheximide to block protein synthesis for a 5-h period in the absence or presence of the proteasome inhibitor MG115. Cells were harvested and lysed at 1-h intervals. Cell lysates containing equal amounts of total protein were analyzed by Western blotting with anti-HA and anti-actin antibodies. (B) HeLa or Cf2Th cells stably expressing the HA-tagged TRIM5αrh variants were stained with an anti-HA antibody, followed by an anti-rat secondary antibody conjugated to Alexa 488. The TRIM5αrh proteins are shown in green, and the nuclei are stained blue with DAPI.
TRIM proteins aggregate into nuclear or cytoplasmic bodies (34), and alteration of the RING, B-box, and/or coiled-coil domains of some TRIM proteins, including TRIM5, have been shown to affect the formation of bodies and localization in the cell (3, 9, 31). We examined the subcellular localization of wild-type TRIM5αrh and the B-box 2 mutants stably expressed in HeLa and Cf2Th cells. The results were similar in the two cell types (Fig. 2B). The wild-type TRIM5αrh protein exhibited distinct cytoplasmic bodies superimposed on a diffuse pattern of cytoplasmic staining. Both B-box 2 mutants exhibited brighter cytoplasmic staining, a finding consistent with higher steady-state levels of expression. A reticular staining pattern in the cytoplasm was observed for the mutants, but cytoplasmic bodies were not evident. Thus, the B-box 2 changes affect the steady-state levels of TRIM5αrh expression and the tendency to form cytoplasmic bodies.
Dependence of higher-order self-association of TRIM5α on the B-box 2 domain.
The decreased propensity of the TRIM5αrh B-box 2 mutants to form cytoplasmic bodies suggested that the alteration of the B-box 2 domain may influence TRIM5α self-association. To address this possibility, we sought to determine whether preformed oligomers of the wild-type or mutant TRIM5αrh proteins would coprecipitate with wild-type TRIM5αrh oligomers. Wild-type TRIM5αrh and B-box 2 mutant proteins were expressed in separate cells, which allows the formation of homo-oligomers (Fig. 1B). Lysates from cells expressing individual TRIM5αrh variants were mixed and used for immunoprecipitation. HA-tagged wild-type TRIM5αrh (w.t.-HA) was efficiently coprecipitated with FLAG-tagged wild-type TRIM5αrh (Fig. 3A, left panel), indicating that the wild-type TRIM5αrh oligomer is capable of efficient self-association. In contrast, R121E-HA did not efficiently coprecipitate with the FLAG-tagged wild-type TRIM5αrh protein. Similarly, only the w.t.-HA protein, but not the ER/RE-HA protein, coprecipitated with the FLAG-tagged wild-type TRIM5αrh protein (Fig. 3A, right panel). These results indicate that preformed wild-type TRIM5αrh oligomers can associate and that both of the studied changes in the B-box 2 domain disrupt this higher-order self-association.
FIG. 3.
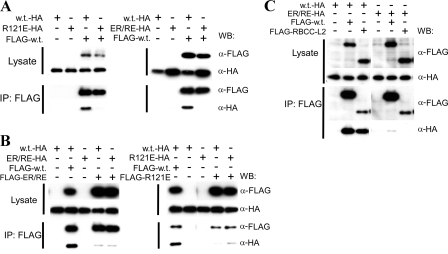
Contribution of the B-box 2 domain to higher-order self-association of TRIM5αrh. (A) The coprecipitation of HA-tagged TRIM5αrh variants with FLAG-tagged TRIM5αrh was examined. 293T cells were transfected transiently with pLPCX plasmids expressing C-terminally HA-tagged wild-type (w.t.) TRIM5αrh, R121E, or ER/RE or N-terminally FLAG-tagged wild-type (w.t.) TRIM5αrh. Cytosolic lysates containing the TRIM5αrh variants were prepared, and the concentration of TRIM5 protein in the lysates was adjusted to account for differences in the levels of expression, as described in Materials and Methods. The adjusted lysates were then mixed in a 1:1 ratio and used for precipitation with an anti-FLAG antibody. The amounts of HA- and FLAG-tagged proteins in the lysates and immunoprecipitates (IPs) were analyzed by Western blotting (WB) with HRP-conjugated anti-HA and anti-FLAG antibodies. (B) The ability of TRIM5αrh ER/RE and R121E to associate with themselves in the coimmunoprecipitation assay was examined. The assay was carried out as described above using N-terminally FLAG-tagged TRIM5αrh ER/RE and R121E in addition to the FLAG-tagged wild-type TRIM5αrh protein (FLAG-w.t.). (C) The requirement for the B30.2(SPRY) domain for the higher-order self-association of TRIM5αrh was examined. An N-terminally FLAG-tagged truncation mutant of TRIM5rh (RBCC-L2), which lacks a B30.2(SPRY) domain (16), was used in the coimmunoprecipitation assay along with HA-tagged wild-type TRIM5αrh and TRIM5αrh ER/RE.
Although the B-box 2 mutant oligomers do not interact with wild-type TRIM5αrh oligomers, the reciprocal changes in the two charged residues in the ER/RE mutant might restore interaction with itself and thus gain some restriction activity. To test this hypothesis, coimmunoprecipitation was performed with HA-tagged and FLAG-tagged ER/RE proteins. As shown in Fig. 3B, ER/RE did not associate with itself any better than it associated with wild-type TRIM5αrh. As expected, R121E also poorly associated with itself in the coimmunoprecipitation assay, as was observed for its hetero-association with wild-type TRIM5αrh.
The ability of B30.2(SPRY) domain sequences to modulate the restriction phenotypes of some B-box 2 mutants (20) raised the possibility that the B30.2(SPRY) domain contributes to the B-box 2-dependent, higher-order self-association of TRIM5α. We tested this hypothesis by using a FLAG-tagged TRIM5αrh mutant, RBCC-L2, which lacks the B30.2(SPRY) domain (16), in the coimmunoprecipitation assay. Although the anti-FLAG antibody precipitated the FLAG-tagged RBCC-L2 protein less efficiently than wild-type TRIM5αrh, a significant amount of HA-tagged wild-type TRIM5αrh coprecipitated with the RBCC-L2 protein (Fig. 3C). The HA-tagged ER/RE mutant did not coprecipitate with either the wild-type TRIM5αrh protein or the RBCC-L2 mutant. Thus, a TRIM5αrh variant lacking the B30.2(SPRY) domain can still associate with other TRIM5αrh proteins in a B-box 2-dependent manner.
B-box 2 domain-dependent recruitment of TRIM5αrh to the HIV-1 capsid.
Higher-order self-association of TRIM5αrh, if compatible with binding to the HIV-1 capsid, might contribute to avidity for the capsid. To achieve a quantitative comparison of the capsid-binding abilities of wild-type and B-box 2 mutant TRIM5αrh proteins, cell lysates containing the highly expressed R121E and ER/RE mutants were diluted with lysates from control cells transduced with the empty LPCX vector prior to their use in the in vitro HIV-1 CA-NC binding assay (39). At input concentrations that more closely resembled that of wild-type TRIM5αrh, both R121E and ER/RE proteins exhibited significantly less association with the HIV-1 CA-NC complexes than wild-type TRIM5αrh (Fig. 4A). Thus, as has been observed for B-box 2 TRIM5αrh mutants with conditional phenotypes (20), some changes in the B-box 2 domain result in decreased TRIM5α binding to the retroviral capsid.
FIG. 4.
Contribution of the B-box 2 domain to TRIM5α recruitment to the capsid. (A) The binding of the wild-type TRIM5αrh protein (w.t.) and the R121E and ER/RE TRIM5αrh mutants to the HIV-1 CA-NC complexes was examined. Cell lysates of 293T cells transiently expressing the C-terminally HA-tagged TRIM5αrh variants were used in the HIV-1 CA-NC binding assay. To compare the binding efficiency quantitatively, the lysates containing the highly expressed R121E and ER/RE proteins were diluted with lysates from 293T cells transiently transfected with the empty pLPCX plasmid to achieve input levels comparable to that of w.t. TRIM5αrh. The top and middle panels show Western blots with an anti-HA antibody to detect the amount of TRIM5αrh proteins in the input and pellet, respectively; the bottom panel shows the amount of HIV-1 CA-NC protein that was pelleted through the 70% sucrose cushion. (B) The ability of wild-type (w.t.) TRIM5αrh to recruit the ΔV1 TRIM5αrh mutant and TRIM5αhu to HIV-1 CA-NC complexes was examined. Cell lysates made from 293T cells transiently transfected with the empty pLPCX plasmid were mixed with HA-tagged wild-type TRIM5αrh, R121E, or ER/RE in a 1:1 ratio. The mixed lysates were then used in the in vitro HIV-1 CA-NC binding assay as controls to demonstrate that they bind the assembled capsid complexes. HA-tagged ΔV1 and TRIM5αhu, which do not bind the capsid well when present alone (6, 21, 36, 39), were mixed with lysates from cells transfected with pLPCX or cells expressing FLAG-tagged wild-type TRIM5αrh, R121E, or ER/RE and subjected to the binding assay. The input amounts of lysates containing FLAG-tagged wild-type TRIM5αrh, R121E, and ER/RE were adjusted so that the amounts of FLAG-R121E and FLAG-ER/RE bound to the HIV-1 CA-NC complexes were at least as great as that of the FLAG-tagged wild-type TRIM5αrh protein. Cell lysates containing HA-tagged TRIM5αrh mixed with lysates from pLPCX-transfected cells were included as a positive control in the TRIM5αhu recruitment experiment. The amounts of HA-tagged and FLAG-tagged proteins present in the input and the pellet were detected by Western blotting with an anti-HA antibody (Roche) and an anti-FLAG antibody (Sigma), respectively. To compensate for the high level of R121E expression, all HA-tagged and FLAG-tagged R121E input and pellet samples were loaded onto the SDS-polyacrylamide gel at half the volume of the other protein samples, except in the pellet sample of the TRIM5αhu recruitment experiment. Due to the different input levels of the FLAG-tagged wild-type TRIM5αrh, R121E, and ER/RE, in some cases the FLAG-w.t. protein is not visible on the Western blot at the exposure shown. (C) The ability of the ER/RE mutant to recruit itself to HIV-1 capsid complexes was examined. A C-terminally HA-tagged mutant containing both the ΔV1 B30.2(SPRY) domain deletion and the ER/RE B-box 2 changes was used in the recruitment assay, as described above. Cell lysates containing HA-tagged wild-type TRIM5αrh, ΔV1, and ΔV1-ER/RE were mixed with pLPCX-transfected cell lysates or lysates containing FLAG-tagged wild-type TRIM5αrh or ER/RE in a 1:1 ratio and used in the capsid binding assay.
To test directly the hypothesis that the higher-order self-association of TRIM5αrh can occur in the context of capsid binding, we sought to determine whether capsid-bound TRIM5αrh could recruit TRIM5α variants that bind poorly to the HIV-1 capsid and whether this recruitment is dependent on the integrity of the B-box 2 domain. Lysates of cells transduced with the empty LPCX vector or cells expressing FLAG-tagged wild-type TRIM5αrh, R121E, or ER/RE proteins were mixed with the lysate from cells expressing an HA-tagged TRIM5αrh mutant, ΔV1-HA. The ΔV1-HA mutant associates poorly with HIV-1 CA-NC complexes due to a 13-amino-acid deletion in the B30.2(SPRY) domain V1 region (Δ332-344) (6, 36). The mixture was then used in the in vitro CA-NC binding assay (39). To achieve similar levels of TRIM5α variants bound to the CA-NC complexes, the input amounts of the TRIM5α B-box 2 mutants were adjusted to be significantly higher than that of the wild-type TRIM5αrh protein. In the presence of FLAG-tagged wild-type TRIM5αrh, ΔV1-HA associated with the HIV-1 CA-NC complexes about fivefold better than ΔV1-HA in the presence of lysates from control LPCX-transduced cells (Fig. 4B, middle panel). Although, under these conditions, both R121E and ER/RE bound the HIV-1 CA-NC complexes efficiently, neither recruited the ΔV1-HA proteins to the capsid complexes above the background level observed for the control LPCX lysates.
Wild-type TRIM5αhu also binds the HIV-1 capsid poorly (21, 39). HIV-1 capsid recruitment experiments were performed with TRIM5αhu in place of ΔV1-HA, with a similar outcome. Wild-type TRIM5αrh recruited TRIM5αhu to the capsid, whereas R121E and ER/RE were inefficient in this regard (Fig. 4B, right panel). These results demonstrate that wild-type TRIM5αrh can promote cooperative HIV-1 capsid binding by recruiting other TRIM5 proteins to the capsid, whereas B-box 2-defective mutants cannot. The results also suggest that the ability to engage in B-box 2-mediated higher-order interactions is also retained in TRIM5αhu; this is not surprising, given the single amino acid difference between the TRIM5αrh and TRIM5αhu B-box 2 domains (38).
Although ER/RE does not apparently self-associate in coimmunoprecipitation experiments, this mutant might recruit itself once bound to the HIV-1 capsid. To test this possibility, an HA-tagged TRIM5αrh mutant that contains the ΔV1 and ER/RE changes was made and used in the HIV-1 capsid recruitment assay. The FLAG-tagged wild-type TRIM5αrh recruited ΔV1-HA more efficiently than ΔV1-ER/RE-HA to the HIV-1 CA-NC complexes (Fig. 4C). This result indicates that an intact B-box 2 function is required on the TRIM5α protein recruited to the HIV-1 capsid complexes. However, the FLAG-tagged ER/RE recruited ΔV1-ER/RE-HA no better than ΔV1-HA.
DISCUSSION
Most residues on the surface of the TRIM5αrh B-box 2 domain can be altered without a significant impact on HIV-1-restricting activity (7). However, changes in one B-box 2 region predicted to be surface exposed on the assembled TRIM5 oligomer resulted in dramatic reductions in restriction (7). In the present study, we utilized two TRIM5αrh mutants altered in this region. One mutant, R121E, is almost completely devoid of HIV-1-restricting ability, whereas the other, ER/RE, exhibits a conditional phenotype. In the context of some TRIM5 variants and particular targeted retroviruses, the effects of the ER/RE changes in the B-box 2 domain on restriction are minimal; in other contexts, the ER/RE changes eliminate restricting ability. We demonstrate that both the R121E and the ER/RE mutants efficiently form homo-oligomers and therefore negotiate the first-order self-association of TRIM5 mediated by the coiled coil and adjacent L2 linker (16, 27, 30). However, both mutants are deficient in establishing higher-order associations among TRIM5α oligomers, a process that appears to be important for the efficient recruitment of TRIM5 proteins to the retroviral capsid. The format of our assays did not allow us to determine whether the recruited TRIM5α oligomers bind the capsid directly or associate only with the bound TRIM5α oligomers. However, we did demonstrate that the B30.2(SPRY) domain is not essential for the higher-order self-association of TRIM5α; thus, in the higher-order TRIM5α complex, the B30.2(SPRY) domain is apparently free to interact with the capsid. If the spatial geometry of the B-box 2-mediated TRIM5-TRIM5 association is compatible with each of the associated TRIM5 oligomers binding the capsid, significant gains in avidity would result. Future studies will be aimed at understanding the structural basis and geometry of the higher-order association of TRIM5 oligomers.
In light of this discovery, the heretofore puzzling conditional nature of some TRIM5α B-box 2 mutant phenotypes (20) can be explained. In these B-box 2 mutants, which include the ER/RE mutant described herein, the degree of functional attenuation is related to the B30.2(SPRY) domain-determined affinity for the retroviral capsid and the restriction potency of the parental TRIM5α protein for the targeted retrovirus (20). The potentiation of capsid binding that results from B-box 2-mediated self-association appears to be essential for restriction when B30.2(SPRY)-mediated interactions with the retroviral capsid are weak. Conversely, a high-affinity B30.2(SPRY) interaction with the capsid can compensate for the deficiency in the avidity of capsid binding resulting from the inability of these B-box 2 mutants to form higher-order structures. From this perspective, B-box 2-mediated TRIM5 self-association and B30.2(SPRY)-mediated capsid binding are complementary mechanisms by which a dense array of capsid-bound TRIM5α proteins can be achieved. The diminished restricting ability of TRIM5α mutants in which one or both of these mechanisms is compromised suggests that achieving adequate density and/or proper geometry of the bound TRIM5α proteins on the capsid is important for restriction. Images of TRIM5α proteins associated with HIV-1 capsids in infected cells are consistent with such a model (4).
Although in some circumstances, the restriction phenotypes of the R121E and ER/RE mutants differed, these mutants were almost indistinguishable in the coimmunoprecipitation and capsid recruitment assays. The assays may have failed to detect quantitative differences that exist between these mutants; alternatively, the additional E120R change in the ER/RE conditional revertant might contribute in a qualitative manner to TRIM5α functions beyond capsid association. Additional studies will be needed to address these possibilities.
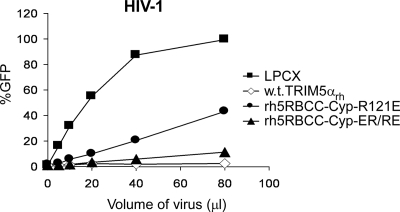
The ability of owl monkey TRIMCyp to restrict HIV-1 infection is only minimally affected by changes in the B-box 2 domain that significantly decrease TRIM5α capsid binding and restriction (6, 20). Similarly, B-box 2 changes exert minimal effects on the HIV-1-restricting ability of rh5RBCC-Cyp, a chimeric protein consisting of the TRIM5αrh RING, B-box 2, and coiled-coil domains fused to CypA (20) (Fig. 5). Dimerization of Cyp A has been shown to be sufficient for moderately potent restriction of HIV-1 (15, 44); the B-box 2-mediated higher-order self-association appears to be dispensable in this context. Cyp A binds monomeric HIV-1 capsid protein with low affinity (23, 48), whereas TRIM5α does not detectably interact with capsid monomers (10). Oligomerization of Cyp A may allow sufficient avidity for the HIV-1 capsid to achieve restriction, whereas TRIM5α oligomers may require the reinforcing B-box 2-mediated interactions to achieve comparable levels of capsid binding and restriction. Moreover, the prolyl isomerase activity of Cyp A may promote uncoating and restriction (15, 44) once sufficient TRIMCyp molecules are bound, whereas TRIM5α may need to recruit additional factors to mediate restriction.
FIG. 5.
HIV-1-restricting ability of TRIM5-Cyp A fusion proteins with changes in the B-box 2 domain. HeLa cells stably expressing wild-type TRIM5αrh or rh5RBCC-Cyp B-box 2 mutants, or cells transduced with the empty LPCX vector, were exposed to increasing amounts of HIV-1-GFP. Infected, GFP-positive cells were counted by FACS. The results of a single experiment are shown; similar results were obtained in a repeat experiment.
A property common to all TRIM proteins is the tendency to aggregate into cytoplasmic or nuclear bodies when overexpressed (34). Cytoplasmic bodies, although not necessary for TRIM5α-mediated restriction (31, 37), may be a manifestation of higher-order self-association in cells overexpressing TRIM5α. That many TRIM proteins form nuclear or cytoplasmic bodies hints that B-box-mediated self-association may be common to many TRIM proteins. The requirement to assemble a large array of TRIM5α on the incoming retroviral capsid may explain the selection of this host restriction factor from the aggregation-prone TRIM family.
Acknowledgments
We thank Yvette McLaughlin and Elizabeth Carpelan for manuscript preparation.
This study was supported by the National Institutes of Health (grants AI063987 and AI076094 and a Center for AIDS Research Award AI06354), the International AIDS Vaccine Initiative, and the late William F. McCarty-Cooper.
Footnotes
Published ahead of print on 17 September 2008.
REFERENCES
- 1.Anderson, J. L., E. M. Campbell, X. Wu, N. Vandegraaff, A. Engelman, and T. J. Hope. 2006. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J. Virol. 809754-9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beenders, B., P. L. Jones, and M. Bellini. 2007. The tripartite motif of nuclear factor 7 is required for its association with transcriptional units. Mol. Cell. Biol. 272615-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borden, K. L., M. N. Boddy, J. Lally, N. J. O'Reilly, S. Martin, K. Howe, E. Solomon, and P. S. Freemont. 1995. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 141532-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, E. M., O. Perez, J. L. Anderson, and T. J. Hope. 2008. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5α. J. Cell Biol. 180549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, T., K. L. Borden, P. S. Freemont, and L. D. Etkin. 1997. Involvement of the rfp tripartite motif in protein-protein interactions and subcellular distribution. J. Cell Sci. 110(Pt. 14)1563-1571. [DOI] [PubMed] [Google Scholar]
- 6.Diaz-Griffero, F., A. Kar, M. Lee, M. Stremlau, E. Poeschla, and J. Sodroski. 2007. Comparative requirements for the restriction of retrovirus infection by TRIM5α and TRIMCyp. Virology 369400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz-Griffero, F., A. Kar, M. Perron, S. H. Xiang, H. Javanbakht, X. Li, and J. Sodroski. 2007. Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5α B-box 2 domain. J. Virol. 8110362-10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz-Griffero, F., X. Li, H. Javanbakht, B. Song, S. Welikala, M. Stremlau, and J. Sodroski. 2006. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology 349300-315. [DOI] [PubMed] [Google Scholar]
- 9.Diaz-Griffero, F., N. Vandegraaff, Y. Li, K. McGee-Estrada, M. Stremlau, S. Welikala, Z. Si, A. Engelman, and J. Sodroski. 2006. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology 351404-419. [DOI] [PubMed] [Google Scholar]
- 10.Dodding, M. P., M. Bock, M. W. Yap, and J. P. Stoye. 2005. Capsid processing requirements for abrogation of Fv1 and Ref1 restriction. J. Virol. 7910571-10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forshey, B. M., U. von Schwedler, W. I. Sundquist, and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 765667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 28380-83. [DOI] [PubMed] [Google Scholar]
- 13.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 10110774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Javanbakht, H., F. Diaz-Griffero, M. Stremlau, Z. Si, and J. Sodroski. 2005. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5α. J. Biol. Chem. 28026933-26940. [DOI] [PubMed] [Google Scholar]
- 15.Javanbakht, H., F. Diaz-Griffero, W. Yuan, D. F. Yeung, X. Li, B. Song, and J. Sodroski. 2007. The ability of multimerized cyclophilin A to restrict retrovirus infection. Virology 36719-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Javanbakht, H., W. Yuan, D. F. Yeung, B. Song, F. Diaz-Griffero, Y. Li, X. Li, M. Stremlau, and J. Sodroski. 2006. Characterization of TRIM5α trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 353234-246. [DOI] [PubMed] [Google Scholar]
- 17.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 10110780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, X., B. Gold, C. O'HUigin, F. Diaz-Griffero, B. Song, Z. Si, Y. Li, W. Yuan, M. Stremlau, C. Mische, H. Javanbakht, M. Scally, C. Winkler, M. Dean, and J. Sodroski. 2007. Unique features of TRIM5α among closely related human TRIM family members. Virology 360419-433. [DOI] [PubMed] [Google Scholar]
- 19.Li, X., Y. Li, M. Stremlau, W. Yuan, B. Song, M. Perron, and J. Sodroski. 2006. Functional replacement of the RING, B-box 2, and coiled-coil domains of tripartite motif 5α (TRIM5α) by heterologous TRIM domains. J. Virol. 806198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, X., B. Song, S. H. Xiang, and J. Sodroski. 2007. Functional interplay between the B-box 2 and the B30.2(SPRY) domains of TRIM5α. Virology 366234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Y., X. Li, M. Stremlau, M. Lee, and J. Sodroski. 2006. Removal of arginine 332 allows human TRIM5α to bind human immunodeficiency virus capsids and to restrict infection. J. Virol. 806738-6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loer, B., R. Bauer, R. Bornheim, J. Grell, E. Kremmer, W. Kolanus, and M. Hoch. 2008. The NHL-domain protein Wech is crucial for the integrin-cytoskeleton link. Nat. Cell Biol. 10422-428. [DOI] [PubMed] [Google Scholar]
- 23.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 731067-1078. [DOI] [PubMed] [Google Scholar]
- 24.Massiah, M. A., J. A. Matts, K. M. Short, B. N. Simmons, S. Singireddy, Z. Yi, and T. C. Cox. 2007. Solution structure of the MID1 B-box2 CHC(D/C)C(2)H(2) zinc-binding domain: insights into an evolutionarily conserved RING fold. J. Mol. Biol. 3691-10. [DOI] [PubMed] [Google Scholar]
- 25.Massiah, M. A., B. N. Simmons, K. M. Short, and T. C. Cox. 2006. Solution structure of the RBCC/TRIM B-box1 domain of human MID1: B-box with a RING. J. Mol. Biol. 358532-545. [DOI] [PubMed] [Google Scholar]
- 26.Meroni, G., and G. Diez-Roux. 2005. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 271147-1157. [DOI] [PubMed] [Google Scholar]
- 27.Mische, C. C., H. Javanbakht, B. Song, F. Diaz-Griffero, M. Stremlau, B. Strack, Z. Si, and J. Sodroski. 2005. Retroviral restriction factor TRIM5α is a trimer. J. Virol. 7914446-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen, J. C. 1998. Gene transfer vectors derived from equine infectious anemia virus. Gene Ther. 51481-1487. [DOI] [PubMed] [Google Scholar]
- 29.Peng, H., G. E. Begg, D. C. Schultz, J. R. Friedman, D. E. Jensen, D. W. Speicher, and F. J. Rauscher III. 2000. Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J. Mol. Biol. 2951139-1162. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 798969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Caballero, D., T. Hatziioannou, F. Zhang, S. Cowan, and P. D. Bieniasz. 2005. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J. Virol. 7915567-15572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perron, M. J., M. Stremlau, M. Lee, H. Javanbakht, B. Song, and J. Sodroski. 2007. The human TRIM5α restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J. Virol. 812138-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 10111827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 202140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saenz, D. T., W. Teo, J. C. Olsen, and E. M. Poeschla. 2005. Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate TRIM5α proteins. J. Virol. 7915175-15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 1022832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song, B., F. Diaz-Griffero, D. H. Park, T. Rogers, M. Stremlau, and J. Sodroski. 2005. TRIM5α association with cytoplasmic bodies is not required for antiretroviral activity. Virology 343201-211. [DOI] [PubMed] [Google Scholar]
- 38.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427848-853. [DOI] [PubMed] [Google Scholar]
- 39.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. USA 1035514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 793139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao, H., B. N. Simmons, S. Singireddy, M. Jakkidi, K. M. Short, T. C. Cox, and M. A. Massiah. 2008. Structure of the MID1 tandem B-boxes reveals an interaction reminiscent of intermolecular ring heterodimers. Biochemistry 472450-2457. [DOI] [PubMed] [Google Scholar]
- 42.Wu, X., J. L. Anderson, E. M. Campbell, A. M. Joseph, and T. J. Hope. 2006. Proteasome inhibitors uncouple rhesus TRIM5α restriction of HIV-1 reverse transcription and infection. Proc. Natl. Acad. Sci. USA 1037465-7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamauchi, K., K. Wada, K. Tanji, M. Tanaka, and T. Kamitani. 2008. Ubiquitination of E3 ubiquitin ligase TRIM5α and its potential role. FEBS J. 2751540-1555. [DOI] [PubMed] [Google Scholar]
- 44.Yap, M. W., G. B. Mortuza, I. A. Taylor, and J. P. Stoye. 2007. The design of artificial retroviral restriction factors. Virology 365302-314. [DOI] [PubMed] [Google Scholar]
- 45.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 10110786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr. Biol. 1573-78. [DOI] [PubMed] [Google Scholar]
- 47.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43(Pt. A)99-112. [DOI] [PubMed] [Google Scholar]
- 48.Yoo, S., D. G. Myszka, C. Yeh, M. McMurray, C. P. Hill, and W. I. Sundquist. 1997. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269780-795. [DOI] [PubMed] [Google Scholar]
- 49.Yu, J. W., T. Fernandes-Alnemri, P. Datta, J. Wu, C. Juliana, L. Solorzano, M. McCormick, Z. Zhang, and E. S. Alnemri. 2007. Pyrin activates the ASC pyroptosome in response to engagement by autoinflammatory PSTPIP1 mutants. Mol. Cell 28214-227. [DOI] [PMC free article] [PubMed] [Google Scholar]