Abstract
Human La protein has been implicated in facilitating internal ribosome entry site (IRES)-mediated translation of hepatitis C virus (HCV). Earlier, we demonstrated that the RNA recognition motif (RRM) encompassing residues 112 to 184 of La protein [La (112-184)] interacts with the HCV IRES near the initiator AUG codon. A synthetic peptide, LaR2C (24-mer), derived from La RRM (112-184), retains RNA binding ability, competes with La protein binding to the HCV IRES, and inhibits translation. The peptide interferes with the assembly of 48S complexes, resulting in the accumulation of preinitiation complexes that are incompetent for the 60S ribosomal subunit joining. Here, nuclear magnetic resonance spectroscopy of the HCV IRES-bound peptide complex revealed putative contact points, mutations that showed reduced RNA binding and translation inhibitory activity. The residues responsible for RNA recognition were found to form a turn in the RRM (112-184) structure. A 7-mer peptide comprising this turn showed significant translation inhibitory activity. The bound structure of the peptide inferred from transferred nuclear Overhauser effect experiments suggests that it is a β turn. This conformation is significantly different from that observed in the free RRM (112-184) NMR structure, suggesting paths toward a better-stabilized mimetic peptide. Interestingly, addition of hexa-arginine tag enabled the peptide to enter Huh7 cells and showed inhibition of HCV IRES function. More importantly, the peptide significantly inhibited replication of the HCV monocistronic replicon. Elucidation of the structural determinant of the peptide provides a basis for developing small peptidomimetic structures as potent anti-HCV therapeutics.
The mechanism of internal initiation of translation of hepatitis C virus (HCV) RNA is unique and fundamentally different from the cap-dependent translation of host cell mRNAs, and thus, the HCV internal ribosome entry site (IRES) offers a potential target for developing novel antiviral therapeutics (6).
The ribosome assembly at the HCV IRES has been shown to be prokaryotic-like and requires a minimal number of initiation factors (23). The HCV IRES has been shown to be capable of binding directly to 40S ribosomal subunit with the help of the ribosomal protein S5 (10, 18). Although, HCV IRES binds to the 40S ribosomal subunit specifically and stably even in the absence of any initiation factors, efficient translation requires some of the canonical initiation factors and noncanonical trans-acting factors, possibly to facilitate ribosome binding and to ensure proper positioning of the initiator AUG (iAUG) codon in the P site (13). Several cellular trans-acting factors have been reported to be critically required for HCV IRES-mediated translation, which includes human La autoantigen (2).
Human La protein was originally identified in the sera from patients with systemic lupus erythematosus and Sjögren's syndrome (28). Earlier, it was reported in the literature that La protein contains three RNA recognition motifs (RRMs), which were putatively located between residues 1 and 100 (previously named RRM1), 101 and 208 (previously, RRM2), and 209 and 300 (previously, RRM3) (12, 24). However, according to the recent nomenclature, the first structured domain in human La is termed “La motif” located between residues 16 and 75 [RRM (16-75)], followed by two RRMs, RRM (112-184) and RRM (230-300) (22, 29). Although, based on the structure determinations, the precise boundaries of the structured cores were found to be slightly different [La motif (7-92), RRM (110-194), RRM (229-327)], they largely encompass the above regions (1, 17).
La becomes associated with the 3′ termini of many newly synthesized small RNAs made by RNA polymerase III, as well as certain small RNAs synthesized by other RNA polymerases (20, 31). The N terminal 80 amino acid (aa) residues termed the La motif and the adjacent RRM part have been shown to be required for high-affinity binding with polymerase III transcripts, where the La motif helps in specific recognition for the polyuridylate (UUUOH) sequence at the 3′ end of the RNA. The C-terminal RRM (230-300) has been shown to have a beta sheet comprising five strands and a long C-terminal helix that binds to the putative RNA binding site (17). The central RRM (112-184) has been shown to possess a classical RRM-type fold containing four stranded beta sheets backed by two alpha helices. It also has an additional beta strand inserted between alpha 2 and beta 4. The helix alpha 3 sheet of RRM (112-184) is predominantly hydrophilic and protrudes away from the body of the domain. The beta sheet surface of the central RRM (112-184) contains five basic residues and no acidic residues, which makes it more basic and suitable for RNA interactions (1).
La protein has been shown to bind several viral and cellular IRES elements and to influence their functions. In fact, La has been implicated as a molecular chaperone to facilitate IRES structure and translational competence (5). La protein has been shown to enhance poliovirus (PV) IRES function and to correct its aberrant translation in reticulocyte lysate (21).
Earlier, we demonstrated that La protein plays an important role in mediating HCV IRES-mediated translation. It interacts at the GCAC motif near the iAUG codon of HCV IRES, which might trigger some conformation alterations that facilitate formation of the functional initiation complex and stimulate internal initiation of translation (24). It also seems to assist the binding of ribosomal protein S5 and probably plays a role in recruitment of 40S ribosomal subunit to the HCV IRES (26). Recently, we have demonstrated that a 24-mer peptide (LaR2C) derived from the C terminus of RRM (230-300) of La protein competes with the cellular La protein binding to the HCV IRES and interferes with the functional initiation complex formation (25). It appears that LaR2C interferes with 48S ribosome complexes, rendering it incompetent for 60S joining during internal initiation of translation of HCV RNA (25).
Here, we characterize the structural and functional domain of the LaR2C peptide. Using nuclear magnetic resonance (NMR) spectroscopy of the RNA-bound peptide, coupled with mutational analysis, we have delineated the minimal sequences within the peptide that are required for binding and inhibition of HCV RNA translation. We have shown the presence of a unique β turn at the N terminus of the peptide, which is more or less sufficient for its function. More importantly, we have demonstrated direct delivery of the arginine-tagged peptide inside Huh7 cells. The study constitutes the first report of a small anti-HCV peptide (7-mer) targeting the HCV IRES and provides the “proof of concept” that a peptide consisting of a minimal RNA binding domain can successfully inhibit HCV RNA translation and replication, which can be exploited as a target to design efficient and more potent antiviral therapeutics against HCV infection in the near future.
MATERIALS AND METHODS
NMR spectroscopy.
Both the peptide and the RNA were dissolved in 20 mM potassium phosphate buffer (pH 7) containing 0.1 M KCl, and 10% D2O. All spectra of LaR2C were recorded with a 500-MHz DRX-500 machine (Bruker Biospin, Switzerland) at 27°C, unless stated otherwise. Total correlation spectroscopy (TOCSY; mixing time, 80 ms) and nuclear Overhauser effect spectroscopy (NOESY) (mixing time, 400 ms) spectra were recorded with solvent suppression achieved using the watergate scheme, and the spinlock in TOCSY experiment was attained by MLEV sequence.
Determination of the RNA-bound structure of the 7-mer peptide.
Mixing time for NOESY was 400 ms, and the same for the TOCSY (with MLEV spinlock) experiment was 60 ms for transferred NOE experiments. The distance between two protons was determined under transferred NOE conditions. NOESY experiments were done at a final concentration of 450 μM 7-mer peptide with 15 μM p383 RNA at 283 K in 20 mM deuterated Tris-HCl buffer (pH 7) containing 100 mM NaCl. The one-dimensional spectrum under these conditions is significantly broadened compared to that of free peptide, without showing any new peaks, indicating fast exchange conditions. The distance between two adjacent protons in the aromatic ring of a tyrosine residue, i.e., 2.48 Å, was considered the standard. It was used to calculate all other distances by comparing their peak volumes. However, the distance constraints were given in terms of strong (<3.5 Å), medium (3.5 to 4.5 Å), and weak (>4.5 Å) and may be considered somewhat of an overestimate due to transferred NOE conditions used. The maximum limit that was kept was 5.0 Å, and the minimum was 2.8 Å. The angle constraints were derived by direct measurement of scalar coupling constants from the one-dimensional spectrum under the conditions mentioned above, and corresponding dihedral angles were derived from Karplus relationships. An upper and lower limit of ±20° was used for angle constraints. The J values were also cross-checked with those obtained from double quantum filter-correlation spectroscopy (DQF-COSY) and the procedure described by Kim and Prestegard (19). A total of 30 distance constraints and 6 ϕ angle constraints (Table 1) were used for structure determination.
TABLE 1.
Restraints for the wt LaR2C-N7 (174-180) peptide structure calculation
| Restraint | No. of peak volumes |
|---|---|
| Total NOE distance restraints | 30 |
| Short range (<3.5 Å) | 4 |
| Medium range (3.5-4.5 Å) | 16 |
| Long range (>4.5 Å) | 10 |
| Dihedral angle restraints (°) | 6 |
| Hydrogen bond restraints (Å) | 0 |
The angle and distance constraints, along with the complete protein sequence, were analyzed by DYANA software to calculate the structures (see supplemental material). Those with target function values of 40.00 or lower (i.e., deviation from the inputs) were combined, and energy was minimized by Discover version 3.0 software in molecular modeler Insight II, version 2005 (Biosym/MSI), using the simple minimization algorithm. Then, the structures were validated by the Research Collaboratory for Structural Bioinformatics (RCSB) validation server, and the statistics are given in Table 2.
TABLE 2.
Structure determination data for wt LaR2C-N7 peptide
| Ramachandran parametera | Plot values |
|---|---|
| No. of nonglycine and nonproline residues | 7 |
| No. of end residues (excluding Gly and Pro) | 2 |
| No. of glycine residues | 0 |
| No. of proline residues | 0 |
| Total no. of residues | 7 |
| Residues in most favored regions (%) | 40.0 |
| Residues in additional allowed regions (%) | 60.0 |
| Residues in generously allowed regions (%) | 0.0 |
| Residues in disallowed regions (%) | 0.0 |
| Deviation from idealized geometry | |
| Bond lengths (Å) | 0.015 |
| Bond angles (°) | 1.9 |
| RMSD from exptl restraints | |
| NOE (Å) | 0.154 |
| Dihedral angle restraints (°) | 13.6 |
| Atomic RMSD | |
| Backbone (all residues) | 0.33 |
| Heavy atoms (all residues) | 1.62 |
RMSD, root mean square deviation.
Peptide synthesis and purification.
The LaR2C peptide (KYKETDLLILFKDDYFAKKNEERK), corresponding to residues 174 to 197 of the La protein, the mutant LaR2C peptide (KYKATDLLILFKDDYFAKKNEERK; the point mutation is in italics), and the nonspecific La peptide (La-Nsp; aa 71 to 98 [16]) (ALSKSKAELMEISEDKTK) were custom synthesized at Sigma Genosys, with 90% purity. The peptides were dissolved in nuclease/protease-free water, and integrity was checked by gel electrophoresis, followed by silver stain analysis.
All other smaller peptides (with or without the six-Arg tag) were synthesized on a 0.1-mmol scale by using a solid-phase peptide synthesis strategy using 9-fluorenylmethoxy carbonyl chemistry and Rink amide resin. Cleavage of the peptide from Rink amide resin and removal of all side chain-protecting groups were achieved in 95% trifluoroacetic acid solution. The crude peptides were purified by reversed-phase high-performance liquid chromatography (Waters Associates) with a Waters C18 column (μBondapak) with linear gradients of water/acetonitrile containing 0.1% trifluoroacetic acid. Peptide masses and purity (>95%) were checked by positive ion mode electrospray ionization mass spectrometry (Waters Inc.).
Plasmids and cells.
Construction of the pRSET-A La plasmid, the pHCV383Luc monocistronic construct, and the bicistronic construct containing the HCV IRES has been described previously (24). The HCV monocistronic replicon construct pFKi383hygubiNS3-3′5.1 used in the study was a generous gift from R. Bartenschlager (9). The mutant La clones were generated by a megapriming PCR method and cloned into the pRSET-A expression vector. The mutants carried substitutions only at the respective amino acid positions (P2 or P4 or P15 or P21/22); all other positions remained unaltered. Thus, the changes were only in RRM (112-184), not in other RRMs. The PV 5′ untranslated region (UTR) or HCV IRES encompassing nucleotides (nt) 18 to 383 [HCV IRES (18-383)] was cloned upstream of the luciferase reporter gene in pCDNA3.1 to generate pCDPVLuc or the pCDHCVLuc monocistronic construct, respectively. Additionally, the HCV IRES (18 to 383) or hepatitis A virus (HAV) 5′ UTR was cloned in between the RLuc and FLuc reporter genes in the pCDNA3.1 vector to generate the respective bicistronic constructs.
Huh7 monolayer cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, and the Huh7 cells harboring the HCV replicon were grown in DMEM with 10% fetal bovine serum, 25 μg/ml hygromycin B (Calbiochem).
Purification of wild-type and mutant La proteins.
The wild-type and the mutant La proteins were overexpressed in Escherichia coli BL21(DE3) cells and purified using a Ni-nitrilotriacetic acid agarose column as described previously (24). Briefly, the cultures were induced with 0.6 mM isopropyl-β-d-thiogalactopyranoside for 4 h at 37°C. The crude extracts were mixed with a Ni-nitrilotriacetic acid agarose slurry (Qiagen) and kept for rocking at 4°C for 4 h. The lysate was loaded onto a column, washed with 50 ml of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 40 mM imidazole), and the bound proteins were eluted with 500 μl of elution buffer containing 500 mM imidazole. The eluted proteins were dialyzed at 4°C for 6 h in dialysis buffer (50 mM Tris [pH 7.4], 100 mM KCl, 7 mM β-mercaptoethanol, 20% glycerol), aliquoted, and stored in a −70°C freezer.
In vitro transcription and translation.
Radiolabeled mRNAs were transcribed in vitro using T7 RNA polymerase (Promega) and [α-32P]UTP. The HCV IRES (18-383) cloned in the pcDNA3 vector was linearized with EcoRI, gel eluted, and transcribed in vitro in the presence of [α-32P]UTP to generate the labeled HCV IRES RNA as described previously (25). The HCV IRES-containing monocistronic construct (HCV luciferase) was linearized with XhoI to prepare HCV Luc RNA, and the HCV bicistronic construct was linearized with PmeI to generate capped bicistronic RNA (Rluc-HCV IRES-Fluc) to use in the in vitro translation studies. The monocistronic construct pCDLuc (containing the luciferase gene in the pCDNA3 backbone) was linearized downstream of the luciferase gene with XhoI, and the template was used to generate capped luciferase RNA by in vitro transcription in the presence of a cap analog using T7 RNA polymerase. The HAV bicistronic construct was linearized downstream of firefly luciferase and used as the template for RNA synthesis as described previously (25). The poly linker sequence of the pGEM-T-easy vector was linearized with SacI to generate the Nsp RNA.
In vitro translation of the capped luciferase mRNA, capped bicistronic RNAs, or uncapped HCV luciferase RNA was carried out in micrococcal nuclease-treated rabbit reticulocyte lysate (RRL; Promega Corporation, WI) as described previously (25).
UV cross-linking experiment.
The purified proteins or the synthetic peptides were incubated with ∼75 fmol of either 32P-labeled HCV IRES RNA or the Nsp RNA at 37°C for 15 min, containing RNA binding buffer (5 mM HEPES [pH 7.6], 25 mM KCl, 2 mM MgCl2, 3.8% glycerol, 2 mM dithiothreitol, 0.1 mM EDTA, and 5 μg tRNA) in a reaction volume of 15 μl; the complex was then subjected to UV-induced cross-linking as described previously (24). The entire reaction mixture was then denatured, followed by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) (for protein) or Tris-Tricine 17% PAGE (for peptide) and analyzed by phosphorimaging.
Filter binding assay.
The α-32P-labeled HCV IRES RNA or the Nsp RNA was incubated with the wild-type La or the mutant La protein or the peptides at 30°C for 15 min in RNA binding buffer (containing 5 mM HEPES [pH 7.6], 25 mM KCl, 2 mM MgCl2, 3.8% glycerol, 2 mM dithiothreitol, and 0.1 mM EDTA). The reaction mixtures were loaded onto nitrocellulose filters equilibrated with 2 ml of RNA binding buffer. The filters were then washed four times with 1 ml of binding buffer and then air dried. The counts retained were measured in a liquid scintillation counter. The graph was plotted with protein or peptide concentration (μM) on the x axis and the percentage of bound RNA as the percentage of counts retained on the y axis.
Real-time RT-PCR.
RNA isolated from the from peptide-treated and -untreated replicon cells was reverse transcribed with HCV 5′ primer using avian myeloblastosis virus reverse transcriptase (RT; Promega) for the amplification of negative strand of HCV RNA. For real-time PCR analysis, the cDNA was used for PCR amplification using a real-time assay mixture (Finnzymes) as per the manufacturer's instruction, and the data were analyzed by using ABI-Prism's real-time PCR machine. Actin was used as an internal control for the above reactions.
Fluorescence microscopy.
Huh7 monolayer cells were grown on a coverslip up to 70% confluence. Before addition of the peptide, cells were washed with phosphate-buffered saline (PBS) twice and then incubated with 1 μM of fluorescein-labeled peptide (dissolved in DMEM) for 3 h. After being washed extensively with PBS, cells were fixed with 3.7% formaldehyde for 30 min at room temperature, followed by washing twice with PBS. Cells were observed with fluorescence microscopy.
RESULTS
NMR spectroscopy of the HCV IRES RNA-bound peptide complex.
Identification of amino acid residues important for the recognition of RNA can be performed by NMR spectroscopy. In this case, since the RNA is relatively large (nt 18 to 383 of the HCV IRES), obtaining the full structure of the RNA-peptide complex (1:1) would have been extremely difficult. Alternatively, the NMR spectrum of the 24-mer peptide was studied, which was relatively simple in the absence and presence of a substoichiometric amount of RNA. It was assumed that under fast exchange conditions, the chemical shifts of peptide protons in the absence of RNA will be the average of free and bound species (the intensity of RNA protons will be insignificant due to substoichiometric presence and broader line width). Thus, a comparison of peptide chemical shifts in the presence of RNA with that of the free peptide is expected to shed light on the residues that may be involved in recognition.
TOCSY provides connectivity between all adjacent protons (three-bond connectivity) within an amino acid unit and hence a fingerprint for the type of amino acid. Figure 1A shows the TOCSY spectrum of the region that connects NH (approximately 7.5 to 9.5 ppm) with αH and other side-chain protons. Out of the expected 23 NH protons, 18 to 19 could be resolved. When a substoichiometric amount of RNA was added, a significant shift of many protons (but not all) was observed. This perhaps indicated either an extensive peptide-RNA interface or a folding of the peptide coupled to RNA binding. Determination of the bound conformation of the peptide and the whole interface is beyond the scope of this work, and we have focused on identifying at least one region that may be involved in recognition. This will allow us to mutate that residue in the whole protein and validate the possibility that the peptide is a good model for studying protein-RNA interaction.
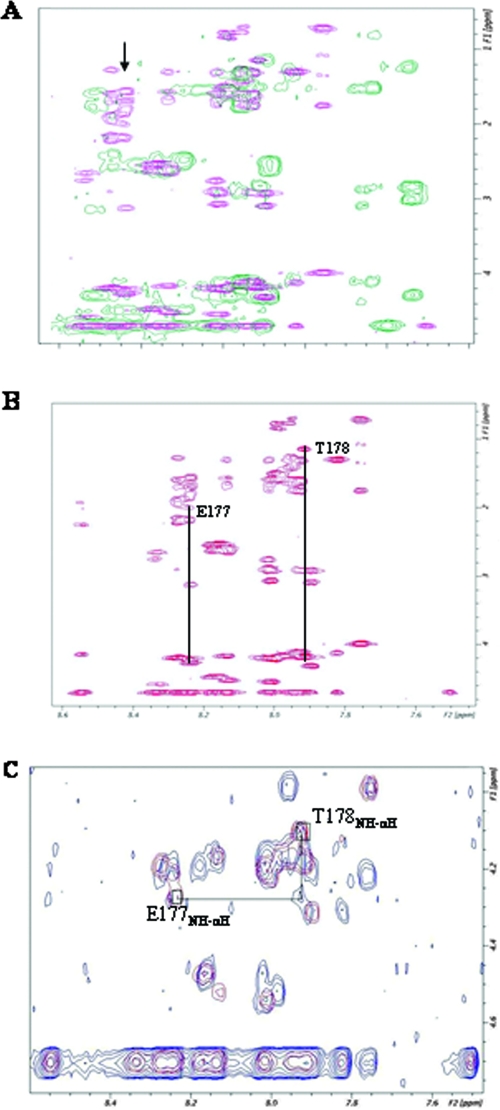
FIG. 1.
NMR analysis of HCV IRES RNA-bound peptide. (A) Overlay of two TOCSY spectra; pink indicates the LaR2C peptide without RNA and the green indicates the La-derived peptide with HCV IRES RNA. The arrow indicates the shifting of the E177 peaks after addition of HCV IRES RNA. All the spectra in this figure were recorded with a Bruker DRX-500 NMR spectrometer. Experimental details are given in Materials and Methods. (B) TOCSY spectrum of the LaR2C peptide with spin system identification of two amino acid residues, labeled with their corresponding one-letter symbols. The subscripts indicate the amino acid position in the peptide. (C) Overlay of TOCSY (red) and NOESY (blue) spectra of the LaR2C peptide and demonstration of the TOCSY-NOESY connectivity between T178 and E177. The boxes identify the location of the NH-αH TOCSY cross peaks for the residues E177 (down field) and T178 (up field).
Among other shifted residues in the TOCSY spectra, one residue at 8.27 ppm shows significant shift upon complex formation (Fig. 1). Chemical shifts and connectivity patterns indicate that this residue is a glutamic acid (no glutamine is present in the peptide). There is only one threonine (position five) in the peptide. Threonine α and β and the methyl protons have very characteristic chemical shifts and can be identified easily. Figure 1B shows the position of T178. T178 is connected to the shifted glutamic acid (E177) by NOE (Fig. 1C), indicating that the glutamic acid at 8.27 ppm is E177. Thus, at least E177 is likely to be involved in the recognition of HCV IRES RNA. Additionally, significant shifts were observed corresponding to Y175 (Tyr) at the N terminus and also Y188 (Tyr) and K192-N193-E194 positions at the C terminus of the LaR2C peptide.
Effect of point mutation within the LaR2C region of the La protein on HCV IRES binding.
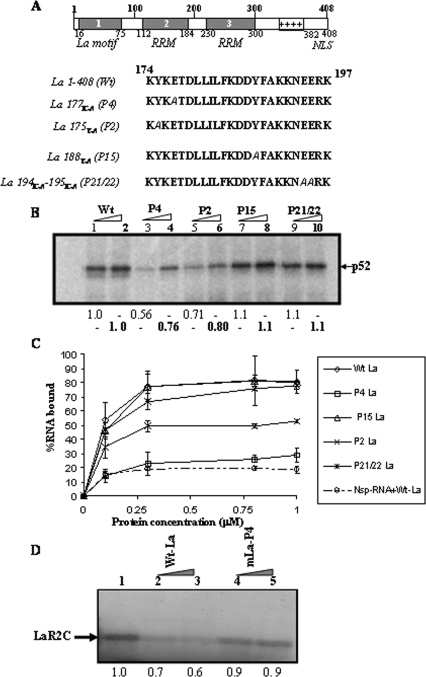
As mentioned above, the chemical shift perturbation can be due to direct interaction or indirect coupled folding events. To determine whether the above-mentioned amino acid residues of the La protein are actually involved in the recognition of HCV IRES RNA, we have generated corresponding point mutations in the full-length protein, using site-directed mutagenesis (Fig. 2A). The RNA binding activities of the mutant La proteins were tested and compared with that of the wild-type (wt) La protein by using a UV cross-linking assay with 32P-labeled HCV IRES RNA. The results showed that mutations La175Y-A and La177E-A (corresponding to the N-terminal P2 and P4 positions of LaR2C peptide, respectively) mostly affected the HCV IRES binding, whereas mutations La188Y-A and La194E-A-195E-A (corresponding to C-terminal positions P15 and P21/22) did not significantly alter the RNA binding ability of the La protein (Fig. 2B).
FIG. 2.
Effect of point mutation in La protein on HCV IRES binding. (A) Schematic representation of the domain organization of human La protein. The residues mutated in full-length La protein (between aa 174 and 197) with their corresponding positions within 24-mer LaR2C peptide are indicated. (B) UV cross-linking analysis. [α-32P]UTP-labeled HCV IRES RNA (∼75 fmol) was UV cross-linked with increasing concentrations (150 and 300 ng) of either wt La protein or the mutants (as indicated at the top of the lanes). The protein-nucleotide complex was resolved in SDS-10% PAGE, followed by phosphorimaging analysis. The position of La protein (p52) is indicated. The band intensities corresponding to La were quantified by densitometry. The numbers below lanes 3, 5, 7, and 9 represent the relative intensities, taking lane 1 (150 ng of protein) as control, whereas the numbers (in boldface) below lanes 4, 6, 8, and 10 represent the relative intensities, taking lane 2 (300 ng of protein) as control. (C) Filter binding assay: α-32P-labeled HCV IRES RNA was bound to increasing concentrations of either wt La or the mutant La proteins (as indicated). Additionally, α-32P-labeled Nsp RNA was also used, along with the wt La protein. The amount of bound RNA was determined by binding to the nitrocellulose filters. The percentage of bound RNA was plotted against the protein concentration (μM). (D) Competition UV cross-linking: [α-32P]UTP-labeled HCV IRES RNA was preincubated with the LaR2C peptide, followed by addition of either wt La protein (lanes 2 and 3) or mutant La protein (P4; lanes 4 and 5) in the reaction mixture for competition. The UV cross-linked complex was treated with RNase and resolved by SDS-15% Tris-Tricine gel. The relative position of the band corresponding to the LaR2C peptide is indicated with an arrow. The numbers below the lanes represent the relative band intensities, taking lane 1 as control.
Additionally, a filter binding assay was performed using 32P-labeled HCV IRES RNA and increasing concentrations of purified recombinant proteins (wt La or the mutants). Considering the midpoint of transition, it appeared that the mutation at the P4 residue significantly affected the RNA binding ability of the La protein. Mutation at the P2 residue also showed a decrease in RNA binding ability, but only to an extent. However, mutation at P15 or P21/22 did not alter the binding ability of the La protein (Fig. 2C). As expected, a nonspecific RNA probe did not show considerable binding with the wt La protein in the same filter binding assay.
Earlier, we showed that the LaR2C peptide can effectively compete with full-length La protein for binding near the iAUG codon within HCV IRES RNA (25). Here, in the competition UV cross-linking experiment, we observed that the full-length wt La protein can also compete out binding of LaR2C with the HCV IRES RNA (Fig. 2D). However, the mutant P4 La protein failed to compete the binding of LaR2C peptide significantly with the HCV IRES RNA (Fig. 2D). The result suggests that the domain of La protein encompassing the amino acid P4 (La177E-A) might be involved in making contact with the HCV IRES RNA near the iAUG, where LaR2C peptide also binds. However, binding of La protein to other sites within HCV IRES RNA might not be affected as much by the above-described mutation.
Effect of mutation in the LaR2C peptide on RNA binding and translation.
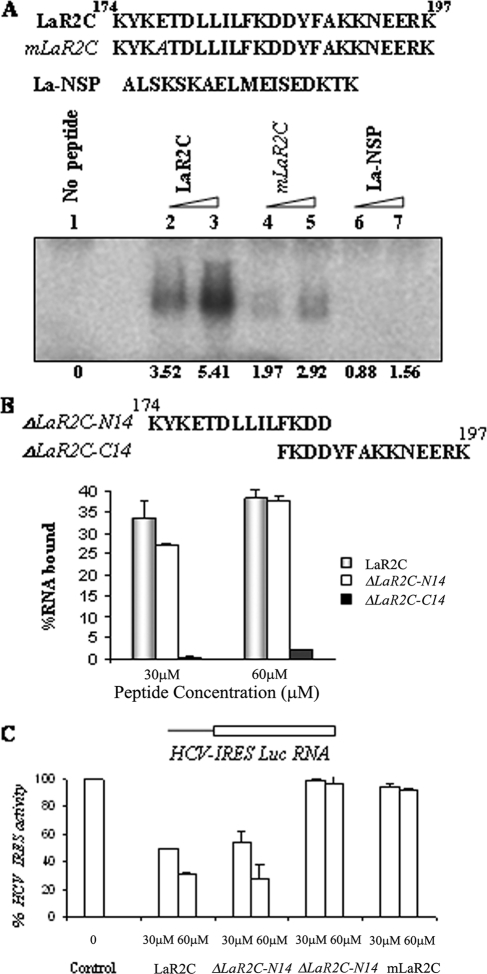
To further determine the role of N-terminal amino acids in the peptide activity, we generated a mutant LaR2C peptide, where E177 was changed from glutamic acid to alanine (Fig. 3A). The RNA binding abilities of the wt and the mutant peptides were tested by UV cross-linking assay. For this purpose, α-32P-labeled HCV IRES RNA was UV cross-linked with increasing concentrations of either wt LaR2C or the mutant LaR2C or an Nsp La peptide. Results showed that the mutation at P4 (La177E-A) did affect the RNA binding ability of the mutant peptide (Fig. 3A). The Nsp La peptide did not show any RNA binding activity (Fig. 3A). To further confirm the RNA binding ability of the peptides, a filter binding assay was performed using increasing concentration of wt and mutant peptides and α-32P-labeled HCV IRES RNA. The results showed a reduced level of saturation for the mutant peptide-RNA complex compared to that of the wt LaR2C peptide, suggesting a critical role for the P4 residue in the activity of the LaR2C peptide (data not shown).
FIG. 3.
Effect of P4 point mutation in LaR2C peptide activity. (A) Schematic representation of the peptide used in the UV cross-linking analysis. The residue mutated in the mutant LaR2C peptide is indicated in italics. [α-32P]UTP-labeled HCV IRES RNA (∼75 fmol) was UV cross-linked with increasing concentrations (30 μM and 60 μM) of LaR2C, mLaR2C, and La-Nsp (NSP). The peptide-nucleotide complex was resolved in 15% Tris-Tricine PAGE followed by phosphorimaging analysis. The band intensities were quantified by densitometry. The numbers below the lanes represent the intensities, taking lane 1 (no peptide) as control. (B) Filter binding assay: 32P-labeled HCV IRES RNA was bound to increasing concentrations of either wt LaR2C peptide or mutant peptide as indicated. The amount of bound RNA was determined by its binding to the nitrocellulose filters. The percentage of bound RNA was graphically represented relative to the peptide concentration (μM). (C) Effect on HCV IRES-mediated translation in vitro. One microgram of uncapped HCV IRES Luc RNA was translated in RRL in the absence (lane 1) or in the presence of increasing concentrations (30 and 60 μM) of either wt LaR2C or mutant peptides (as indicated). The relative FLuc activities were represented as percentages of the control reaction (expressed as 100%).
Interestingly, results from a similar filter binding assay suggest that deletion of N-terminal amino acids almost abrogated the RNA binding activity of the peptide (ΔLaR2C-C14), whereas retention of only 14 amino acids in the N terminus (ΔLaR2C-N14) appeared to be sufficient for significant RNA binding activity (Fig. 3B).
Additionally, the effect of mutation in the peptide was tested in the in vitro translation assays with RRLs using uncapped monocistronic RNA containing HCV IRES upstream of the firefly luciferase gene. Results showed significant decreases in HCV IRES-mediated translation of HCV Luc RNA in the presence of increasing concentrations (50% inhibition at 30 μM and 70% inhibition at 60 μM) of wt LaR2C peptide. However, a similar concentration of mutant peptide failed to inhibit the HCV IRES function significantly (Fig. 3C). Also, it appears that the C-terminal truncated peptide ΔLaR2C-N14 retained the translation inhibitory activity compared to the wt peptide control. However, deletion of the N-terminal amino acids (ΔLaR2C-C14) resulted in abrogation of the translation inhibitory activity (Fig. 3C).
Characterization of conformation of the N-terminal seven-residue peptide.
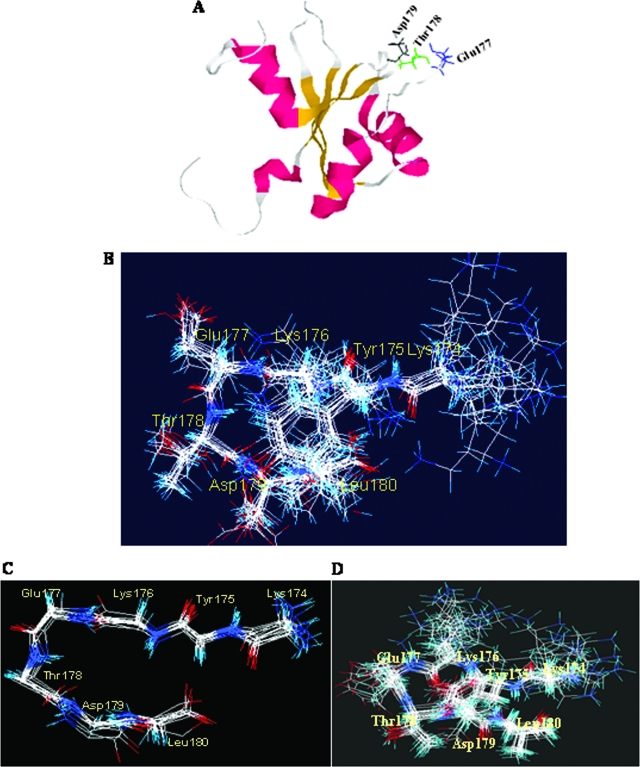
The N-terminal part of the LaR2C (174-196) has been shown to constitute the β4 sheet and β4′ strand of RRM (101-200) (1). Interestingly, the residues of the peptide responsible for RNA recognition were mapped to a turn in the context of the RRM (112-184) NMR structure. Based on previous NMR structure information (PDB identification no. 1S79), when the LaR2C peptide was modeled, these critical N-terminal amino acids were found to be located in a similar turn that appears to be exposed for RNA binding (Fig. 4A).
FIG. 4.
Structural characterization of the LaR2C-N7 peptide. (A) Structural model of LaR2C in the context of structural model of RRM (101-200) of La protein (PDB identification no. 1S79). The helix regions are pink, β sheets are yellow, and turns and random coils are gray. The wt LaR2C-N7 sequence lies in the gray region (aa 174 to 180). The glutamic acid (Glu177) is blue, threonine (Thr178) is green, and aspartic acid (Asp179) is black. (B) Superimposition of the best 16 structures (backbone) of 7-mer peptide (residues 174 to 180) under RNA bound conditions, simulated using Dyana software (version 1.5; Peter Guentert and Kurt Wuthrich, Zurich, Switzerland). The amino acid residues are represented by three-letter codes and are numbered corresponding to their positions in RRM (101-200). (C) Backbone structure of RNA bound LaR2C-N7. (D) Sixteen superimposed structures from the same region of the RRM (101-200) structure for comparison. The amino acid residues are represented by three-letter codes and are numbered from the amino terminus starting from 174.
Since the RRM (112-184) structural model suggests that the N-terminal seven (N7) residues completely cover the turn that sticks out of the globular structure of the domain (Fig. 4A), we were interested in determining the structure and function of the seven-residue peptide.
The HCV IRES RNA bound structure of the LaR2C-N7 structure was determined by NMR spectroscopy. Under bound conditions, the peptide gave several new NOEs and change of value for J for several amide protons in addition to significant line broadening (data not shown). This indicates that conformational parameters derived from this experiment indeed reflect the bound form. The sequence KETD forms a β turn as the distance between the Cα atoms of K and D is less than 7 Å (Fig. 4B). However, the Ramachandran angles do not fall within any defined turn category. The KETD sequence in the RRM (112-184) structure is also a β turn, but the conformational parameters do not fall strictly into any well-defined category (Fig. 4C). Although the structures in two contexts are similar, there are noticeable differences in Ramachandran angles. Thus, there could be significant remodeling of the structure upon binding of La to the RNA (Fig. 4D).
The smaller peptide (LaR2C-N7) inhibits HCV IRES-mediated translation in vitro.
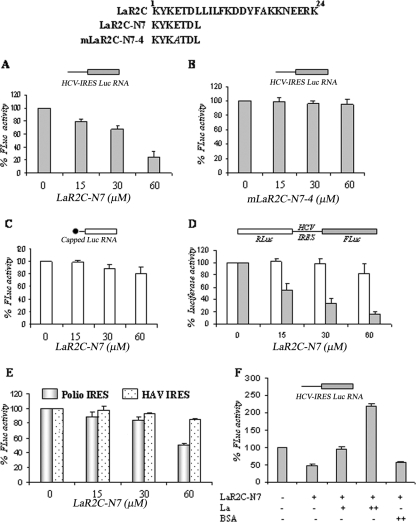
After determining the preference for beta turn conformation in the N-terminal seven residues, we were interested in determining whether the 7-mer peptide comprising these amino acids would also inhibit HCV IRES-mediated translation. For this purpose, we used monocistronic RNA containing HCV IRES upstream of the reporter luciferase gene in the in vitro translation assays in the presence or absence of increasing concentrations of LaR2C-N7 peptide. Interestingly, the smaller peptide (LaR2C-N7) also showed translation inhibitory activity which was only slightly weaker than the that of the 24-mer peptide (Fig. 5A). However, mutation at the P4 position of the LaR2C-N7 completely abrogated its translation inhibitory activity (Fig. 5B). Interestingly, the LaR2C-N7 peptide did not show significant inhibition of capped luciferase RNA (representing cap-dependent translation), suggesting the specificity of the inhibition (Fig. 5C). Also, LaR2C-N7 showed selective inhibition of HCV IRES-mediated translation in the context of HCV bicistronic RNA (Fig. 5D). Furthermore, the LaR2C-N7 peptide failed to inhibit IRES-mediated translation of HAV but showed significant inhibition of PV IRES function at a higher concentration (Fig. 5E).
FIG. 5.
Effect of LaR2C-N7 on HCV IRES-mediated translation in vitro. (A and B) Above the panels is a schematic representation of the wt 7-mer peptide (LaR2C-N7) and the mutant 7-mer peptides, along with the 24-mer wt LaR2C peptide. The mutated residue is indicated in italics. One microgram of HCV IRES Luc monocistronic RNA was translated in RRL in the absence or presence of increasing concentrations (15, 30, and 60 μM) of either the wt LaR2C-N7 or the mutant mLaR2C-N7-4 peptide. Respective luciferase activities were measured and plotted against different peptide concentrations. The relative FLuc activities are represented as percentages of the control reaction (expressed as 100%). Results represent an average of three independent experiments. (C) Similarly, 1 μg of capped luciferase RNA was translated in RRL in the absence or presence of increasing concentrations (15, 30, 60 μM) of LaR2C-N7 peptide, and luciferase activities were plotted against different concentrations of the wt LaR2C-N7 peptide. The relative FLuc activities were represented as percentages of the control reaction (expressed as 100%). Results represent an average of three independent experiments. (D) One microgram of capped bicistronic RNA was translated in RRL in the absence or presence of increasing concentrations (15, 30, 60 μM) of LaR2C-N7 peptide, and luciferase activities were plotted against different concentrations of the wt LaR2C-N7 peptide. The relative RLuc and FLuc activities were represented as a percentage of the respective control reactions (expressed as 100%). Results represent an average of three independent experiments. (E) One microgram of either PV-luciferase monocistronic RNA or capped HAV-bicistronic RNA (containing Rluc-HAV-Fluc, in order) translated in the absence (lane 1) and presence of increasing concentrations (15, 30, 60 μM) of the LaR2C-N7 peptide. The translation of the firefly luciferase activities was measured and plotted relative to the peptide concentrations for the respective constructs as indicated. The relative FLuc activities were represented as percentages of the control reaction (expressed as 100%). Results represent an average of three independent experiments. (F) One microgram of HCV IRES Luc monocistronic RNA was translated in RRL in the absence or presence of wt LaR2C-N7 peptide (40 μM). Increasing concentrations (25 ng and 50 ng) of purified wt La protein or bovine serum albumin (50 ng) were added to the reaction mixtures as indicated below the lanes. Respective luciferase activities were measured and plotted in the graph. The relative FLuc activities were represented as percentages of the control reaction (expressed as 100%). Results represent an average of three independent experiments.
More importantly, addition of increasing concentrations of purified wt La protein showed significant rescue of the suppressive effect of LaR2C-N7 (Fig. 5F). However a similar concentration of bovine serum albumin protein was not able to rescue the inhibition. Addition of increasing concentrations of recombinant La protein (25 ng and 50 ng) in the reaction mixture (in the absence of peptide) showed dose-dependent stimulation in HCV IRES-mediated translation (data not shown) as observed previously (24). The result reinforces the idea that the LaR2C-N7 peptide-mediated inhibition of translation could be due to competition with the endogenous La protein. Taken together, the results suggest that the turn at the N terminus of the LaR2C peptide could be critical for its RNA binding, as well as for translation inhibitory activity.
The arginine-tagged LaR2C-N7 peptide inhibits HCV IRES function in vivo.
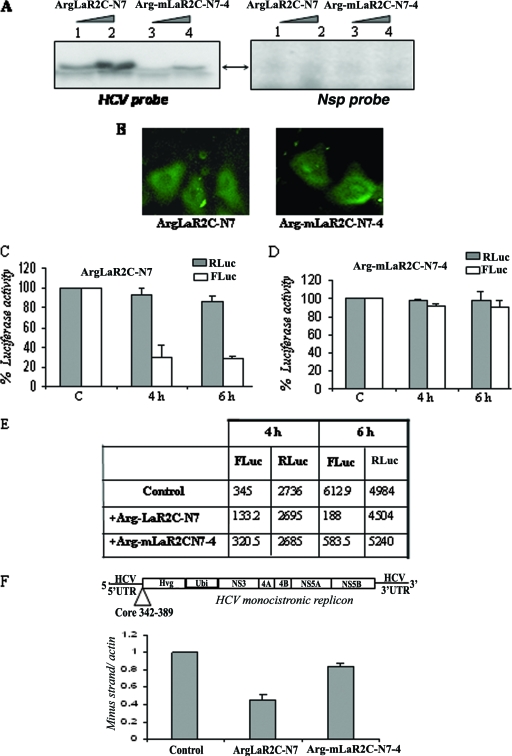
To determine whether the LaR2C-N7 peptide would be equally effective at inhibiting HCV IRES-mediated translation in vivo in Huh7 cells; we explored delivery of the peptides inside the cells, with the help of a hexa-arginine fusion tag. Arginine-tagged peptide has the ability to be internalized into mammalian cells when it is supplied exogenously in the medium (11). The RNA binding ability of the arginine-tagged LaR2C-N7 peptide was demonstrated by UV cross-linking experiment using 32P-labeled HCV IRES RNA. The mutant mLaR2CN7-4 (arginine-tagged) peptide did not show appreciable RNA binding activity. A nonspecific RNA probe was also used as negative control in the experiment (Fig. 6A).
FIG. 6.
Effect of arginine-tagged LaR2C-N7 on HCV IRES function in Huh7 cells. (A) UV cross-linking. Increasing concentrations (2 μM and 4 μM) of hexa-arginine-tagged peptides, wt ArgLaR2C-N7, or the Arg-mLaR2C-N7-4 mutant were UV cross-linked with [α-32P]UTP-labeled HCV IRES RNA or a nonspecific RNA probe and analyzed by SDS-17% Tris-Tricine gel analysis, followed by phosphorimaging. (B) Fluorescein-tagged hexa-arginine peptides (both the wt and the mutant) were incubated with Huh7 cells for 3 h and washed extensively with PBS, and observed with a fluorescence microscope. Left panel shows wt ArgLaR2C-N7 and the right panel shows mutant Arg-mLaR2C-N7-4 peptide. (C and D) Huh7 monolayer cells were transfected with 2 μg of HCV bicistronic DNA (containing Rluc-HCV IRES-Fluc, in order). After 3 h of transfection, cells were overlaid with 2 μM of either wt or mutant Arg-LaR2C-N7 peptide. Cells were harvested at different time points (as indicated) and lysed, and luciferase activities were measured using the dual luciferase assay system. The RLuc (gray bar) and FLuc (white bar) activities were plotted as (fold) change increases or decreases in the presence of the peptide with respect to the corresponding control (in the absence of peptide) taken as 100. (E) Absolute values of RLuc and FLuc activities (in relative light units) of a representative experiment are presented in the table. (F) Schematic representation of the HCV monocistronic replicon RNA (9). The monolayer Huh7 cell harboring the above-mentioned replicon was overlaid with either the wt 7-mer (ArgLaR2C-N7) or mutant 7-mer (Arg-mLaR2C-N7-4) peptide (4 μM each), added twice, at 0 and 12 h. RNA was isolated at 24 h and subjected to cDNA synthesis. The HCV negative strand was detected using real-time PCR. Data were normalized with actin control, and negative strand synthesis was expressed as (fold) change compared to that of control cells (in the absence of peptide).
To investigate the internalization of the arginine-tagged peptides, we used fluorescein-labeled hexa-arginine-tagged peptides and found that both the peptides are internalized in Huh7 cells (Fig. 6B). To determine the effect of these peptides on HCV IRES function inside the cells, we used these arginine-tagged peptides in Huh7 cells. Huh7 monolayer cells were first transfected with the pcDNA3-HCV bicistronic construct and incubated for 3 h, washed, and layered with medium containing 2 μM of Arg-LaR2C-N7 peptide and incubated further for either 4 h or 6 h. Similarly, as a negative control, another set of dishes was layered with mutant peptides (Arg-mLaR2C-N7-4). After incubation with the peptide, the cells were washed and then lysed, and the luciferase (FLuc and RLuc) activities were measured. The relative luciferase activities were represented so that RLuc represents cap-dependent translation, and FLuc represents HCV IRES-mediated translation. The results showed a significant decrease (inhibition up to 70%) in the HCV IRES-mediated translation over that of the control, when cells were incubated with the Arg-LaR2C-N7 peptide. However, the mutant (Arg-LaR2C-N7-4) did not show appreciable inhibitory effect (Fig. 6C and D). The absolute levels of RLuc and FLuc activities of a representative experiment are presented in the table (Fig. 6E). Taken together, these results indicated that LaR2C-N7 does compete with the interaction of cellular La protein with HCV IRES RNA and exerts a dominant negative effect to inhibit HCV IRES-mediated translation in vivo.
Additionally, to determine whether this peptide would inhibit HCV replication as well, we treated Huh7 cells harboring the HCV monocistronic replicon (9) with either wt (Arg-LaR2CN7) or mutant (Arg-mLaR2C-N7-4) peptide for 24 h. The peptides (4 μM) were added twice over 12-h intervals. As the measure of negative strand synthesis indicates replication of HCV RNA (4), we have quantitated it by real-time RT-PCR. The results showed an almost 50% decrease in levels of HCV negative strand RNAs compared to that of the untreated cells when 4 μM of ArgLaR2C-N7 peptide was used (Fig. 6F). However, the mutant peptide (Arg-mLaR2C-N7-4) did not show an appreciable decrease in negative strand synthesis (Fig. 6F). At lower concentration of the peptide (2 μM), the inhibition was around 30%, and at higher concentration (10 μM), considerable increase in the inhibitory activity was observed (data not shown).
Taken together, the results provide the “proof of concept” that the LaR2C-N7 peptide might be effective against HCV IRES function and consequently inhibit replication of HCV RNA in Huh7 cells.
DISCUSSION
HCV infection is a major public health problem with limited established therapeutic options. The most common established therapy is interferon α, which is not effective in the majority of cases. Newer experimental therapies include nucleoside analogs, protease inhibitors, and polymerase inhibitors, but there is a growing need for agents that are directed against new targets.
Human La protein plays an important role in HCV infection. Earlier studies indicated that for HCV IRES RNA, which lacks 3′UUUOH, several subdomains of La protein contribute to RNA recognition. The central RRM (112-184) binds strongly to HCV IRES in the context of iAUG, whereas the C-terminal RRM (230-300) and the hydrophobic domain have also been shown to contribute to HCV IRES binding perhaps at other sites (2, 5, 24, 26). Earlier, the La motif was shown to indirectly influence the RNA binding ability of the La protein via other RRMs (16). However, we have demonstrated that the RRM (112-184) of La protein binds directly to HCV IRES RNA at the GCAC motif near the iAUG codon (24, 25, 26). We have also demonstrated that the LaR2C peptide (corresponding to aa 174 to 196) interacts with the GCAC motif near the iAUG codon in the SLIV region and acts in a dominant negative manner by competing with the endogenous La protein binding at this site (25, 26). Thus, La-HCV IRES interaction could be a potential target for drug design. In fact, previously, Izumi et al. (16) also reported a short peptide, LAP (11-28), derived from the La motif, which could selectively inhibit IRES-mediated translation of HCV and PV.
Our results are consistent with those of the study by Izumi et al. (16) that show a single-point mutation in the La motif of the full-length La protein can completely abrogate the RNA binding activity. Both of these studies indicate that perhaps the secondary structure and the tertiary interactions between the domains influence La protein folding, which might contribute to RNA binding. Thus, a point mutation at any of the hot spots (the La motif or RRM) could have drastic consequences for the La protein structure and its RNA binding ability.
The inhibitory activities of the peptides may not be attributed to their RNA-binding ability alone. In fact, in our earlier publication (25), we mentioned that in addition to HCV RNA binding, the LaR2C peptide might also interfere with the binding of some other cellular protein factors to HCV IRES RNA, and thus, the inhibitory effect of the peptide could be the combination of both effects. It is possible that the mutation at P4 in the LaR2C peptide might have affected both of these possibilities and hence have drastically reduced the inhibitory activity. It is worth mentioning that the LAP peptide (as reported previously) did not show HCV IRES RNA binding but still could inhibit the translation effectively (16).
La protein has been shown to interact with the HCV 3′ UTR and influence viral RNA replication (7). It would be interesting to determine whether the LaR2C peptide or its smaller derivative (LaR2C-N7) dislodges La binding to HCV 3′ UTR as well. In that case, the inhibitory effect of the peptide on HCV RNA replication should have been more pronounced. However, at this point we cannot comment on whether the 50% inhibition of HCV RNA replication achieved by the LaR2C-N7 peptide (4 μM) is merely the consequence of translation inhibition. Future experiments would address this issue, using higher concentration of the peptide, and also test the efficacy of this peptide at protecting cells from HCV infection in a cell culture model.
Although a large peptide has been used as a drug in other viruses, e.g., against human immunodeficiency virus (T-20), in general the potential of drug development increases sharply as the molecular weight of the peptide decreases. In this study, we have demonstrated that a small 7-mer peptide (hexa-arginine tagged) corresponding to an exposed turn can significantly inhibit HCV IRES-mediated translation in cell culture at a significantly lower concentration (2 μM) than that required for inhibition of translation in vitro. The hexa-arginine tagged 7-mer peptide is also capable of inhibiting HCV RNA replication. The fact that the effect of the peptide (at 4 μM concentration) on HCV replication is only 50% could be due to its relative instability during long incubation. But designing peptidomimetic structures through mimicking this turn might improve both the affinity and the in vivo stability (bioavailability).
The bound structure of the peptide inferred from transferred NOE experiments suggests it is a β turn but falls into no defined category (30). The NMR structure of the unliganded domain indicates this conformation is a β turn as well. However, the Ramachandran angles of i + 1 and i + 2 residues are different from the prescribed values of any type of β turn (15). Even though the domain structure and the bound peptide structure are both β turns and fall into any of the defined categories, they are themselves different. This suggests conformational change of the turn upon binding to target RNA. Thus, stabilization of the bound β turn conformation in a peptidomimetic structure by suitable residues may enhance binding.
Specificity is a crucial issue in designing peptidomimetic structures, as well as other therapeutic entities. Lack of inhibition of IRES function with E4 mutations (mLaR2C-N7-4) strongly suggests a highly specific mode of inhibition. In addition, the fact that the LaR2C-N7 peptide did not inhibit HAV IRES suggests the specificity of its inhibitory activity. It is possible that La protein is not as critical for HAV-IRES function. On the other hand, La protein has been shown to interact and enhance IRES-mediated translation of PV RNA. However, LaR2C-N7 peptide was not as effective against PV IRES-mediated translation, again indicating a high degree of selectivity. Domains other than RRM (112-184) have been identified with this stimulatory function in PV (5). Also, the LaR2C-N7 peptide can selectively inhibit HCV IRES-mediated translation in vivo at 2 μM concentration without affecting cap-dependent translation, suggesting specificity of the approach.
One notable point here is that in the in vitro translation assays, approximately 60 μM concentration of 7-mer peptide was necessary to achieve around 70% inhibition. However, the hexa-arginine-tagged 7-mer peptide was found to be more effective, and similar levels of inhibition were achieved at much lower concentration (5 to 10 μΜ) (data not shown). Even in the in vivo assay, 2 to 4 μM hexa-arginine-tagged peptide was sufficient to achieve 50% inhibition. It is possible that the hexa-arginine residues might have played some unintended positive roles by contributing to the net positive charge of the peptide and, thus, enhanced its RNA binding ability, thereby increasing the inhibitory activity as well. Also, in the in vivo situation, in the context of properly folded HCV IRES RNA in the presence of other trans-acting factors, the inhibitor could be more effective.
The field of therapeutic peptide analogs is in its infancy. Many of the problems associated with the use of therapeutic peptides are gradually being solved. In fact, a number of peptidomimetic structures are currently being tried as antiviral agents (8, 14). One of the best approaches in the design of effective peptidomimetic structure is to replace naturally occurring amino acids with synthetic amino acids that stabilize the interacting conformation guided by the structure of the peptide. This not only reduces the entropic cost of binding to the receptor (disorder-to-order) but also stabilizes the small peptides from proteolysis and degradation (3). Design of the peptidomimetic inhibitor BILIN-2061 against HCV protease was possible because it is based on structural relationship studies, availability of the crystal structure of the protease, replacement of the natural amino acids, etc. (27, 32). Availability of the structure of this small peptide will be helpful in developing more stable peptidomimetic structures with higher affinity for HCV IRES, so that it could effectively inhibit IRES-dependent translation at much lower concentrations while increasing the bioavailability and solving the stability issues.
Supplementary Material
Acknowledgments
We thank Akio Nomoto, Nahum Sonenberg, and Ellie Ehrenfeld for plasmid constructs. We also thank Ralf Bartenschlager and Volker Vogt for providing the Huh7 cells harboring the HCV replicon. We also gratefully acknowledge our laboratory members for their help and discussions.
This work was supported by a grant from the Department of Biotechnology and Life Science Research Board, India, to S.D. and Council of Scientific and Industrial Research (CSIR), India, to S.R. T.M. and A.M. were supported with predoctoral fellowships from CSIR, India.
Footnotes
Published ahead of print on 1 October 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alfano, C., D. Sanfelice, J. Babon, G. Kelly, A. Jacks, S. Curry, and M. R. Conte. 2004. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 11323-329. [DOI] [PubMed] [Google Scholar]
- 2.Ali, N., G. J. M. Pruijn, D. J. Kenan, J. D. Keene, and A. Siddiqui. 2000. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 27527531-27540. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee, R., G. Basu, Chene, and P. S. Roy. 2002. Aib-based peptide backbone as scaffolds for helical peptide mimics. J. Pept. Res. 6088-94. [DOI] [PubMed] [Google Scholar]
- 4.Carriere, M., V. Pene, A. Breiman, F. Conti, S. Chouzenoux, E. Meurs, M Andrieu, P. Jaffray, L. Grira, O. Soubrane, P. Sogni, Y. Calmus, S. Chaussade, A. R. Rosenberg, and P. Podevin. 2007. A novel, sensitive, and specific RT-PCR technique for quantitation of hepatitis C virus replication. J. Med. Virol. 79155-160. [DOI] [PubMed] [Google Scholar]
- 5.Costa-Mattioli, M., Y. Svitkin, and N. Sonenberg. 2004. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 246861-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dasgupta, A., S. Das, R. Izumi, A. Venkatesan, and B. Barat. 2004. Targeting internal ribosome entry site (IRES)-mediated translation to block hepatitis C and other RNA viruses. FEMS Microbiol. Lett. 15189-199. [DOI] [PubMed] [Google Scholar]
- 7.Domitrovich, A. M., K. W. Diebel, N. Ali, S. Sarker, and A. Siddiqui. 2005. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology 33572-86. [DOI] [PubMed] [Google Scholar]
- 8.Dragovich, P. S., T. J. Prins, R. Zhou, E. L. Brown, F. C. Maldonado, S. A. Fuhrman, L. S. Zalman, T. Tuntland, C. A. Lee, A. K. Patick, D. A. Matthews, T. F. Hendrickson, M. B. Kosa, B. Liu, M. R. Batugo, J. P. Gleeson, S. K. Sakata, L. Chen, M. C. Guzman, J. W. Meador III, R. A. Ferre, and S. T. Worland. 2002. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 6. Structure-activity studies of orally bioavailable, 2-pyridone-containing peptidomimetics. J. Med. Chem. 451607-1623. [DOI] [PubMed] [Google Scholar]
- 9.Frese, M., K. Barth, A. Kaul, V. Lohmann, V. Schwärzle, and R. Bartenschlager. 2003. Hepatitis C virus RNA replication is resistant to tumor necrosis factor-α. J. Gen. Virol. 841253-1259. [DOI] [PubMed] [Google Scholar]
- 10.Fukushi, S., M. Okada, J. Stahl, T. Kageyama, F. B. Hoshino, and K. Katayama. 2001. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem. 27620824-20826. [DOI] [PubMed] [Google Scholar]
- 11.Futaki, S., T. Suzuki, W. Ohashi, T. Yagami, S. Tanaka, K. Ueda, and Y. Sugiura. 2001. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 2765836-5840. [DOI] [PubMed] [Google Scholar]
- 12.Goodier, J. L., H. Fan, and R. J. Maraia. 1997. A carboxy-terminal basic region controls RNA polymerase III transcription factor activity of human La protein. Mol. Cell. Biol. 175823-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellen, C. U. T., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 151593-1612. [DOI] [PubMed] [Google Scholar]
- 14.Huang, Z., M. G. Murray, and J. A. Secrist. 2006. Recent development of therapeutics for chronic HCV infection. Antiviral Res. 71351-362. [DOI] [PubMed] [Google Scholar]
- 15.Hutchinson, E. G., and J. M. Thornton. 1994. A revised set of potentials for beta-turn formation in proteins. Protein Sci. 32207-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izumi, R. E., S. Das, B. Barat, S. Raychaudhuri, and A. Dasgupta. 2004. A peptide from autoantigen La blocks poliovirus and hepatitis C virus cap-independent translation and reveals a single tyrosine critical for La RNA binding and translation stimulation. J. Virol. 783763-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacks, A., J. Babon, G. Kelly, I. Manolaridis, P. D. Cary, S. Curry, and M. R. Conte. 2003. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure 11833-843. [DOI] [PubMed] [Google Scholar]
- 18.Kieft, J. S., K. Zhou, R. Jubin, and J. A. Doudna. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7194-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, Y., and J. H. Prestegard. 1989. Measurement of vicinal couplings from cross peaks in COSY spectra. J. Magn. Reson. 849-13. [Google Scholar]
- 20.Maraia, R. J., and R. V. Intine. 2001. Recognition of nascent RNA by the human La antigen: conserved and divergent features of structure and function. Mol. Cell. Biol. 21367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 673798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohndorf, U. M., C. Steegborn, R. Knijff, and P. Sondermann. 2001. Contributions of the individual domains in human La protein to its RNA 3′-end binding activity. J. Biol. Chem. 27627188-27196. [DOI] [PubMed] [Google Scholar]
- 23.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. T. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1267-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pudi, R., S. Abhiman, N. Srinivasan, and S. Das. 2003. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by specific interaction of independent regions of human La autoantigen. J. Biol. Chem. 27812231-12240. [DOI] [PubMed] [Google Scholar]
- 25.Pudi, R., S. S. Ramamurthy, and S. Das. 2005. A peptide derived from RNA recognition motif 2 of human la protein binds to hepatitis C virus internal ribosome entry site, prevents ribosomal assembly, and inhibits internal initiation of translation J. Virol. 799842-9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pudi, R., P. Srinivasan, and S. Das. 2004. La protein binding at the GCAC site near the initiator AUG facilitates the ribosomal assembly on the hepatitis C virus RNA to influence internal ribosome entry site-mediated translation. J. Biol. Chem. 27929879-29888. [DOI] [PubMed] [Google Scholar]
- 27.Steinkuhler, C., G. Biasiol, M. Brunetti, A. Urbani, U. Koch, R. Cortese, A. Pessi, and R. De Francesco. 1998. Product inhibition of the hepatitis C virus NS3 protease Biochemistry 238899-8905. [DOI] [PubMed] [Google Scholar]
- 28.Tan, P. L., M. Blumenstein, S. Yeoman, and J. D. Watson. 1989. B cell lymphokines in human systemic lupus erythematosus. Ann. Rheum Dis. 48941-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teplova, M., Y. R. Yuan, A. T. Phan, L. Malinina, S. Ilin, A. Teplov, and D. J. Patel. 2006. Structural basis for recognition and sequestration of UUU(OH) 3′ termini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol. Cell 2175-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilmot, C. M., and J. M. Thornton. 1988. Analysis and prediction of the different types of beta-turn in proteins J. Mol. Biol. 5221-232. [DOI] [PubMed] [Google Scholar]
- 31.Wolin, S. L., and T. Cedervall. 2002. The La protein. Annu. Rev. Biochem. 71375-403. [DOI] [PubMed] [Google Scholar]
- 32.Yao, N., P. Reichert, S. S. Taremi, W. W. Prosise, and P. C. Weber. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure 151353-1363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.