Abstract
Cymbidium ringspot virus (CymRSV) satellite RNA (satRNA) is a parasitic subviral RNA replicon that replicates and accumulates at the cost of its helper virus. This 621-nucleotide (nt) satRNA species has no sequence similarity to the helper virus, except for a 51-nt-long region termed the helper-satellite homology (HSH) region, which is essential for satRNA replication. We show that the accumulation of satRNA strongly depends on temperature and on the presence of the helper virus p19 silencing suppressor protein, suggesting that RNA silencing plays a crucial role in satRNA accumulation. We also demonstrate that another member of the Tombusvirus genus, Carnation Italian ringspot virus (CIRV), supports satRNA accumulation at a higher level than CymRSV. Our results suggest that short interfering RNA (siRNA) derived from CymRSV targets satRNA more efficiently than siRNA from CIRV, possibly because of the higher sequence similarity between the HSH regions of the helper and CIRV satRNAs. RNA silencing sensor RNA carrying the putative satRNA target site in the HSH region was efficiently cleaved when transiently expressed in CymRSV-infected plants but not in CIRV-infected plants. Strikingly, replacing the CymRSV HSH box2 sequence with that of CIRV restores satRNA accumulation both at 24°C and in the absence of the p19 suppressor protein. These findings demonstrate the extraordinary adaptation of this virus to its host in terms of harnessing the antiviral silencing response of the plant to control the virus parasite satRNA.
Satellite RNAs (satRNAs) are linear or circular RNAs that require a helper virus to supply all trans-acting factors for replication. satRNAs have little or no similarity to the sequence of the helper virus, and they are not required for the accumulation of helper virus. There are two classes of satRNAs: large RNAs that generally encode a single nonstructural protein and small RNAs with no functional coding capacity. Several satRNAs are associated with helper viruses from different plant virus families (27).
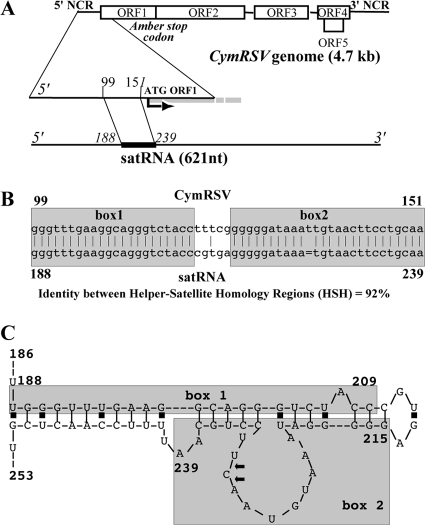
satRNAs also are associated with tombusviruses (7, 10, 23, 25). Three tombusvirus satRNAs have been identified so far: satRNAs B1 and B10 are associated with Tomato bushy stunt virus (7), and satRNA is associated with Cymbidium ringspot virus (CymRSV). CymRSV satRNA was the first tombusvirus satRNA that was discovered (14, 23) and also is the best characterized. CymRSV satRNA is a 621-nucleotide (nt) RNA lacking any messenger activity in vivo and in vitro, and its replication and accumulation require trans-acting factors provided by the helper virus and the host (6, 10, 23). During the replication process, double-stranded RNA (dsRNA) replicative forms often are present at high levels in the infected cells (10). There is no sequence similarity between satRNA and the helper virus genome except in a small region, 51 nt in length, which has ∼90% similarity to the CymRSV 5′-end untranslated region (also called the helper-satellite homology region, or HSH) (23). This region is located at positions 99 to 151 in the CymRSV genome and at positions 188 to 239 in the satRNA sequence (Fig. 1A and B). satRNA contains cis-acting RNA domains essential for replication; these signals are located throughout the entire satRNA sequence, including the HSH region (8, 9).
FIG. 1.
Schematic representation of the CymRSV-satRNA homology regions. satRNA is represented by a thin black line, except for the HSH region (thick black line) located between positions 188 and 239, and is referred to as the satRNA genome. (A) The corresponding region in CymRSV is located between positions 99 and 151 in the 5′-end virus noncoding region (NCR). (B) The HSH regions are indicated by pale gray boxes. (C) Secondary structure of the satRNA HSH region. Viral and satellite noncoding regions are indicated by thick black lines, while viral ORFs are indicated by white open boxes. Arrows indicate the cleavage sites.
Full-length satRNA transcribed in vitro can replicate, accumulate, and spread systemically in Nicotiana benthamiana plants when coinoculated with helper virus. In addition to the original helper virus (CymRSV), satRNA replication can be supported by other members of the Tombusvirus genus as well, including different strains of Tomato bushy stunt virus, Carnation Italian ringspot virus (CIRV), and Cucumber Bulgarian latent virus (M. Russo, personal communication). Surprisingly, the heterologous helper viruses seem to support the satRNA more efficiently than the natural helper virus (J. Burgyán, unpublished results).
In addition to the satRNA, other subviral entities such as defective interfering RNAs (DI RNAs) have been found to be associated with CymRSV. DI RNAs are shortened forms of viral genomes that are defective for all viral genes involved in transmission, replication, and encapsidation. The presence of DI RNAs in virus-infected plants dramatically suppresses virus accumulation and leads to persistent attenuated symptoms. To date, there is no single, generally accepted mechanism that accounts for DI RNA interference with the accumulation of helper viruses and the corresponding symptom attenuation. Mechanisms for DI RNA symptom modulation have been proposed that involve competition with the helper virus for trans-acting replication factors, specific interaction with virus-encoded products, or the activation of posttranscriptional gene silencing (26-28).
In contrast to DI RNA, satRNA does not seem to be involved in either virus accumulation or symptom modulation (J. Burgyán and M. Russo, personal communication). Interestingly, satRNA replication suppresses DI RNA accumulation in N. benthamiana transgenic plants expressing satRNA (24). In contrast, the simultaneous inoculation of DI RNA and satRNA with the helper CymRSV results in a decrease or elimination of satRNA by short DI RNA due to competition (J. Burgyán, L. Rubino, and M. Russo, unpublished data). Importantly, both DI RNA and satRNA contain the conserved HSH sequence that is indispensable for the replication of both satRNA and DI RNA. We have previously shown that DI RNAs are strong inducers of RNA silencing, which plays an important role in the evolution and accumulation of DI RNA (28). This observation raised the question of whether RNA silencing also has an important role in satRNA accumulation. RNA silencing is more active at higher temperatures (29), so the idea that RNA silencing affects satRNA accumulation was supported by an earlier report that high temperatures (>26°C) strongly inhibited satRNA accumulation (6). The role of RNA silencing in satRNA-virus interaction also has been reported very recently for the Turnip crinkle virus system (17). These findings prompted us to analyze the role of RNA silencing in satRNA replication and accumulation.
RNA silencing is a highly conserved gene inactivation mechanism in eukaryotes, and endogenous silencing pathways are important in gene regulation at the transcriptional, RNA stability, and translational levels (4). Although it operates through diverse pathways, RNA silencing relies on a set of core processes. RNA silencing is triggered by highly structured single-stranded RNA or dsRNA molecules that are processed into 21 to 24-nt short interfering RNA (siRNA) or microRNA (miRNA) duplexes by the RNase III-type DICER enzymes. These short RNAs then are incorporated into a ribonucleoprotein complex termed the RNA-induced silencing complex (RISC) (30).
In higher plants, the AGO1 protein is the slicer component of RISC (22), and AGO1 recruits small silencing-related RNAs such as miRNAs, trans-acting siRNAs, transgene-derived siRNAs (3), and virus-derived siRNAs (21, 34). Virus-induced RNA silencing is triggered by dsRNA intermediates of cytoplasmically replicating viruses, by the RDR1- or RDR6-dependent formation of dsRNA, or by structured regions of viral RNAs (13, 20). This silencing-based antiviral response by the plant leads to the sequence-specific degradation of viral RNA (21); in turn, this often results in attenuated viral symptoms and low virus titer.
In this study, we describe how the invading virus can harness the silencing machinery of the host plant to control the accumulation of virus parasite satRNA, which is associated with tombusvirus infection. We found that the suppression of RNA silencing at a low temperature or in the presence of a silencing suppressor protein markedly facilitates the accumulation of satRNA. Using sensor constructs with the HSH sequence conserved in both helper virus and satRNA, we show that this sequence is the target site of helper virus-triggered RNA silencing. In addition, the accumulation of viral siRNA, which can guide the RISC to target satRNA, also was detected. The role of RNA silencing in satRNA accumulation was further supported by the observation that CIRV, which is a more efficient helper for satRNA, has altered nucleotides in its HSH sequence and was unable to generate satRNA-targeting RISC. Moreover, the introduction of altered nucleotides into the HSH region of the CymRSV helper dramatically enhanced satRNA accumulation when it was coinoculated with the modified helper virus.
MATERIALS AND METHODS
Plant material, in vitro transcription, and CymRSV mutant clones.
Plasmids containing CymRSV, Cym19stop, CIRV, CIRV19stop, and satRNA cDNA clones have been described previously (5, 6, 12, 24). The p19-encoding open reading frames (ORFs) were mutated in both Cym19stop and CIRV19stop constructs; therefore, these mutant viruses were not able to express p19 protein. All constructs were linearized with SmaI, ethanol precipitated, and transcribed in vitro using T7 RNA polymerase. N. benthamiana plants were inoculated by helper and satRNA in vitro transcripts as described previously (24).
The Cym19stop-HSH-mut and CymRSV-HSH-mut constructs were obtained by introducing C145T and T147A mutations using the QuikChange site-directed mutagenesis kit (Stratagene) by following the manufacturer's instructions.
Protoplasts were prepared from fully expanded leaves, and transfection was performed as described previously (11). Protoplasts were transfected with full-length CIRV and CymRSV genomic RNA and with satRNA in vitro transcripts and were kept for 24 h at 24°C under fluorescent lighting.
Agrobacterium tumefaciens infiltration and GFP constructs.
The agroinfiltration of Cym19stop- and CIRV19stop-infected plants was performed as described previously (21). Tobacco etch virus HC-Pro in a binary construct was described previously (16), and the green fluorescent protein (GFP)-HSH launching binary vector was obtained by excising the 133-bp MscI/NaeI DNA fragment from a satRNA cDNA clone (9) and cloning it into an SmaI-linearized 35S-GFP plasmid (21) (see Fig. 4A). GFP-box2(+) and GFP-box2(−) were obtained by the site-directed mutagenesis of the 35S-GFP plasmid (21) using the oligonucleotides HSHbox2(+)P (5′-GAGCTCGAATTTCCCCGATAAATGTAACTTCCTGCAAGGGATCGTTCAAA-3′) and HSHbox2(+)N (5′-GTTTGAACGATCCCTTGCAGGAAGTTACATTTATCGGGGAAATTCGAGCTC-3′). GFP HSHbox2(−) was constructed using the oligonucleotides HSHbox2(−)P (5′-GAGCTCGAATTTCCCCTTGCAGGAAGTTACATTTATCGGGATCGTTCAAAC-3′) and HSHbox2(−)N (5′-GTTTGAACGATCCCGATAAATGTAACTTCCTGCAAGGGGAAATTCGAGCTC-3′). The extra bases introduced into the 35S-GFP plasmid are underlined.
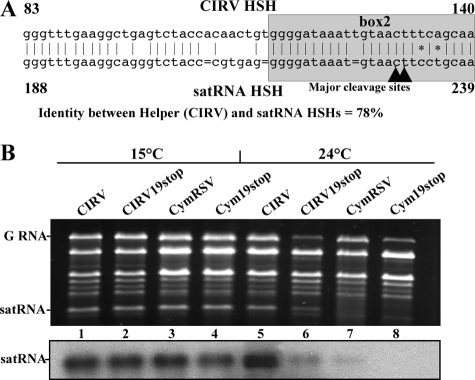
FIG. 4.
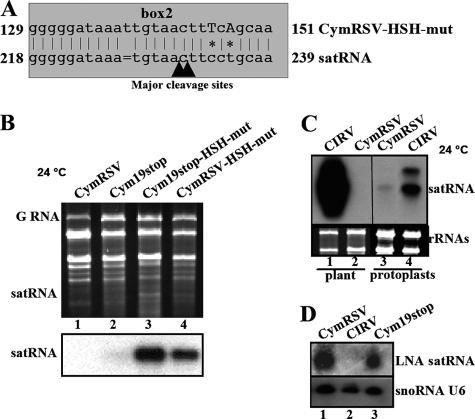
Replication of satRNA in the presence of CIRV as the virus helper. (A) Sequence alignment of CIRV and satRNA HSH regions. Asterisks indicate the two mismatches, and the bars show the gaps in the satRNA HSH sequence; the major cleavage sites in the corresponding satRNA HSH region are indicated by arrowheads. (B) Accumulation of satRNAs in the presence of different helper viruses at different temperatures. RNA samples from plants coinoculated with satRNA and CIRV, CIRV19stop, CymRSV, and CymRSV19stop were analyzed in ethidium bromide-stained agarose gels (upper panel) and in Northern blot analyses of the satRNA accumulation in the same samples (lower panel). GRNA, genomic RNA.
RNA analysis.
RNA was extracted from plants and protoplasts and analyzed as described previously (28). RNA in nondenaturing 1.2% agarose gels was stained with ethidium bromide, and RNA in formaldehyde-denaturing gels was transferred to nylon membranes and detected using specific probes. The probes complementary to the 5′-end satRNA from nt 1 to 188 and 3′-end satRNA from nt 300 to 621 were amplified by PCR using a 5′-fused oligonucleotide containing the T7 RNA polymerase promoter sequence. The transcribed probes were used to detect plus-stranded satRNA. GFP from nt 608 to 750 was PCR amplified with a reverse oligonucleotide 5′ fused by the T7 RNA polymerase promoter and was used to detect GFP mRNA. A probe against the 0.3-kb 3′-noncoding region (21) was used to detect plus-stranded CymRSV RNA. Riboprobes were generated by in vitro transcription with the StripEZ RNA kit (Ambion) in the presence of [α-32P]ATP. The locked nucleic acid (LNA) probe, 5′-GAT AAA TGT AAC TTC CTG CAA-3′, was from Exiqon (Denmark) and was used to detect the CymRSV-derived satRNA targeting siRNA. The DNA oligonucleotide probe was complementary to small nuclear RNA U6. The secondary structures of RNA sequences were predicted using mfold version 3.2 (35). The relative amounts of virus helper RNA, satRNA, or rRNA accumulation were estimated by Quantity One software (Bio-Rad).
3′-End rapid amplification of cDNA ends (3′-RACE) and sequence analysis of satRNA and cleaved GFP-HSH sensors.
Total RNA (5 μg) was ligated with a 3′-end adapter oligonucleotide (20). Ligated RNA was reverse transcribed using the specific oligonucleotide that was complementary to the 3′-end adapter. It was amplified by PCR using the oligonucleotide that was used for the reverse transcription as a reverse primer; the forward primer either was an oligonucleotide homologue of nt 1 to 18 of the satRNA cDNA clone (in the case of the 3′-end analysis of satRNA) or an oligonucleotide homologue of nt 608 to 630 of the GFP ORF (in the case of the 3′-end analysis of GFP-HSH cleavage products). The amplified products then were cloned into SmaI-linearized pUC18 and sequenced.
RESULTS
Accumulation of satRNA in the presence of CymRSV is temperature and p19 dependent.
It has been shown previously that the accumulation of satRNA is often below the detection level when inoculated with CymRSV as a helper virus in N. benthamiana plants that are grown at a high temperature (6). To better understand the effect of temperature on satRNA accumulation, we analyzed this phenomenon in greater detail. N. benthamiana plants with six to eight leaves were coinoculated with CymRSV and satRNA in vitro transcripts and were grown at 28, 24, or 20°C. RNA was extracted from the systemically infected leaves when symptoms appeared (6 to 10 days postinfection [dpi]). As expected, the growing temperature dramatically affected the level of the accumulation of the helper virus due to the activation of RNA silencing (29). The level of viral RNA in the plants grown at 28°C was barely detectable by Northern blotting, while plants grown at 24 or 20°C had markedly higher levels of viral RNA (Fig. 2A). The accumulation of satRNA showed an even stronger dependence on temperature: at 24°C, the level of satRNA was just above the detection limit, but at 20°C there were high levels of satRNA (Fig. 2A). These results suggested that the inhibition of satRNA accumulation and/or replication is the consequence of activated RNA silencing.
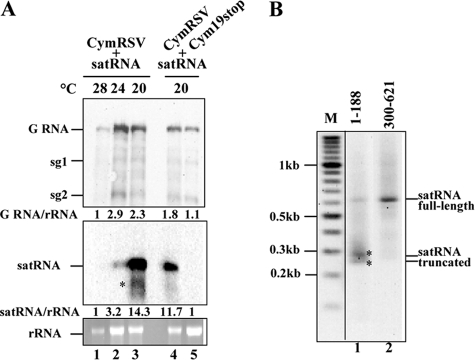
FIG. 2.
Accumulation of satRNA in the presence of helper viruses. (A) Temperature- and p19-dependent accumulation of satRNA. RNA preparations from N. benthamiana plants coinoculated with satRNA and CymRSV or Cym19stop and incubated at different temperatures (28, 24, and 20°C) were submitted to Northern blot analysis with probes specifically raised against the viral RNA (upper panel) or the 5′ part of satRNA (middle panel). Ethidium bromide-stained 25S rRNA was used as a loading control (lower panel). G, genomic; sg, subgenomic. (B) Southern blot analysis of satRNA 3′-RACE products obtained from total RNA extractions from plants infected with CymRSV and satRNA and incubated at 20°C. Probes used for hybridization specifically recognized the indicated regions of satRNA. M indicates the lane containing the molecular size marker. Asterisks indicate the truncated forms of satRNA.
To better clarify the role of RNA silencing in this phenomenon, we compared the helper efficiency of wild-type CymRSV to that of a helper that lacked the silencing suppressor p19 (Cym19stop). Plants were coinoculated with satRNA and CymRSV or Cym19stop and kept at 20°C. RNA was extracted from the first leaves that showed systemic infection symptoms (10 dpi). The accumulation of the genomic RNA of CymRSV and Cym19stop was similar (Fig. 2, upper); however, satRNA was detected only in the presence of the wild-type helper (Fig. 2, lower), which expresses p19. The lack of satRNA accumulation using the Cym19stop mutant helper virus further supports the idea that the RNA silencing mechanism is involved in this phenomenon.
RNA silencing induced by helper virus targets satRNA.
The coinfection of plants with CymRSV and satRNA often, but not always, resulted in the appearance of satRNA-specific RNA molecules shorter than full-length satRNA (Fig. 2A, lane 3). We hypothesized that these short RNA molecules could be the product of cleaved satRNA targeted by satRNA-specific siRNA-programmed RISCs. To clarify this, 3′-RACE analysis was performed on the sample shown in lane 3 of Fig. 2A. The amplified products were submitted to Southern blot analysis using two specific riboprobes targeting the 5′ and the 3′ ends of the satRNA (nt 1 to 188 and nt 300 to 621, respectively). The amplified 650-nt-long product corresponded to full-length satRNA (Fig. 2B, lanes 1 and 2), which was detected using either the 5′-specific or the 3′-specific probe. Interestingly, the 5′-end-specific probe also detected shorter molecules containing satRNA sequences; these corresponded to amplified products in the 250- to 290-bp range (Fig. 2B, lane 1). The low-molecular-weight PCR products were cloned into the pUC plasmid and sequenced, and the sequences indicated that the 3′ end of these shorter-than-unit-length satRNA-specific RNAs was part of the HSH region. These shorter RNA species could be cleavage products of full-length satRNA. Alternatively, they could be products of prematurely terminated satRNA transcription. To differentiate between these possibilities, a 133-nt satRNA fragment (from positions 173 to 306) containing the HSH region was cloned into a binary vector downstream of the GFP reporter ORF (GFP-HSH) (21) (Fig. 3A). The sensor plasmid was delivered into CymRSV19stop-infected N. benthamiana leaves by Agrobacterium-mediated transient expression. In order to prevent silencing by the sense transgene (15) and also to prevent transitive gene silencing (18, 31), the strong silencing suppressor HC-Pro from Tobacco etch virus was codelivered with GFP-HSH. Note that HC-Pro does not suppress the cleavage activity of siRNA-programmed RISC, because it has been shown that it inhibits RNA silencing by double-stranded siRNA sequestration before RISC assembly (16). The decreased expression of GFP from the GFP-HSH sensor was observed 3 days after agroinfiltration in Cym19stop-infected plants but not in noninfected or CIRV19stop-infected plants (Fig. 3B). GFP-HSH sensor mRNA levels also were analyzed in RNA extracts 3 days after agroinfiltration, and Northern blot analysis showed that the level of GFP-HSH sensor mRNA was reduced in CIRV19stop-infected plants (Fig. 3C). The level of GFP-HSH sensor mRNA was below the detection limit in Cym19stop-infected plants, demonstrating the efficiency of sensor mRNA degradation mediated by helper virus-activated RNA silencing.
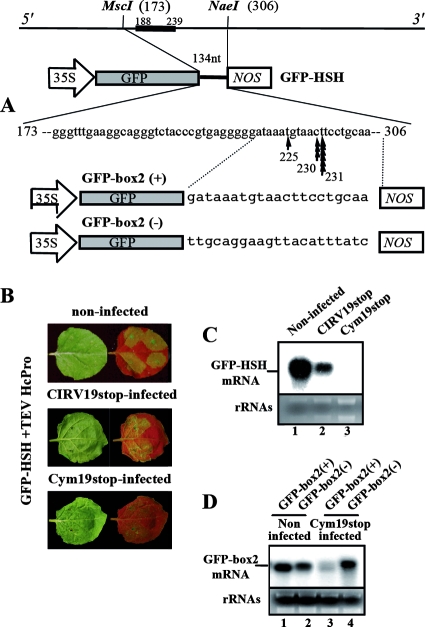
FIG. 3.
CymRSV-mediated targeting of GFP-HSH sensor mRNAs. (A) Schematic representation of GFP-HSH sensor construct and the cleavage sites in sensor mRNA when it is expressed in CymRSV-infected plants. The arrows indicate the site of identified cleavages in the sensor mRNA. The numbers below the arrows indicate the cleavage sites in the satRNA genome sequence. Sensor constructs containing the two orientations of HSH box2 are indicated as GFP-box2(+) and GFP-box2(−), respectively. NOS, nopalin synthase. (B) Expression of the GFP-HSH sensor by agroinfiltration in N. benthamiana plants. GFP-HSH mRNA was expressed in the presence of Tobacco etch virus HC-Pro into noninfected, CIRV19stop-infected, and Cym19stop-infected plants. Leaves were viewed at 3 days after inoculation under visible and long-wavelength UV illumination. (C) Northern blot analysis of RNA samples extracted from plants shown in panel B. RNA samples were extracted from agroinfiltrated leaves at 2 dpi and subjected to RNA blot analysis using a [α-P32]ATP-labeled probe raised against the GFP ORF. The 25S and 16S RNAs were used as a loading control for total RNA. (D) Northern blot analysis of GFP-box2(+) and GFP-box2(−) expression in Cym19stop-infected N. benthamiana plants. RNA samples were extracted and analyzed as described in the legend to panel C.
Based on sequence homology between the satRNA and helper genome, we expected two putative target sites in the common HSH region (termed box1 and box2) (Fig. 1B). In box1, the sequence homology is 100%, and in box2, the helper genome contains one extra base (Fig. 1B). There is a short (5-nt) nonhomologous stretch between the two homologous boxes. As expected, the 3′-RACE analysis of the cleaved sensor construct confirmed our prediction: all sequenced clones showed the cleavage sites in the highly homologous box2, and 10 of 11 clones showed a cleavage product at positions 230 and 231 of the satRNA sequence. This indicates a very narrow hot spot for cleavage (Fig. 3A). These results strongly suggest that the low level of satRNA accumulation is the consequence of helper virus-induced satRNA silencing. Surprisingly, we did not find cleavage in box1 in spite of 100% homology between satRNA and CymRSV in this region. However, the predicted secondary structure of the HSH region of satRNA may explain this observation. The identified target sites in box2 are located in a predicted single-stranded region, while the nucleotides in the box1 sequence are in a predicted base-paired structure (Fig. 1C) that could make this region more resistant to RISC-mediated cleavage (1). To better localize the putative target site in box2, a new sensor construct containing only the last 21 nt of box2 (GFP-box2) (Fig. 3A) was generated in both plus and minus orientations and expressed in the recovered leaves of Cym19stop-infected plants. The mRNA of GFP-box2(+) and GFP-box2(−) accumulated at high levels in noninfected N. benthamiana; however, GFP-box(+) accumulated at much lower levels in the recovered leaves of plants previously infected by Cym19stop (Fig. 3D). This finding suggests that the satRNA box2(+) sequence is an efficient target site for Cym19stop-induced RNA silencing. Surprisingly, the level of GFP-box(−) sensor RNA was almost unaffected, suggesting that the complementary strand of satRNA is not an efficient target of helper virus-induced RNA silencing.
CIRV and CIRV19stop support satRNA replication more efficiently than CymRSV and the p19-deficient mutant.
The Tombusvirus genus includes many species that share replication, genome organization, and expression mechanisms as well as sequence similarity (26). In addition, all of the tombusvirus species can support the replication of satRNA (M. Russo, personal communication). Our results suggested that one antisense viral siRNA generated from the viral HSH region of CymRSV cleaved either satRNA or an artificial mRNA containing the satRNA box2 sequence. This phenomenon requires that the viral siRNA and the target sequence are perfectly or nearly perfectly complementary (2), which is the case for CymRSV and satRNA (Fig. 1B). CIRV is a tombusvirus that is an efficient helper of satRNA replication (Burgyán, unpublished), so we analyzed the HSH sequences of satRNA and CIRV. The homology is only 78%, compared to 92% homology for satRNA and CymRSV, and there is no perfectly homologous 21-nt stretch in the HSH region (Fig. 4A). Moreover, two mismatches are located just downstream of the putative target sites (nt 230 to 231) (Fig. 4A). The presence of the mismatches in that region could destroy the siRNA target site generated from the CIRV genome and may explain why CIRV is an efficient helper for satRNA accumulation. In order to test this hypothesis, CIRV and CIRV19stop (33) were compared to CymRSV and Cym19stop as helpers for satRNA accumulation at different temperatures.
As previously reported by Szittya et al. (29), virus-induced RNA silencing is almost completely abolished at 15°C. The helper viruses supplied with p19 (i.e., CIRV and CymRSV) induce strong symptoms that quickly result in plant death at 15°C. In RNA extracted from these plants, the ratio of the helper virus to the accumulated satRNA was estimated to be 1:1 and 2:1 for CIRV and CymRSV, respectively (Fig. 4B, lanes 1 and 3). Moreover, the mutant viruses lacking p19 also were able to support the replication of satRNA to achieve the same ratios (Fig. 5B, lanes 2 and 4). Conversely, at 24°C, a temperature at which RNA silencing is enhanced (29), the accumulation of satRNA in the presence of CymRSV was just above the detection limit, and the suppressor mutant Cym19stop helper was not able to support the accumulation of satRNA at a detectable level (Fig. 4B, lanes 7 and 8). However, in the presence of CIRV, the satRNA accumulated at high levels, and the suppressor mutant CIRV19stop also supported the accumulation of satRNA, although less efficiently (Fig. 4B, lanes 5 and 6). These findings further support our hypothesis that CymRSV-induced RNA silencing targets subviral satRNA, since the inhibition of RNA silencing by low temperature or by the presence of the p19 silencing suppressor enhances satRNA accumulation. Moreover, when the helper virus is CIRV, a virus that contains mismatches in the common HSH region, the effect of RNA silencing is much less pronounced.
FIG. 5.
satRNA accumulation in the presence of wild-type-infected and mutant helper virus-infected plant and protoplasts and the detection of satRNA targeting viral siRNAs. (A) Alignment of the HSH box2 sequences after the introduced mutations in positions 145 and 147, which were introduced in both CymRSV and Cym19stop (CymRSV-HSH-mut and Cym19stop-HSH-mut, respectively). Capital letters in boldface indicate the newly introduced bases; asterisks indicate the newly generated mismatches with the corresponding satRNA region. (B) Accumulation of satRNA in the presence of different helper viruses. RNA samples extracted from plants coinoculated with satRNA in the presence of the indicated helper virus were analyzed in an ethidium bromide-stained 1.2% agarose gel (upper panel) and by Northern blot analysis (lower panel). GRNA, genomic RNA. (C) Northern blot analysis of the accumulation of satRNA in N. benthamiana plant and protoplasts coinoculated with satRNA and helper viruses. (D) Accumulation of satRNA targeting viral siRNA in different helper virus-infected plants. Small RNAs were separated in 15% denaturing polyacrylamide gel electrophoresis, blotted, and hybridized with a 32P-labeled LNA probe designed against the putative antisense siRNA at nt 130 to 151 of the CymRSV genome. As a loading control, the accumulated small nucleolar RNA U6 detected by a specific 32P-labeled oligonucleotide-DNA probe.
Replacement of CymRSV HSH box2 sequence with CIRV HSH box2 sequence restores satRNA replication at 24°C and in the absence of p19.
The replication and accumulation of satRNA in the presence of different helper viruses could be influenced by factors or mechanisms other than RNA silencing. We wished to determine whether the HSH sequences were solely responsible for the observed differences in satRNA accumulation with the two helpers (CymRSV and CIRV). In order to determine this, we replaced the HSH box2 sequences of CymRSV and Cym19stop with the CIRV sequence to obtain the constructs CymRSV-HSH-mut and Cym19stop-HSH-mut (Fig. 5A). Importantly, the mutations in positions 145 and 147 of the CymRSV genome are predicted to disrupt the putative base pairing between the helper viral siRNA and the satRNA target sequence close to the presumed cleavage site in the HSH region of satRNA (Fig. 5A). The mutations in the helper virus genomes did not affect the replication or accumulation of viral genomes significantly compared to that of wild-type viruses (Fig. 5B). Moreover, the sequence analysis of the mutant progeny did not show any reversion to the wild-type sequence (data not shown). satRNA coinoculated with CymRSV-HSH-mut and Cym19stop-HSH-mut into N. benthamiana plants accumulated at a high level (Fig. 5B, lanes 3 and 4), while the helper viruses carrying the wild-type HSH box2 sequences did not support the accumulation of satRNA at 24°C (Fig. 5B, lanes 1 and 2).
The accumulation of satRNA also could be influenced by cell-to-cell movement, which is facilitated by the movement protein provided by the helper virus. Therefore, we also tested the efficiency of the two helpers, CymRSV and CIRV, in protoplasts in order to exclude the effects of movement on satRNA accumulation. N. benthamiana protoplasts were prepared and infected by satRNA and helper virus in vitro transcripts, and the satRNA accumulation was analyzed by Northern blotting. The results demonstrated that, similarly to our observations in plants, satRNA accumulation was much higher in the presence of CIRV than CymRSV, indicating that the helper-dependent accumulation of satRNA is not a consequence of altered cell-to-cell movement (Fig. 5C).
These findings further supported the idea that helper-activated RNA silencing controls the accumulation of satRNA. If this is true, there should be virus-derived siRNAs with perfect or near-perfect complementarity to the putative HSH box2 target site. The accumulation of HSH box2-targeting siRNA in CymRSV-infected plants was demonstrated using an LNA oligonucleotide probe with the satRNA HSH box2 sequence (Fig. 5D). We have shown previously that LNA probes are able to detect small RNAs very specifically, because they anneal to the cRNA sequence with higher stability (32).
DISCUSSION
The satRNA of CymRSV is a parasitic molecule that replicates with the aid of a helper virus. An optimal balance between the helper and the parasite satRNA is required for the long-term coexistence of the two molecules. This balance could be controlled by the availability of proteins provided by the helper, which are required for viral RNA replication. satRNA accumulates very efficiently in the helper virus-infected cells, demonstrating that it competes very successfully with the helper genome for replication factors. Because satRNA needs helper-encoded proteins, satRNA in the virus-infected plant cell negatively regulates helper genome accumulation without completely outcompeting the helper virus. However, the ratio between satRNA and helper genome accumulation can vary significantly depending on growth conditions (see, for example, Fig. 4B). These findings suggest that other mechanisms also play a role in maintaining a balance. We hypothesized that helper virus-induced gene silencing could be such a mechanism controlling the accumulation of satRNA by targeting the HSH sequence in the satRNA genome, a sequence that is absolutely required for replication (8, 26).
satRNA accumulates more efficiently when RNA silencing is inhibited by low temperature or in the presence of silencing suppressor protein.
We have shown previously that virus-induced RNA silencing is strongly inhibited at 15°C (29), and we have also observed that satRNA accumulates more efficiently at a lower temperature (6). The current investigation into the effect of temperature on satRNA clearly demonstrated that high temperatures (28 to 24°C), which enhance the activity of RNA silencing, strongly inhibit the accumulation of satRNA in the presence of the natural helper CymRSV. An important role for RNA silencing in satRNA accumulation was further supported by the observation that the silencing suppressor mutant Cym19stop helper failed to support satRNA accumulation at a detectable level at 24°C. However, when the heterologous helper CIRV was used, the accumulation of satRNA was not influenced dramatically by temperature, confirming that satRNA replication itself was not affected significantly by temperature. Importantly, satRNA accumulation was easily detectable, even at 24°C, when it was coinoculated with the p19 suppressor mutant CIRV19stop, although in this case, the incorporation of virus-derived siRNAs into RISC is not inhibited by the p19 silencing suppressor.
Helper virus-induced RNA silencing cleaves satRNA, and the cleavage depends on the homology of the HSH regions of satRNA and its helper.
This study showed that the accumulation of satRNA molecules was downregulated in CymRSV- or Cym19stop-infected plants. In the latter case, satRNA was able to accumulate at detectable levels only at a lower temperature, suggesting that RNA silencing-mediated cleavage was very efficient at normal temperatures. To better understand the molecular basis of this observation, we used a sensor construct (GFP-HSH) with the HSH region of the satRNA molecule. The sensor construct was cleaved in the HSH region, and the sequencing of the cleavage products indicated that the cleavage site was in the second homology region (box2) of the HSH sequence (Fig. 1B). Interestingly, we never observed cleavage in the first homology region (box1) of HSH in spite of 100% homology between the CymRSV HSH and satRNA HSH regions. A closer inspection of the putative secondary structure of the satRNA HSH region could explain this surprising observation. Although there are a few mismatches between the helper and satRNA in the box2 region, this sequence is located in a predicted single-stranded region of the folded satRNA (Fig. 1C). The RISC-mediated cleavage of target RNA is more efficient when the target sequence is located in a single-stranded or weakly structured region (1, 21). Thus, viral siRNA-programmed RISC has better access to box2 than to box1. The sensor mRNA, containing only the last 21 nt of the box2 target sequence, also was cleaved efficiently, which helped to identify the target sequence in the HSH region of satRNA. Knowing the precise target sequence allowed us to predict the sequence of the viral siRNA that targets the satRNA. Indeed, this viral siRNA was detected using an LNA probe containing the target sequence of box2. As expected, the satRNA-targeting siRNA was present in CymRSV-infected plants but not in CIRV-infected plants (Fig. 5D). The observed high efficiency of single siRNAs raises the possibility to generate very effective artificial siRNAs for practical purposes. However, the accessibility of the target site for the specific siRNA-containing RISC should be studied experimentally.
The mutational analysis of the box2 sequence of the CymRSV HSH region also supported a role for RNA silencing in controlling satRNA accumulation. Introducing two mismatches (the same mismatches as those in the CIRV sequence) into the box2 sequence to create the CymRSV-HSH-mut helper significantly enhanced the level of satRNA after coinoculation. We also found that the siRNA, which specifically targets the box2 sequence of satRNA, was undetectable in CymRSV HSH-mut-infected plants (data not shown). Moreover, these two mismatches are located just downstream of the cleavage site. In other words, the virus-derived siRNA, which should target the satRNA box2 sequence, has the mismatches in the 5′ half of the siRNA that is incorporated into the RISC. The lack of cleavage is in line with what has been found for miRNAs in plants. Indeed, Parizzotto et al. (2004) (22a) showed that mi171 activity displays unequal tolerance to a 5′-symmetrical mismatch and a 3′-symmetrical mismatch.
These findings further highlight the crucial role of the box2 sequence in satRNA helper function via the activation of RNA silencing. In addition, these results demonstrate that the sequence differences in other parts of the viral genomes of CymRSV and CIRV do not play an important role in the level of accumulation of satRNA.
Helper virus controls satRNA accumulation at the cellular level.
The accumulation of viral RNA, including subviral parasite satRNA, in virus-infected plant tissues also could be influenced by cell-to-cell movement. Cell-to-cell movement is facilitated by the movement protein, which is provided by the helper virus. In order to better understand the effect of the helper virus in satRNA accumulation, we analyzed helper virus function in virus- and satRNA-infected protoplasts, which lack the cell-to-cell movement of viral RNA. The results confirmed that the observed difference between the two helpers, CymRSV and CIRV, in controlling satRNA accumulation is not the consequence of the altered movement of satRNA molecules.
Taken together, these findings provide evidence of the extraordinary adaptation of a virus to its host plant. They also provide a better understanding of the molecular mechanisms the virus uses to harness an antiviral plant response to control the accumulation of its own parasite. Harnessing the antiviral silencing plant response for the benefit of a virus invader also was reported for infection by the Cauliflower mosaic virus (19). The 35S polycistronic transcript of this dsDNA plant virus contains an extensively structured leader sequence, and the siRNA derived from this leader induces the downregulation of an endogenous plant gene. Finally, our results also demonstrate the high level of complexity of the intimate plant-virus interaction, which still needs to be studied further.
Acknowledgments
This research was supported by grants from the Hungarian Scientific Research Fund (OTKA, NK60352), the SIROCCO EU project (LSHG-CT-2006-037900), and the bilateral research program CNR-MTA. V.P. was a recipient of EMBO ASTF 92-2007.
Footnotes
Published ahead of print on 24 September 2008.
REFERENCES
- 1.Ameres, S. L., J. Martinez, and R. Schroeder. 2007. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130101-112. [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe, D. 2004. RNA silencing in plants. Nature 431356-363. [DOI] [PubMed] [Google Scholar]
- 3.Baumberger, N., and D. C. Baulcombe. 2005. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 10211928-11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodersen, P., and O. Voinnet. 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22268-280. [DOI] [PubMed] [Google Scholar]
- 5.Burgyan, J., P. D. Nagy, and M. Russo. 1990. Synthesis of infectious RNA from full-length cloned cDNA to RNA of cymbidium ringspot tombusvirus. J. Gen. Virol. 711857-1860. [DOI] [PubMed] [Google Scholar]
- 6.Burgyán, J., and M. Russo. 1988. Studies on the replication of a satellite RNA associated with cymbidium ringspot virus. J. Gen. Virol. 693089-3092. [Google Scholar]
- 7.Celix, A., E. Rodriguez-Cerezo, and F. Garcia-Arenal. 1997. New satellite RNAs, but no DI RNAs, are found in natural populations of tomato bushy stunt tombusvirus. Virology 239277-284. [DOI] [PubMed] [Google Scholar]
- 8.Chernysheva, O. A., and K. A. White. 2005. Modular arrangement of viral cis-acting RNA domains in a tombusvirus satellite RNA. Virology 332640-649. [DOI] [PubMed] [Google Scholar]
- 9.Dalmay, T., and L. Rubino. 1995. Replication of cymbidium ringspot virus satellite RNA mutants. Virology 2061092-1098. [DOI] [PubMed] [Google Scholar]
- 10.Dalmay, T., and L. Rubino. 1994. The nature of multimeric forms of cymbidium ringspot tombusvirus satellite RNA. Arch. Virol. 138161-167. [DOI] [PubMed] [Google Scholar]
- 11.Dalmay, T., L. Rubino, J. Burgyán, A. Kollar, and M. Russo. 1993. Functional analysis of cymbidium ringspot virus genome. Virology 194697-704. [DOI] [PubMed] [Google Scholar]
- 12.Dalmay, T., M. Russo, and J. Burgyán. 1993. Repair in vivo of altered 3′ terminus of cymbidium ringspot tombusvirus RNA. Virology 192551-555. [DOI] [PubMed] [Google Scholar]
- 13.Ding, S. W., and O. Voinnet. 2007. Antiviral immunity directed by small RNAs. Cell 130413-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallitelli, D., and R. Hull. 1985. Preparation of complementary DNA by direct synthesis on plant virus RNAs from agarose gels. J. Virol. Methods 11141-144. [DOI] [PubMed] [Google Scholar]
- 15.Johansen, L. K., and J. C. Carrington. 2001. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lakatos, L., T. Csorba, V. Pantaleo, E. J. Chapman, J. C. Carrington, Y. P. Liu, V. V. Dolja, L. F. Calvino, J. J. Lopez-Moya, and J. Burgyán. 2006. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 252768-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manfre, A. J., and A. E. Simon. 2008. Importance of coat protein and RNA silencing in satellite RNA/virus interactions. Virology 379161-167. [DOI] [PubMed] [Google Scholar]
- 18.Moissiard, G., E. A. Parizotto, C. Himber, and O. Voinnet. 2007. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA 131268-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moissiard, G., and O. Voinnet. 2006. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl. Acad. Sci. USA 10319593-19598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Molnár, A., T. Csorba, L. Lakatos, E. Varallyay, C. Lacomme, and J. Burgyán. 2005. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 797812-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantaleo, V., G. Szittya, and J. Burgyán. 2007. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J. Virol. 813797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi, Y., A. M. Denli, and G. J. Hannon. 2005. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19421-428. [DOI] [PubMed] [Google Scholar]
- 22a.Parizotto, E. A., P. Dunoyer, N. Rahm, C. Himber, and O. Voinnet. 2004. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 182237-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubino, L., J. Burgyán, F. Grieco, and M. Russo. 1990. Sequence analysis of cymbidium ringspot virus satellite and defective interfering RNAs. J. Gen. Virol. 711655-1660. [DOI] [PubMed] [Google Scholar]
- 24.Rubino, L., J. C. Carrington, and M. Russo. 1992. Biologically active cymbidium ringspot virus satellite RNA in transgenic plants suppresses accumulation of DI RNA. Virology 188429-437. [DOI] [PubMed] [Google Scholar]
- 25.Rubino, L., and M. Russo. 1995. Characterization of resistance to cymbidium ringspot virus in transgenic plants expressing a full-length viral replicase gene. Virology 212240-243. [DOI] [PubMed] [Google Scholar]
- 26.Russo, M., J. Burgyán, and G. P. Martelli. 1994. Molecular biology of tombusviridae. Adv. Virus Res. 44381-428. [DOI] [PubMed] [Google Scholar]
- 27.Simon, A. E., M. J. Roossinck, and Z. Havelda. 2004. Plant virus satellite and defective interfering RNAs: new paradigms for a new century. Annu. Rev. Phytopathol. 42415-437. [DOI] [PubMed] [Google Scholar]
- 28.Szittya, G., A. Molnar, D. Silhavy, C. Hornyik, and J. Burgyán. 2002. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szittya, G., D. Silhavy, A. Molnar, Z. Havelda, A. Lovas, L. Lakatos, Z. Banfalvi, and J. Burgyán. 2003. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 22633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomari, Y., and P. D. Zamore. 2005. Perspective: machines for RNAi. Genes Dev. 19517-529. [DOI] [PubMed] [Google Scholar]
- 31.Vaistij, F. E., L. Jones, and D. C. Baulcombe. 2002. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14857-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Válóczi, A., C. Hornyik, N. Varga, J. Burgyan, S. Kauppinen, and Z. Havelda. 2004. Sensitive and specific detection of microRNAs by Northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 32e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vargason, J., G. Szittya, J. Burgyán, and T. M. Hall. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115799-811. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, X., Y. R. Yuan, Y. Pei, S. S. Lin, T. Tuschl, D. J. Patel, and N. H. Chua. 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 203255-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]