Abstract
Four conserved RNA stem-loop structures designated SL47, SL87, SL248, and SL443 have been predicted in the hepatitis C virus (HCV) core encoding region. Moreover, alternative translation products have been detected from a reading frame overlapping the core gene (core+1/ARFP/F). To study the importance of the core+1 frame and core-RNA structures for HCV replication in cell culture and in vivo, a panel of core gene silent mutations predicted to abolish core+1 translation and affecting core-RNA stem-loops were introduced into infectious-HCV genomes of the isolate JFH1. A mutation disrupting translation of all known forms of core+1 and affecting SL248 did not alter virus production in Huh7 cells and in mice xenografted with human liver tissue. However, a combination of mutations affecting core+1 at multiple codons and at the same time, SL47, SL87, and SL248, delayed RNA replication kinetics and substantially reduced virus titers. The in vivo infectivity of this mutant was impaired, and in virus genomes recovered from inoculated mice, SL87 was restored by reversion and pseudoreversion. Mutations disrupting the integrity of this stem-loop, as well as that of SL47, were detrimental for virus viability, whereas mutations disrupting SL248 and SL443 had no effect. This phenotype was not due to impaired RNA stability but to reduced RNA translation. Thus, SL47 and SL87 are important RNA elements contributing to HCV genome translation and robust replication in cell culture and in vivo.
Hepatitis C virus (HCV) infection causes a wide spectrum of clinical manifestations, ranging from a healthy carrier state to acute and chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (17). More than 170 million people worldwide are chronically infected with the virus, which is responsible for more than 100,000 cases of liver cancer per year (39). A vaccine against the virus is not available at present, and therapeutic approaches are still limited (13, 18).
HCV is classified within the genus Hepacivirus of the family Flaviviridae (51). Seven major HCV genotypes and more than 100 subtypes have been identified (42). The single-stranded positive-sense RNA genome of HCV (∼9.6 kb in length) is flanked at both termini by conserved, highly structured nontranslated regions (NTRs) and encodes a polyprotein precursor of about 3,000 amino acids (4, 37). The NTRs are required for RNA translation and replication. An internal ribosome entry site (IRES) residing in the 5′ NTR controls translation initiation, which is influenced by multiple transacting cellular factors via interaction either with the IRES sequence or core encoding sequences (20, 33, 40). Furthermore, long-range RNA-RNA interactions between the IRES and distally located sequences residing in the core coding region or 3′ NTR, as well as interactions with the viral-protein core, nonstructural protein 4A (NS4A), NS4B, and NS5A, appear to modulate IRES activity.
In the N-terminal region, the polyprotein, is processed by cellular proteases to yield the structural-protein core (C) and envelope proteins 1 and 2 (E1 and E2), which are required for the formation of infectious virus particles. C-terminal to E2 is the hydrophobic p7 protein, required for efficient assembly and release of infectious virions (22, 45). The remainder of the polyprotein is cleaved into NS2, NS3, NS4A, NS4B, NS5A, and NS5B. NS2 is a cysteine protease that, in conjunction with NS3, mediates cleavage at the NS2-NS3 site; in addition, it is involved in an early stage of virion morphogenesis, prior to the assembly of infectious virus (21, 22). NS3 carries a serine-type protease activity in its N-terminal domain and a nucleoside triphosphatase/helicase activity in the C-terminal two-thirds. NS4A is the NS3 protease cofactor, and NS4B contributes to the formation of the membrane-associated replication complex (15). NS5A is a phosphoprotein that plays a crucial role in RNA replication and assembly (1, 46, 47). NS5B is the RNA-dependent RNA polymerase (RdRp). The replicase complex, likely consisting of NS3 to NS5B and cellular proteins, copies the viral positive strand into a negative-strand intermediate serving as a template for the synthesis of excess progeny RNA genomes. Due to the absence of a 3′-5′ exonuclease proofreading activity of the NS5B RdRp, HCV RNA replication is error prone, causing a high degree of genetic variability of the virus.
Computer-assisted comparative analyses of diverse HCV sequences revealed a high conservation of the core encoding region that is more conserved than necessary to maintain the amino acid sequence of the core protein. This is due to the suppression of synonymous substitutions (19, 44). Ina and colleagues (19) suggested that an overlapping gene might constrain the core sequence, whereas Smith and Simmonds (44) concluded that suppression may be due to constraints imposed by RNA secondary structures identified within the core region.
In 1998, the first evidence for the expression of a protein from an alternate open reading frame (ORF) overlapping the core encoding region in the +1 frame (hence designated the core+1 ORF) was presented (58). This alternate ORF contains at least 124 codons and is present in all seven HCV genotypes (57). It lacks an in-frame AUG start codon and appears to be translated by ribosomal frameshifting or by internal translation initiation. In cell extracts and in transfected cells, a 17-kDa protein (p17), known as ARFP (alternative reading frame protein), F (frameshift), or core+1 (to indicate the position), was shown to be synthesized from the initiator codon of the HCV type 1 (HCV-1) (genotype 1a) polyprotein sequence by a +1 frameshift occurring in the A-rich core encoding region (codons 8 to 11) (Fig. 1) (11, 52, 57, 60). However, expression of this form of the core+1 protein is dependent on conditions supporting cytoplasmic expression and is limited to the HCV-1 isolate, which carries a stretch of 10 consecutive A residues within codons 8 to 11 (52, 54). In addition, a +1 frameshift at codon 42 of HCV-1b, followed by a rephrasing into the core ORF at stop codon 144, was observed in Escherichia coli (8). Moreover, internal translation initiation occurring at internal methionine codons 85/87 and resulting in a shorter form of the core+1 protein has been observed in cell culture (53, 55). This protein is expressed independently of the A-rich sequence and the polyprotein translation initiator codon and has been detected under conditions supporting either nuclear or cytoplasmic transcription. Furthermore, in the absence of codons 85/87, the core+1 codon 26 was also found to function as an internal translation initiation site (3).
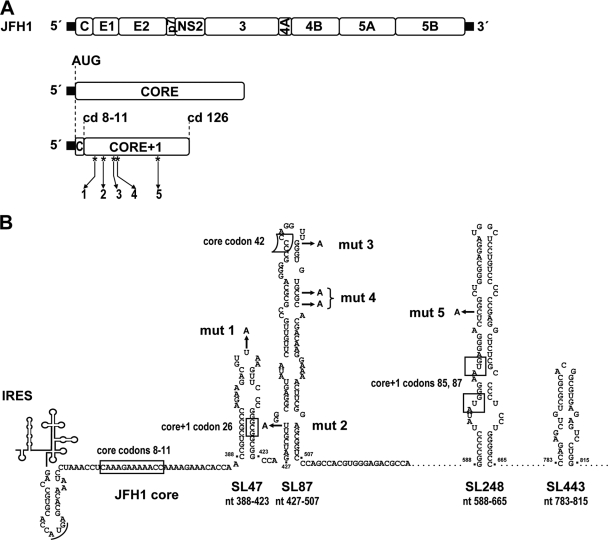
FIG. 1.
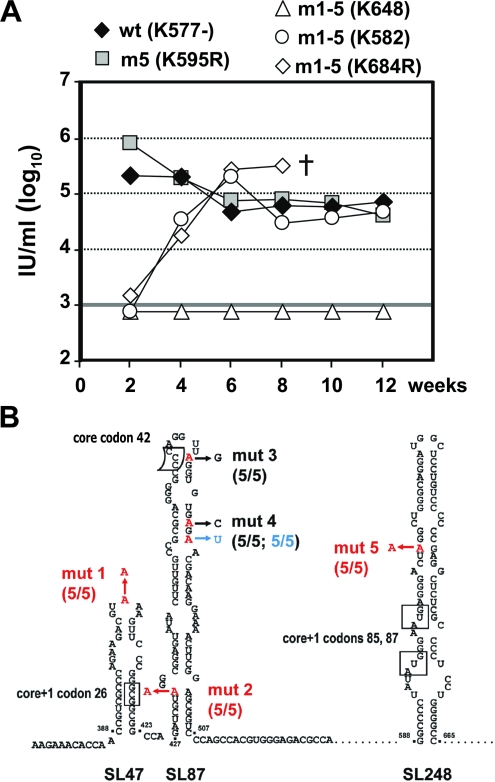
Predicted RNA structures in the core encoding region and positions of introduced mutations. (A) A schematic representation of the parental wt JFH1 genome is shown at the top. The 5′ and 3′ NTRs are indicated with thick black lines. The core encoding region and the core+1 ORF are drawn below. The positions of the five mutations introduced into the latter are depicted by asterisks and arrows. The positions of the polyprotein initiator AUG; of codons (cd) 8 to 11, which are implicated in core+1 translation; and of the termination codon 126 of the core+1 ORF are highlighted with dotted lines. (B) Schematic representation of the positions of the nonsense substitutions mut 1 to mut 5 (indicated by arrows) relative to the predicted core RNA secondary structures SL47, SL87, SL248, and SL443 of JFH1. All five nucleotide substitutions introduce a stop codon into the core+1 ORF and, with the exception of mut 1, in addition a mismatch into the putative stem of the respective RNA structure. The frameshift sites reported for core+1 translation at core codons 8 to 11 in genotype 1a and codon 42 in genotype 1b, as well as the internal translation initiation core+1 codons 26 and 85/87, are marked with boxes. The polyprotein initiator AUG is underlined. Note that none of the five nucleotide substitutions affects the amino acid sequence of the core encoding region. The nucleotide numbers refer to the JFH1 isolate.
Converging data from several laboratories suggest that core+1 is expressed during natural infection (2, 10, 29, 52, 57, 60). However, the biological role of this protein remains unknown. Previous studies using the subgenomic replicon system (lacking the region encoding the structural proteins) have shown that the core+1 protein is not required for viral replication in cell culture (6, 32). However, these experiments cannot exclude the possibility of an effect of the protein during the early or late stage of the HCV replication cycle, such as entry, assembly, and egress.
Four highly conserved RNA secondary structures, designated stem-loop 47 (SL47) (nucleotides [nt] 388 to 423 with respect to the 5′ end of the JFH1 genome), SL87 (nt 427 to 507), SL248 (nt 588 to 665), and SL443 (nt 783 to 815) (44, 48, 49), have been predicted in the core encoding region and overlapping the core+1 ORF (Fig. 1B). A recent study has provided the first evidence for the role of SL87 in virus production, but it is not clear at which stage of the HCV replication cycle this RNA structure is required (34).
In this study, we investigated the importance of the core+1 ORF and RNA elements SL47, SL87, SL248, and SL443 for HCV production. By using the highly efficient cell culture system based on the JFH1 HCV isolate (24, 56) and the chimeric H77/JFH1 genome (38), we demonstrate that core+1 has no role in the production of infectious HCV in vitro and in vivo, whereas SL47 and SL87 are vital cis-acting RNA elements contributing to RNA translation.
MATERIALS AND METHODS
Cell culture.
The Huh7-derived cell clones Huh7.5 and Huh7-Lunet were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Karlsruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum. Most transfection experiments were performed with Huh7-Lunet cells, which support high-level HCV RNA replication and have superior properties for immunofluorescence microscopy (14). Huh7.5 cells were primarily used in infection experiments because of their high level of CD81 expression (7).
Plasmid construction.
All nucleotide numbers refer to the JFH1 (GenBank accession no. AB047639) (24) and H77 (GenBank accession no. AF009606) (28) isolates. Plasmids pFK-JFH1wt and pFK-H77/C3/JFH1wt have been described previously (38, 56). Plasmid pFK-JFH1wt/ΔGDD has an in-frame deletion of 10 amino acids (MLVCGDDLVV) spanning the GDD motif of the NS5B RdRp active site (underlined). This plasmid was constructed by replacing the NsiI-EcoRV fragment derived from pFK-JFH1wt (nt 5298 to 9237) with the corresponding fragment derived from pSGR-JFH1/ΔGDD (23). Nucleotide substitutions were introduced by PCR-based site-directed mutagenesis with oligonucleotides carrying the desired nucleotide exchange(s). The amplified DNA fragments were analyzed by automated nucleotide sequencing using an ABI 310 sequencer (Applied Biosystems, Darmstadt, Germany). Substitutions generating nonsense mutations in the core+1 ORF without affecting the core amino acid sequence were, in the case of pFK-JFH1wt (Fig. 1B), as follows: mut 1, T406A (Leu codon 22→TAA); mut 2, T433A (Leu codon 31→TAG); mut 3, G472A (Trp codon 44→TAG); mut 4, C479A (Cys codon 46→TGA); and mut 5, C613A (Ser codon 90→TAG). In the case of pFK-H77/C3/JFH1wt, the following mutations were inserted: mut 1, C407A (Ser codon 22→TAA); mut 2, T434A (Leu codon 31→TAG); mut 3, G473A (Trp codon 44→TAG); and mut 4, C480A (Cys codon 46→TGA). The additional substitutions C481A and C482G were introduced into pFK-JFH1wt and pFK-H77/C3/JFH1wt, respectively, without affecting core amino acid 47 (Arg) (CGC→AGA and CGC→AGG, respectively). The substitutions affecting the core RNA structures SL47, SL87, SL248, and SL443, but not the core amino acid sequence, were as follows: mut SL47, T391G, C392A, C394G, C403T, T406C, and G415C; mut SL87, C455A, C457A, A458C, G460C, C463G, A467C, G469C, T470C, and G472C; mut SL248, C613G, A622T, A625T, C629T, C631G, C632T, G634A, T635A, C636G, C637T, C640G, A643T, and C646G; and mut SL443, C784G, A785C, T790A, C793G, G796C, and A806C. Overlapping PCR fragments were generated by using the template pFK-JFH1wt or pFK-JFH1wt/ΔGDD and with the oligonucleotide primer pairs specified in Table S1 in the supplemental material. For mut 1 + 2, mut 3 + 4, mut SL47, and mut SL87, DNA fragments were combined by PCR using the outer primers ST7 and A1394-JFH1 and, after restriction with AgeI and SexAI, were inserted into pFK-JFH1wt or pFK-JFH1wt/ΔGDD. For mut 5, mut SL248, and mut SL443, DNA fragments were combined by PCR using the outer primers S343-JFH1 and A1817-JFH1 and, after restriction with SexAI and BsiWI, were inserted into pFK-JFH1wt or pFK-JFH1wt/ΔGDD. Plasmid pFK-JFH1wt or pFK-JFH1wt/ΔGDD was also used as a template to successively insert substitutions mut 1 + 2, mut 3 + 4, and mut 5; mut SL47 and mut SL87; or mut SL248 and mut SL443. For simultaneous introduction of substitutions of mut 1-4 into pFK-H77/C3/JFH1wt, overlapping PCR fragments were generated with primer pairs S438-mut1,2,3,4-H77 and A1150-H77 or S-T7 and A452mut1,2,3,4-H77. Fragments were combined using the outer primers S-T7 and A1150-H77 and, after restriction with AgeI and NruI, were inserted into pFK-H77/C3/JFH1wt. mut 5 was not inserted into pFK-H77/C3/JFH1wt because nucleotide changes created by mut 5 would affect the core amino acid sequence.
In vitro transcription and RNA transfection.
For in vitro transcription, HCV constructs were linearized with MluI, extracted with phenol and chloroform, precipitated with ethanol, and dissolved in RNase-free water. One hundred nanograms of linearized plasmid DNA per microliter was used for in vitro transcription as described recently (25). Cells were detached by trypsin treatment, washed with phosphate-buffered saline (PBS), counted, and resuspended in Cytomix (50) containing 2 mM ATP and 5 mM glutathione at 1 × 107 cells per ml in the case of Huh7-Lunet and at 1.5 × 107 cells per ml in the case of Huh7.5. Ten micrograms of in vitro-transcribed RNA was mixed with 400 μl of the cell suspension and electroporated with a Gene Pulser system (Bio-Rad, Munich, Germany) in a cuvette with a gap width of 0.4 cm (Bio-Rad) at 960 μF and 270 V. The cells were immediately transferred to 20 ml of complete DMEM and subsequently seeded as required for the assay. In most experiments, transfection efficiencies were monitored by cotransfection of 2.5 μg of in vitro-transcribed JFH1 replication-defective subgenomic reporter replicon.
Preparation of total RNA and Northern hybridization.
Total RNA was prepared by single-step acid guanidinium thiocyanate-phenol-chloroform extraction as described previously (12). Ten to 20 μg of total RNA from Huh7-Lunet cells was denatured by treatment with 5.9% glyoxal, 50% dimethyl sulfoxide, and 10 mM sodium phosphate buffer (pH 7.0) for 1 h at 50°C. Samples were separated by denaturing agarose gel electrophoresis, and RNA was transferred to positively charged nylon membranes (Hybond-N+; Amersham Biosciences, Freiburg, Germany) with 50 mM NaOH and cross-linked by UV irradiation. Prior to hybridization, the membrane was stained with methylene blue and cut ∼0.5 cm below the 28S rRNA band. The upper strip containing the HCV replicon RNA was hybridized with a 32P-labeled negative-sense riboprobe complementary to the JFH1 NS5A-NS5B region (nt 6273 to 9678) to detect viral positive-strand RNA. The lower strip, which was hybridized with a β-actin-specific antisense riboprobe, was used to normalize for different loading. Signals were quantified by phosphorimaging by using a Molecular Imager FX scanner (Bio-Rad), and the number of HCV molecules was determined by comparison with a serial dilution of in vitro transcripts of genomic replicon RNAs mixed with 5 μg of total RNA from naive Huh7 cells loaded in parallel onto the gel.
Indirect immunofluorescence.
Electroporated cells were seeded onto glass coverslips in 24-well plates at a density of 2 × 104 to 6 × 104 cells per well. At 24, 48, and 72 h posttransfection (p.t.), the cells were fixed with 500 μl of 4% paraformaldehyde for 10 min at room temperature and permeabilized by 5 min of incubation in 500 μl of 0.5% Triton X-100 in PBS. Staining of NS5A was performed by using culture supernatant of the 9E10 hybridoma (30) at a dilution of 1:1,000 in PBS with 5% normal goat serum for 45 min. Bound primary antibodies were detected by using goat antibodies conjugated to Alexa-Fluor 488 at a dilution of 1:1,000 in PBS containing 5% normal goat serum for 30 min in the dark. DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride) (Molecular Probes, Karlsruhe, Germany) for 1 min. Finally, the cells were mounted on glass slides with Fluoromount G (Southern Biotechnology Associates, Birmingham, AL).
Virus titration in cell culture supernatants.
HCV was titrated essentially as described elsewhere (30). In brief, Huh7.5 target cells were seeded into 96-well plates at a density of 1 × 104 cells per well in a total volume of 200 μl complete DMEM. Twenty-four hours later, serial dilutions of filtered virus-containing cell culture supernatants harvested from transfected cells were added (eight wells per dilution). Infectivity titers were determined by using hybridoma supernatant of the NS5A-specific monoclonal antibody 9E10 (30) and expressed as the base 10 logarithm of 50% tissue culture infective doses (TCID50) per ml of supernatant.
Quantification of HCV core protein.
Intracellular HCV core protein amounts were quantified using the Trak-C enzyme-linked immunosorbent assay (ELISA) kit (Ortho Clinical Diagnostics, Neckargemünd, Germany) according to the instructions of the manufacturer. Huh7-Lunet cells were electroporated with the respective pFK-JFH1/ΔGDD transcript and resuspended in 20 ml culture medium, and 2-ml aliquots were seeded into each well of a six-well plate. Cells were lysed at various time points p.t. by addition of 0.35 ml ice-cold PBS containing 1% Triton X-100, 1/10,000 volume aprotinin (1 U/ml), 1/1,000 volume leupeptin (4 mg/ml), and 1/100 volume phenylmethylsulfonyl fluoride (100 mM), and the lysates were cleared at 18,000 × g for 5 min. Depending on the time of harvest, samples were diluted 1:5 or higher and processed for ELISA. Colorimetric measurements were performed using a Sunrise colorimeter (Tecan Trading AG, Switzerland).
Alb-uPA+/+ SCID mice.
The mouse study was conducted at Ghent University Hospital with protocols approved by the Ethical Committee and Animal Ethics Committee of the Ghent University Faculty of Medicine. Transgenic SCID mice that overexpressed the uPA gene under the control of an albumin promoter (Alb-uPA+/+) were used for xenotransplantation with primary human hepatocytes as described elsewhere (35, 36). The mice were inoculated by intraperitoneal injection with 1.0 × 106 TCID50 of JFH1 wild type (wt), JFH1 mut 1-5, or JFH1 mut 5 virus.
Preparation of virus stocks for intraperitoneal injection of uPA+/+-SCID mice.
Virus stocks were generated by electroporation as described above, and the supernatants of transfected cells were harvested 24, 48, 72, and 96 h p.t. After filtration through 0.45-μm-pore-size filters, the virus particles contained in 30-ml aliquots of the supernatants were concentrated by ultracentrifugation through a self-generating iodixanol gradient. Virus-containing fractions were collected and further concentrated using Centricon Plus-70 centrifugal filter devices (Millipore, Schwalbach, Germany) according to the manufacturer's protocol.
RNA quantification by real-time qRT-PCR.
Viral RNA was isolated from mouse serum using the Nucleo Spin RNA virus kit (Macherey-Nagel, Düren, Germany) as recommended by the manufacturer. The amount of HCV RNA was determined by quantitative reverse transcription-PCR (qRT-PCR)-based amplification using the HCV TaqMan48 assay (Roche) and comparison to serially diluted in vitro transcripts as described recently (25).
Preparation of total RNA from mouse serum, amplification of HCV RNA by RT-PCR, and sequence analysis of cloned amplicons.
HCV RNA was isolated from 150 μl of serum taken from mouse K582 12 weeks postinoculation with mut 1-5 virus. Viral RNA was isolated by using the Nucleo Spin RNA Virus Kit (Macherey-Nagel, Düren, Germany) as recommended by the manufacturer. RT-PCR was done with the Expand-RT system (Roche, Mannheim, Germany) as recommended by the manufacturer using primer A9482 (5′-GGA ACA GTT AGC TAT GGA GTG TAC C-3′) for cDNA synthesis. HCV sequences were amplified by using the Expand Long Template PCR Kit (Roche) according to the instructions of the manufacturer and primers S59-EcoRI (5′-AA GAA TTC TGT CTT CAC GCA GAA AGC GCC TAG-3′) and A1743-MluI (5′-AAA CGC GTC CTA TCC GGA AAG CCT CGA TGT TG-3′). The amplicons were inserted into pFK-I389Luc-EI/NS3-3′/JFH1-dg after restriction with EcoRI and MluI. Sequence analysis was performed using primer A974 (5′-GCA GCC TCG AGC TGC CAA GTG ATG-3′).
RESULTS
Mutational ablation of core+1 translation and core RNA secondary structures delays the kinetics of HCV production.
To investigate the importance of the core+1 ORF and the core RNA structural elements for HCV production, we simultaneously introduced into the core+1 ORF of the JFH1 genome a series of nonsense mutations predicted to abolish translation of this ORF and affecting the secondary structure of all four stem-loop elements residing in the same region (see Fig. S1A and B in the supplemental material). These mutations did not affect the amino acid sequence of the overlapping core region, which was a constraint that limited our selection for the insertion of these nonsense mutations to five distinct positions (Fig. 1B). More specifically, mut 1, introduced at codon 22 (nt 406), is predicted to abolish translation of the core+1/F protein synthesized by a frameshift at core codons 8 to 11 (11, 52, 57, 60) and in parallel alters the sequence, but not the structure, of the terminal loop of SL47 (see Fig. S1A in the supplemental material). mut 2, inserted at codon 31 (nt 433), is predicted to terminate core+1 translation directly downstream of the internal initiator codon 26 (3) and disrupts one base pairing in the lower stem of SL87 (see Fig. S1A in the supplemental material). mut 3 and mut 4, introduced at codons 44 (nt 472) and 46 (nt 479), respectively, are predicted to abolish translation of the core+1 product generated by frameshift in the context of codon 42 (8), as well as of all the core+1 proteins initiated upstream, and in parallel alters the upper half of SL87 (see Fig. S1A in the supplemental material). mut 4 was designed as an additional block of any possible translational read-through and reversion of the three upstream stop codons. Finally, mut 5, inserted at codon 90 (nt 613), is predicted to terminate translation of the core+1 product initiated at the internal methionine codons 85/87 (53, 55), as well as of all the core+1 proteins initiated upstream, and in addition disrupts one base pairing within the stem of SL248 (see Fig. S1B in the supplemental material).
In the first set of experiments, we attempted to verify that the introduced mutations abolished the expression of the core+1 protein(s). However, despite extensive attempts, we were unable to detect these proteins even in cells that had been transfected with the wt JFH1 genome or an H77/JFH1 chimera (see below). We therefore constructed a plasmid that expresses JFH1 core+1-green fluorescent protein (GFP) by design (translation of the core+1 ORF initiates at an artificially inserted methionine codon at nt 384) under the control of the HCMV promoter. By using transient-transfection assays, we confirmed by GFP-specific Western blotting that insertion of nonsense mutation 5 abolished expression of the chimeric protein (not shown).
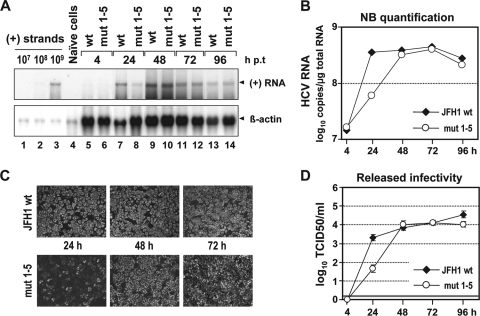
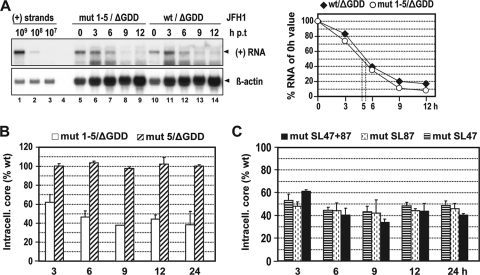
Replication fitness of the JFH1 wt genome or the mutant carrying all five mutations (mut 1-5) was determined by electroporation of in vitro transcripts into Huh7-Lunet cells. Viral RNA replication, expression of viral proteins, and release of infectious HCV particles were determined by Northern hybridization, immunofluorescence, and TCID50 assays, respectively, at various time points p.t. As shown in Fig. 2A and B, at 24 h p.t., the RNA amounts of mut 1-5 were about sixfold lower than those of the wt, whereas at all later time points, the replication rates of the mutant and the wt were comparable. This delay of replication kinetics was confirmed by immunofluorescence analysis of NS5A expression at 24, 48, and 72 h p.t. (Fig. 2C). Moreover, the release of infectivity of mut 1-5 was also significantly lower (about 50-fold) than that of the wt at 24 h p.t. but reached wt levels at later time points (Fig. 2D). These results show that the combination of nonsense mutations predicted to terminate translation of the core+1 ORF and affecting SL47, SL87, and SL248 leads to a delay of replication kinetics. The same results were obtained upon transfection of mut 1-5 into Huh7.5 cells, excluding cell clone-specific effects (data not shown). In addition, to extend our study to another genotype, mut 1 to 4 were inserted into the core encoding region of subtype 1a in the context of the H77/JFH1 chimera (38). Insertion of mut 5 was not possible because of alteration of the core coding region. We chose this H77/JFH1 chimera because expression of the 17-kDa core/core+1 protein was first described in a genotype 1a isolate (31, 52, 57, 60). Also, with this mutant, the same delay of replication kinetics was observed (data not shown). We therefore concluded that the nonsense mutations predicted to terminate core+1 translation and affecting core RNA SL47 and SL87 slow down the kinetics of RNA replication concomitant with a delay in viral-protein and infectious-particle production. Our results also suggest that the effects of the mutations are conserved among HCV-1a and -2a, and likely other genotypes.
FIG. 2.
A combination of mut 1 to 5 delays the kinetics of JFH1 RNA replication and virus production. (Α) Time course of HCV RNA replication after electroporation of Huh7-Lunet cells with the JFH1 wt genome or the mutant carrying all five nucleotide substitutions (mut 1-5). Transfected cells were harvested at the indicated time points (hours) p.t., and HCV positive-strand (+) RNA was detected in total RNA by Northern hybridization. A comparable amount of total RNA from naïve Huh7-Lunet cells served as a negative control (lane 4). Beta-actin served as a loading control (bottom); known amounts of in vitro transcripts spiked with total RNA from naïve cells as a carrier were used for quantification (lanes 1 to 3). (B) Quantification of the Northern blot by phosphorimaging. The JFH1 wt and JFH1 mut 1-5 RNA copy numbers per microgram of total RNA were calculated by comparison with the controls loaded in parallel. The values were normalized for differences in loading by using β-actin signals and were plotted on a logarithmic scale. (C) Indirect immunofluorescence analysis of HCV NS5A in Huh7-Lunet cells transfected with JFH1 wt or JFH1 mut 1 to 5 24, 48, and 72 h p.t. Nuclei were counterstained with DAPI. (D) Kinetics of release of infectivity from JFH1 wt or JFH1 mut 1-5-transfected Huh7-Lunet cells. Supernatants harvested at given time points p.t. were analyzed by TCID50 limiting-dilution assay. A representative experiment of three independent repetitions is shown. The error bars indicate the variability of the TCID50 assay. The cutoff of the assay (∼2 TCID50/ml) is indicated by the thick horizontal line.
Given the high mutation rate of HCV replication, we tested the possibility of (pseudo)reversion of mut 1-5. First, viral RNA present in cells 5 days p.t. was cloned and the nucleotide sequence was determined in multiple regions of the genome (core, 5′ half of E1, NS3, and NS5A). No reversion to wt and no conserved second-site mutation was found (not shown). Second, infected cells were passaged for 4 weeks (as described previously) (25), the core region was amplified by RT-PCR and cloned, and the sequences of four cDNA clones were analyzed. We found that mut 1 to 5 were retained (not shown). These results argue against (pseudo)reversion of mut 1 to 5 in Huh7 cells.
Core+1 does not play a role in the HCV replication cycle in cell culture.
The results described thus far do not allow discrimination of whether the impaired RNA replication is due to the lack of core+1 expression or to the alterations of stem-loop elements in the core encoding region. We therefore generated cell lines stably overexpressing the core+1 protein. However, these cells supported the HCV replication cycle (infection, replication, and virus production) to the same extent as the parental cell lines that did not express core+1 (see Fig. S2A to D in the supplemental material). Analogous results were obtained in transient-replication assays in which we coexpressed core+1 proteins from bicistronic replicons (see Fig. S2E to H in the supplemental material), arguing that core+1 proteins do not affect HCV RNA replication in cis or in trans.
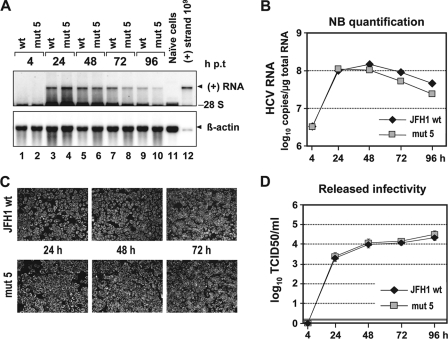
To corroborate this conclusion, we generated a JFH1 mutant carrying a nonsense mutation that terminates core+1 translation from all reported initiation sites (mut 5). As shown in Fig. 3A and B, this mutant replicated with kinetics indistinguishable from those of the wt. Moreover, the kinetics of accumulation of viral protein, as well as the kinetics of release of infectious virus particles, were comparable to those of the wt (Fig. 3C and D). From these results, we conclude that core+1 does not play a role in HCV replication and virus production in cell culture.
FIG. 3.
Blocking expression of all core+1 proteins does not affect HCV RNA replication and virus production. (Α) RNA replication of the JFH1 wt and mut 5 genomes in Huh7-Lunet cells. Total RNAs from cells transfected with constructs specified at the top were prepared at the indicated time points p.t., and HCV RNA was detected by Northern hybridization. The positions of viral (+) RNA, 28S rRNA, and β-actin mRNA are indicated. Total RNA of naïve cells (lane 11) and a known amount of a size-matched in vitro transcript that was spiked with total RNA from naïve cells as a carrier (lane 12) were loaded in parallel. (B) HCV RNA copy numbers were quantified as described in the legend of Fig. 2B. (C) Indirect immunofluorescence analysis of NS5A accumulation in transfected cells at the time points indicated. Nuclei were counterstained using DAPI. (D) Release of infectivity from transfected cells at the indicated time points p.t. by TCID50 assay. A representative result of three independent repetitions is shown. The error bars demonstrate the variability of the TCID50 assay. The cutoff of the assay is indicated by the thick horizontal line.
Important role of SL47 and SL87 in viral RNA replication.
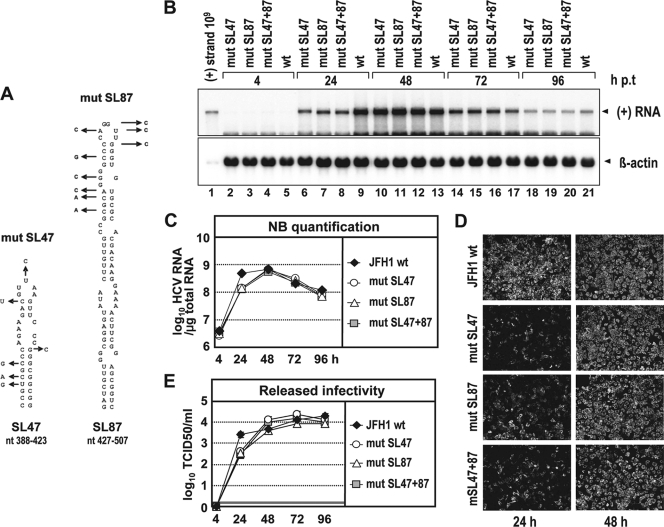
Since nonsense mutations in the core+1 ORF altering RNA structures in the core encoding region impair replication kinetics and because mut 5 did not show an altered phenotype, we assumed that SL47 and/or SL87 is crucial for efficient RNA replication. To address this hypothesis experimentally, we generated a set of extensive nucleotide substitutions (Fig. 4A) in the context of the JFH1 genome. The substitutions were designed to alter RNA secondary structures without affecting the core amino acid sequence. Specifically, the mutations introduced into SL47 were predicted to alter almost the complete stem-loop structure, whereas the mutations within SL87 were predicted to affect the upper half of the stem-loop (see Fig. S1A in the supplemental material). Interestingly, all three groups of mutations affecting either SL47 (mut SL47), SL87 (mut SL87), or both stem-loops (mut SL47 + 87) reduced JFH1 RNA replication about fourfold at 24 h p.t., as shown by quantitative analysis of the Northern blot (Fig. 4B and C). Comparable to the previous observations, replication of the mutated genomes reached wt level at later time points, arguing again for a delayed replication kinetics. In confirmation, the analogous delay was found when transfected cells were analyzed by NS5A-specific immunofluorescence (Fig. 4D) and for release of infectious virus particles, where infectivity titers were reduced about 10-fold at 24 h p.t. (Fig. 4E). These results confirm that stem-loop structures SL47 and SL87 are required for efficient replication in cell culture.
FIG. 4.
Mutation analysis of SL47 and SL87 with respect to RNA replication, expression of viral proteins, and virus production. (A) Schematic representation of the predicted secondary structures of SL47 and SL87 of JFH1 (see Fig. 1B). Introduced mutations are indicated with arrows that point to the substituting nucleotide. Note that these mutations do not affect the amino acid sequence of the core. (B) RNA replication of JFH1 genomes specified at the top after electroporation of Huh7-Lunet cells. Total RNA was harvested at the indicated time points and analyzed by Northern hybridization. For further details, see the legend to Fig. 2A. (C) HCV RNA copy numbers were quantified by phosphorimaging as specified in the legend to Fig. 2B. (D) Indirect immunofluorescence analysis of NS5A accumulated in transfected cells at the indicated time points. Nuclei were counterstained by DAPI. (E) Release of infectivity from transfected cells was determined by TCID50 assay for the indicated time points p.t. A representative result of three independent repetitions is shown. The error bars demonstrate the variability of the TCID50 assay. The cutoff of the assay is indicated by the thick horizontal line.
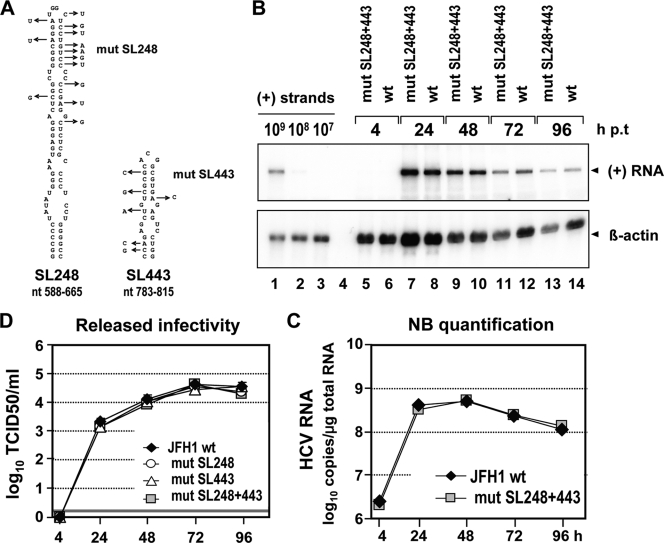
Integrity of RNA secondary structures SL248 and SL443 is dispensable for virus proliferation.
As the first two predicted RNA structures in the core encoding region were shown to be important for HCV replication, we wanted to assess whether the other two highly conserved RNA stem-loops, SL248 and SL443, also play roles in the viral replication cycle (Fig. 1B). For this purpose, a set of nucleotide substitutions was inserted into each of the two stem-loops either separately or simultaneously (Fig. 5A). The substitutions were designed to disrupt RNA secondary structure without affecting the core amino acid sequence. Whereas the mutations within SL248 are predicted to affect the upper half of the stem-loop structure, the mutations within SL443 are predicted to alter the structure completely (see Fig. S1B and C in the supplemental material). Northern hybridization and quantification by phosphorimaging revealed that substitutions affecting both stem-loops had no significant effect on JFH1 RNA replication (Fig. 5B and C). Consistently, the mutations did not significantly alter released infectivity (Fig. 5D). These data show that the structural integrity of SL248 and SL443 residing in the core encoding region is dispensable for virus proliferation in cell culture.
FIG. 5.
Integrity of SL248 and SL443 is dispensable for JFH1 RNA replication and virus release. (A) Schematic diagram of SL248 and SL443 (see Fig. 1B). Substitutions designed to affect SL248 and SL443 integrity but not the core amino acid sequence are indicated by arrows. (B) RNA replication of JFH1 genomes specified at the top after transfection into Huh7-Lunet cells. Total RNA of transfected cells harvested at the indicated time points was prepared and analyzed by Northern hybridization. Known amounts of in vitro transcripts that were spiked with total RNA from naïve cells were loaded in parallel (lanes 1 to 3). (C) HCV RNA copy numbers were quantified by phosphorimaging as described in the legend to Fig. 2B. (D) Kinetics of release of infectivity from cells transfected with either the wt or a mutant as determined by TCID50 assay. A representative result of three independent repetitions is shown. The error bars demonstrate the variability of the TCID50 assay. The cutoff of the assay is indicated by the thick horizontal line.
Disruption of SL47 and SL87 does not impair viral-RNA stability.
In an attempt to investigate how SL47 and SL87 contribute to HCV replication, we examined their possible impact on the stability of the viral RNA genome. For this purpose, we used a JFH1 variant that carries a 10-codon deletion in the NS5B gene (ΔGDD) abolishing RdRp activity. This construct design allowed investigation of RNA stability independently of replication. The mutations described above disrupting the stem-loop structures were inserted into this JFH1/ΔGDD vector, and in vitro transcripts were transfected into Huh7-Lunet cells. At various time points, total RNA was prepared, and HCV RNA was detected by Northern hybridization and quantified by phosphorimaging. An example of this analysis in which the RNA stability of mut 1-5 was analyzed in comparison to the JFH1/ΔGDD wt is shown in Fig. 6A. We found that both RNAs decayed with the same half-life. Moreover, RNAs of all the other mutants carrying more extensive nucleotide substitutions in SL47 and SL87 had half-lives indistinguishable from that of the wt (see Fig. S3 in the supplemental material).
FIG. 6.
Mutations affecting SL47 and SL87 do not affect RNA stability but reduce RNA translation. (A) RNA stability as determined by Northern hybridization and phosphorimaging (left and right, respectively). The replication-defective constructs specified at the top were transfected into Huh7-Lunet cells, and total RNA was prepared at the indicated time points p.t. HCV-specific signals (arrowhead) were detected by Northern hybridization. Beta-actin served as a loading control, and known amounts of in vitro transcripts (lanes 1 to 3) were loaded in parallel to determine RNA copy numbers. HCV RNA levels detected immediately after transfection (0 h) were set to 100%. The dotted vertical lines indicate transcript half-lives. (B) HCV core protein expression kinetics in Huh7-Lunet cells transfected with the replication-defective (ΔGDD) constructs specified at the top. Core protein amounts were quantified at the indicated time points by a core-specific ELISA. (C) Same approach as in panel B, but using different mutants (specified at the top) in the context of a replication-defective ΔGDD construct. In panels B and C, the values determined for the mutated genomes are expressed as percentages of those obtained with the wt/ΔGDD construct. The mean values from two independent experiments are given; error bars represent standard deviations.
SL47 and SL87 enhance RNA translation.
Having excluded the possibility that RNA stability accounts for the detrimental effect of substitutions affecting SL47 and SL87, we explored the possibility that these RNA structures contribute to RNA translation. Also, in this case, we analyzed the mutations in the context of the JFH1/ΔGDD construct to exclude effects that are due to RNA replication. RNA translation was determined by quantification of intracellular core protein levels in Huh7-Lunet cells transfected with either the wt or one of the stem-loop mutants. Core protein amounts were quantified 3, 6, 9, 12, and 24 h p.t. and expressed as percentages of the wt level. The values were normalized to the transfection efficiency by measuring luciferase activity that was expressed from a cotransfected replication-defective subgenomic reporter RNA. As shown in Fig. 6B, mut 1 to 5, which affect both SL47 and SL87, reduced core protein about twofold at all time points. As expected from previous results, mut 5 did not significantly alter the translation efficiency. Likewise, extensive nucleotide substitutions affecting SL47 and SL87 separately (mut SL47 and mut SL87, respectively) or simultaneously (mut SL47 + 87) also significantly impaired core expression at all time points (Fig. 6C). These data show that RNA elements SL47 and SL87 play an important role in translation of the HCV genome and that the impaired replication kinetics observed with mutations affecting either of these stem-loops are due, at least in part, to perturbations of RNA translation.
SL87 structure integrity but not core+1 expression is required for robust replication in vivo.
To analyze the in vivo relevance of core+1 expression and core RNA structure integrity, we took advantage of the uPA-SCID mouse model, in which immunodeficient transgenic mice are xenografted with primary human hepatocytes, which are then permissive for HCV infection (25, 35). Mice were inoculated with 1.0 × 106 TCID50 of JFH1 wt (animal K577−), JFH1 mut 1-5 (animals K648, K582, and K684R), or JFH1 mut 5 (animal K595R) virus. HCV RNA of the wt and mut 5 viruses rapidly increased in mouse sera and by week 2 after infection had already reached high levels (∼2 × 105 and 8 × 105 IU/ml, respectively). In contrast, at week 2 HCV RNA remained below the limit of detection in all three mice inoculated with mut 1-5 (Fig. 7A). In the sera of two animals (K582 and K684R), viremia increased and reached wt levels with a 2- to 4-week delay compared to the wt at week 6 and thereafter (∼2 × 105 and 3 × 105 IU/ml, respectively). In contrast, the third animal (K648) inoculated with mut 1-5 did not become infected and remained RNA negative until week 12, when most of the animals were sacrificed. These results suggest that core+1 expression is dispensable whereas SL47 and SL87 are required for robust JFH1 proliferation in vivo. The fact that mut 5, which contains a stop codon blocking expression of all known forms of core+1 proteins, is viable provides strong evidence that this protein(s) is also dispensable for replication in vivo.
FIG. 7.
In vivo viability of JFH1 mut 1-5 and mut 5 viruses in uPA-SCID mice. (A) Time course of HCV infection of mice inoculated with 1.0 × 106 TCID50 of JFH1 wt (animal K577−), JFH1 mut 5 (animal K595R), or JFH1 mut 1-5 (animals K648, K582, and K684R) virus. The viral-RNA loads in sera were determined at the indicated time points by qRT-PCR. Most of the mice were sacrificed at week 12 postinjection, except for K684R, which died (†) prematurely after week 8. (B) Results from sequence analysis of five cDNA clones obtained from serum of mut 1-5-inoculated mouse K582. Mutations introduced into mut 1-5 and mut 5 are highlighted in red. The arrows point to nucleotides identified in HCV genomes isolated from K582 serum. Retained mutations are indicated in red, reversions to wt are highlighted in black, and the pseudoreversion in blue. Note that mut 1, 2, and 5 were conserved in all five cDNA clones whereas mut 3 reverted to wt. The upstream mutation in mut 4 reverted to wt, and the downstream mutation restored base pairing.
Given the delayed increase of viremia of mut 1-5, we assumed that second-site mutations eventually compensating for the defects of these stem-loop structures might have accumulated. We therefore amplified the sequence encoding core to the N-terminal region of E2 by RT-PCR from virus contained in serum of mouse K582 at week 12 postinoculation. This was the only sample available because mouse K648 remained RNA negative and mouse K864R died prematurely, and neither adequate serum nor liver tissue could be recovered from the cadaver. Five cDNA clones derived from K582 serum were sequenced and aligned with the parental mut 1-5 genome. As summarized in Fig. 7B, mut 1, 2, and 5 were fully conserved in all five cDNA clones. In contrast, in all five clones, mut 3 had reverted to wt. In the case of the double mut 4, the upstream adenosine substitution had also reverted to wt in all sequenced clones whereas the downstream adenosine substitution was replaced by uridine. All three mutations together restored base pairing of the upper stem-loop structure of SL87, arguing that this RNA element plays a very important role in robust replication in vivo. Attempts to reclone viral genomes from mice inoculated with mut 5 or the wt were not successful due to a very limited amount of serum. However, the fact that mutation 5, which disrupts translation of all known forms of core+1, was retained in the revertants of mut 1-5-inoculated mice clearly shows that these proteins are not required for productive HCV replication in the xenografted animals.
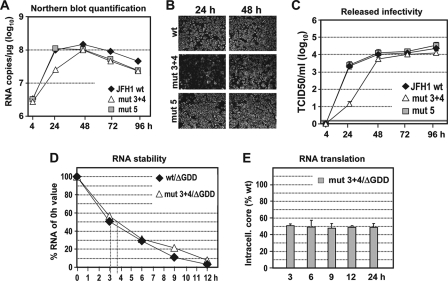
To support this conclusion, we constructed the double mutant mut 3 + 4 and determined RNA replication, as well as virus production, in cell culture. By using Northern hybridization, we found that replication of mut 3 + 4 was clearly delayed compared to that of mut 5 and the wt (Fig. 8A). We also observed an appropriately reduced accumulation of NS5A in cells 24 h p.t., but no longer at 48 h p.t (Fig. 8B). Virus titers released from mut 3 + 4-transfected cells were reduced about 100-fold at early time points p.t., whereas later the titers were comparable to those of the wt and mut 5 (Fig. 8C). Consistent with results obtained with the other mutants, the impairment of replication was not due to lower RNA stability but to reduced RNA translation (Fig. 8D and E).
FIG. 8.
Replication properties of mut 3 + 4 in cell culture. (A) Replication kinetics of mut 3 + 4 in comparison to JFH1 wt and mut 5 in Huh7-Lunet cells as determined by Northern hybridization. The blot (not shown) was quantitated by phosphorimaging as described in the legend to Fig. 2B. (B) Indirect immunofluorescence analysis of NS5A accumulation in transfected cells 24 and 48 h p.t. (C) Kinetics of release of infectious virus particles as determined by TCID50 assay. A representative experiment of three independent repetitions is shown. The error bars indicate the variability of the TCID50 assay. The cutoff of the assay (∼2 TCID50/ml) is indicated by the thick horizontal line. (D) RNA stability as determined by Northern hybridization and phosphorimaging. The replication-defective constructs specified at the upper right were transfected into Huh7-Lunet cells, and total RNA was analyzed by Northern hybridization. The blots (not shown) were quantified as described in the legend to Fig. 6A. (E) HCV core protein expression kinetics in Huh7-Lunet cells transfected with mut 3 + 4, which, due to an inactivating mutation in the NS5B RdRp, cannot replicate. Core protein amounts were quantified as described in the legend to Fig. 6B and C and normalized to the values obtained with the wt/ΔGDD construct. Mean values from two independent experiments are shown; error bars represent standard deviations.
In summary, these data demonstrate that SL87 is a very important RNA element required for robust RNA translation and replication in cell culture and in vivo. In addition, we have shown that all known forms of core+1 are dispensable for all steps of the viral replication cycle in these experimental systems.
DISCUSSION
Although specific antibodies and cellular immune responses against core+1 antigens have been detected in HCV patients (2, 10, 29, 52, 57, 60), the biological importance of the core+1 ORF and protein(s) derived from it is still unknown. Previous studies with subgenomic replicons suggested that core+1 protein(s) is not required for viral replication (6, 32). However, cumulative data associating elevated levels of core+1 expression with the development of hepatocellular carcinoma in HCV patients (9) and studies describing lower rates of HCV genome replication in tumor cells than in nontumor cells indicate that core+1 might exert a negative effect on viral replication. However, even under various experimental conditions of stable or transient coexpression, we did not observe such an effect, arguing that at least in cell culture, HCV replication is not affected by core+1 protein(s). In agreement with this observation, we found that insertion of a nonsense mutation predicted to terminate core+1 translation at codon 90 (mut 5) and in parallel disrupting one base pairing within the stem of SL248 did not affect viral-RNA replication, RNA stability, expression of viral proteins, and virus release. A termination codon at position 90 would abolish translation of the core+1 product initiated at the internal methionine codons 85/87 (53, 55), as well as expression of all core+1 proteins reported to initiate upstream of codon 90 (frameshift products associated with codons 8 to 11 [11, 52, 57, 60] and 42 [8] and products arising from internal initiation at codon 26 [3]). The data thus show that expression of core+1 is dispensable for virus production in cell culture. However, we could not exclude effects of core+1 protein(s) on host cell factors or conditions that contribute to HCV replication in vivo and that are not detectable in Huh7 cells. Moreover, not every inactivation of viral functions leads to a phenotype in cell culture, as shown, e.g., for herpes simplex virus (41).
For these reasons, we studied the viability of the core+1 mutants in vivo by inoculating uPA-SCID mice xenografted with primary human hepatocytes. Consistent with our in vitro data, the JFH1 mut 5 virus exhibited replication kinetics like those of the wt in inoculated mice.
Apart from serving as an ORF for core and core+1, four highly conserved RNA stem-loop structures reside in the same coding region. By using a reverse-genetics approach, we found that disruption of the structural integrity of SL47 and SL87 inhibited RNA replication about 4- to 6-fold and reduced titers of infectious virus about 10- to 50-fold. In contrast, mutations affecting the more downstream RNA structures SL248 and SL443 had no effect. The difference in RNA replication between the wt and the SL47 or SL87 mutant were detectable only at early time points (24 h p.t.), whereas at later time points this difference vanished. This peculiar kinetics is due to the fact that RNA replication of JFH1 is most likely limited by the host cell (5). For this reason, more subtle impairments of replication are detectable only at early time points, while later, when replication becomes limited by the host cell, RNA amounts from an impaired genome accumulate to wt levels. This property is a general limitation of the highly replication-competent JFH1 isolate that should be kept in mind when analyzing more subtle replication phenotypes with this HCV genome.
An analogous delay of viremia was observed when the mut 1-5 genome was inoculated into uPA SCID mice with human liver xenografts. Sequence analysis of genomes isolated 12 weeks postinoculation demonstrated that the mutations affecting the upper stem-loop structure of SL87 had reverted in such a way that base pairing of the stem was restored. Thus, SL87 plays a very crucial role in HCV replication, most likely by enhancing RNA translation. However, the mutation disrupting translation of all known forms of core+1 protein(s) (mut 5) was stable, clearly showing that this protein(s) is irrelevant for viral replication in vivo.
We note that during prolonged passage of mut 1-5 in cell culture, all mutations were stable and no reversion was found over a 4-week observation period. In contrast, in inoculated mice, mut 1-5 was strongly attenuated, and the slow increase in viremia is consistent with the emergence of revertants. This discrepancy between results in cell culture and in vivo is most likely due to the host cell environment. For instance, cell factors contributing to SL87-dependent RNA translation/replication may be more abundant in Huh7 cells than in xenografted human liver cells, and therefore, SL87 mutants would be less impaired in cell culture. Alternatively, inhibitory factors binding to SL87 might be expressed to a lower level in Huh7 cells. More detailed studies, especially of possible cellular interaction partners with SL87, are required to address this possibility.
While this study was ongoing, McMullan and colleagues reported the characterization of a series of nonsense mutations disrupting translation of the core+1 ORF and altering RNA structure in the core coding region (34). These mutations correspond to mut 1-4 described in our study (codons 22/31/44/46 of the core+1 frame) and were tested in the context of an H77/JFH1 and a J6/JFH1 chimeric genome. However, mut 1 to 4 do not affect internal initiation of core+1 translation from codons 85/87. By including mut 5 in our study, we assessed the effect of a block of expression of all reported forms of the core+1 protein(s). Moreover, in contrast to the study by McMullan and coworkers, which focused on SL47 and SL87 only (designated SLV and SLVI, respectively, in their study), we also characterized the roles of the two downstream RNA elements SL248 and SL443 for HCV proliferation. In both studies, SL87 was found to be required for robust replication, but interestingly, our extensive substitution analysis revealed SL47 as an equally important RNA element. While we found that mutations disrupting SL47 and/or SL87 delay replication kinetics only transiently, McMullan et al. reported that mut 1 to 4 exhibited a dramatic and long-lasting defect in replication. During cell culture passage of the mutants, a mixture of reversions emerged in the case of the H77/JFH1 chimera concomitant with an increase in viral replication. Moreover, upon infection of a chimpanzee with this mutant, virus titers in the serum of the animal were low, the mutant appeared to be attenuated, and infection was controlled several weeks earlier than infection with a wt 1a isolate. In contrast, we found no evidence for adaptation in cell culture, and all mutations in mut 1-5 were conserved by week 4 p.t. This result is not too surprising, because in contrast to the study by McMullan et al., mut 1-5 was much less impaired in replication, and therefore, selective pressure was most likely not strong enough to enrich for revertants. However, upon infection of mice, mut 1-5 was clearly attenuated, and high-level viremia was delayed about 4 weeks compared to the wt. Thus, as in to the study by McMullan and coworkers, we also observed an attenuation of the mutant and a selection for revertants in vivo.
Our data cannot exclude the possibility that core+1 proteins perform a function that is not measurable in our cell culture or in vivo mouse model. The highly permissive nature of the Huh7.5 and Huh7-Lunet cells may be related to marked differences in the host environment relative to other liver cell lines and primary human hepatocytes that are naturally infected by HCV. Therefore, should core+1 proteins affect cellular genes that are already mutated in Huh7 subclones (and thus contribute to their permissiveness), core+1 mutants would behave like the wt in those cells. In addition, the JFH1 strain is unique among all HCV isolates and not necessarily representative, because it is the only isolate that replicates without requiring replication-enhancing mutations. JFH1 may contain genetic alterations that provide advantages for viral replication but at the same time make functions exerted by core+1 proteins obsolete. Finally, since the only in vivo model available to us is the xenografted Alb-uPA+/+ SCID mouse, our finding that lack of core+1 protein expression has no detectable effect on virus replication does not exclude the possibility that these proteins may have some other role, e.g., in immune evasion or pathogenesis. In fact, in the study by McMullan and colleagues, the mutant virus was clearly attenuated in the chimpanzee, with low-level viremia and rapid clearance (34).
Interestingly, our data and those of McMullan and coworkers can explain the high degree of evolutionary constraint on sequence alterations in the core region encompassing RNA elements SL47 and SL87 (44, 48, 49). These two stem-loops have been predicted by in silico analyses and supported by RNase cleavage studies. Most importantly, in this study, we show that SL47 and SL87 are not required for RNA stability but play important roles in RNA translation. The integrity of these RNA elements may be important for the regulation of HCV IRES activity either through a long-range RNA-RNA interaction(s) or through binding of a cellular protein(s) in the core encoding region. More specifically, both SL47 and SL87 may be involved in the stimulation of IRES function, e.g., by altering inhibitory interactions between the 5′ NTR and the core region (16, 59). In addition, a sequence at the 5′ end of SL87 (nt 428 to 442) anneals to nt 24 to 38 of the 5′ NTR, and this interaction restrains IRES-dependent translation (27). SL47 may also be involved in interaction with host cell factors, such as nonstructural 1-associated protein 1 (NSAP1), which is highly homologous to human heterogeneous nuclear ribonucleoprotein and which has been shown to enhance IRES translation (26). Finally, it is also possible that SL47 and SL87 are components of genome scale ordered RNA structure and may thus contribute to the conformation of the RNA genome, possibly modulating double-stranded RNA recognition that is associated with innate antiviral defense and virus persistence (43). We note that simultaneous disruption of the structural integrity of SL47 and SL87 impairs RNA translation and replication to the same extent as disruption of only one of these stem-loops. Assuming that the stem-loops participate in RNA-RNA or RNA-protein interactions, the disruption of only one of the stem-loops may be sufficient to block this interaction, and thus, no further impairment of translation/replication will be observed with mutations disrupting the second stem-loop.
In conclusion, we have shown that expression of the core+1 ORF plays no role in HCV replication and virus production in cell culture and xenografted mice. We confirmed that the core structures SL47 and SL87 are important cis-acting RNA elements required for HCV genome translation. These two stem-loop elements are crucial determinants for robust virus proliferation in cell culture and in vivo, thus providing an explanation for their striking evolutionary conservation.
Supplementary Material
Acknowledgments
We are grateful to Takaji Wakita for the JFH1 isolate, to Charles Rice for Huh7.5 cells and the 9E10 monoclonal antibody, and to Didier Trono and Dirk Lindemann for provision of the lentiviral transduction system. We also thank Ulrike Herian for excellent technical assistance and Sandra Bühler for editorial assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (BA1505/2-1 and Sonderforschungsbereich 638, and Teilprojekt A5, both to R.B.), a concerted action grant from UGent no. 01G00507, and Interuniversity Attraction Poles Program P6/36-HEPRO. P.M. is supported by a postdoctoral fellowship from the Research Foundation Flanders (FWO-Vlaanderen).
Footnotes
Published ahead of print on 17 September 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Appel, N., M. Zayas, S. Miller, J. Krijnse-Locker, T. Schaller, P. Friebe, S. Kallis, U. Engel, and R. Bartenschlager. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bain, C., P. Parroche, J. P. Lavergne, B. Duverger, C. Vieux, V. Dubois, F. Komurian-Pradel, C. Trepo, L. Gebuhrer, G. Paranhos-Baccala, F. Penin, and G. Inchauspe. 2004. Memory T-cell-mediated immune responses specific to an alternative core protein in hepatitis C virus infection. J. Virol. 7810460-10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baril, M., and L. Brakier-Gingras. 2005. Translation of the F protein of hepatitis C virus is initiated at a non-AUG codon in a +1 reading frame relative to the polyprotein. Nucleic Acids Res. 331474-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 6371-180. [DOI] [PubMed] [Google Scholar]
- 5.Binder, M., D. Quinkert, O. Bochkarova, R. Klein, N. Kezmic, R. Bartenschlager, and V. Lohmann. 2007. Identification of determinants involved in initiation of hepatitis C virus RNA synthesis by using intergenotypic replicase chimeras. J. Virol. 815270-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 2901972-1974. [DOI] [PubMed] [Google Scholar]
- 7.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 7613001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulant, S., M. Becchi, F. Penin, and J. P. Lavergne. 2003. Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J. Biol. Chem. 27845785-45792. [DOI] [PubMed] [Google Scholar]
- 9.Branch, A. D., D. D. Stump, J. A. Gutierrez, F. Eng, and J. L. Walewski. 2005. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin. Liver Dis. 25105-117. [DOI] [PubMed] [Google Scholar]
- 10.Branch, A. D., J. L. Walewski, J. A. Gutierrez, E. J. Fernandez, G. Y. Im, D. T. Dieterich, T. D. Schiano, S. H. Sigal, M. L. Schilsky, and M. E. Schwartz. 2003. HCV alternate reading frame proteins (ARFPs) may be virulence factors that help the virus survive adverse conditions. Hepatology 38468A-469A. [Google Scholar]
- 11.Choi, J., Z. Xu, and J. H. Ou. 2003. Triple decoding of hepatitis C virus RNA by programmed translational frameshifting. Mol. Cell. Biol. 231489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162156-159. [DOI] [PubMed] [Google Scholar]
- 13.Feld, J. J., and J. H. Hoofnagle. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436967-972. [DOI] [PubMed] [Google Scholar]
- 14.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 775487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honda, M., R. Rijnbrand, G. Abell, D. Kim, and S. M. Lemon. 1999. Natural variation in translational activities of the 5′ nontranslated RNAs of hepatitis C virus genotypes 1a and 1b: evidence for a long-range RNA-RNA interaction outside of the internal ribosomal entry site. J. Virol. 734941-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36S21-S29. [DOI] [PubMed] [Google Scholar]
- 18.Houghton, M., and S. Abrignani. 2005. Prospects for a vaccine against the hepatitis C virus. Nature 436961-966. [DOI] [PubMed] [Google Scholar]
- 19.Ina, Y., M. Mizokami, K. Ohba, and T. Gojobori. 1994. Reduction of synonymous substitutions in the core protein gene of hepatitis C virus. J. Mol. Evol. 3850-56. [DOI] [PubMed] [Google Scholar]
- 20.Jang, S. K. 2006. Internal initiation: IRES elements of picornaviruses and hepatitis C virus. Virus Res. 1192-15. [DOI] [PubMed] [Google Scholar]
- 21.Jirasko, V., R. Montserret, N. Appel, A. Janvier, L. Eustachi, C. Brohm, E. Steinmann, T. Pietschmann, F. Penin, and R. Bartenschlager. 2008. Structural and functional characterization of non-structural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. [DOI] [PMC free article] [PubMed]
- 22.Jones, C. T., C. L. Murray, D. K. Eastman, J. Tassello, and C. M. Rice. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 818374-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 1251808-1817. [DOI] [PubMed] [Google Scholar]
- 24.Kato, T., A. Furusaka, M. Miyamoto, T. Date, K. Yasui, J. Hiramoto, K. Nagayama, T. Tanaka, and T. Wakita. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J. Med. Virol. 64334-339. [DOI] [PubMed] [Google Scholar]
- 25.Kaul, A., I. Woerz, P. Meuleman, G. Leroux-Roels, and R. Bartenschlager. 2007. Cell culture adaptation of hepatitis C virus and in vivo viability of an adapted variant. J. Virol. 8113168-13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, J. H., K. Y. Paek, S. H. Ha, S. Cho, K. Choi, C. S. Kim, S. H. Ryu, and S. K. Jang. 2004. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 247878-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, Y. K., S. H. Lee, C. S. Kim, S. K. Seol, and S. K. Jang. 2003. Long-range RNA-RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA 9599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277570-574. [DOI] [PubMed] [Google Scholar]
- 29.Komurian-Pradel, F., A. Rajoharison, J. L. Berland, V. Khouri, M. Perret, M. Van Roosmalen, S. Pol, F. Negro, and G. Paranhos-Baccala. 2004. Antigenic relevance of F protein in chronic hepatitis C virus infection. Hepatology 40900-909. [DOI] [PubMed] [Google Scholar]
- 30.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 31.Lo, S. Y., M. Selby, M. Tong, and J. H. Ou. 1994. Comparative studies of the core gene products of two different hepatitis C virus isolates: two alternative forms determined by a single amino acid substitution. Virology 199124-131. [DOI] [PubMed] [Google Scholar]
- 32.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Salas, E., A. Pacheco, P. Serrano, and N. Fernandez. 2008. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 89611-626. [DOI] [PubMed] [Google Scholar]
- 34.McMullan, L. K., A. Grakoui, M. J. Evans, K. Mihalik, M. Puig, A. D. Branch, S. M. Feinstone, and C. M. Rice. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl. Acad. Sci. USA 1042879-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercer, D. F., D. E. Schiller, J. F. Elliott, D. N. Douglas, C. Hao, A. Rinfret, W. R. Addison, K. P. Fischer, T. A. Churchill, J. R. Lakey, D. L. Tyrrell, and N. M. Kneteman. 2001. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 7927-933. [DOI] [PubMed] [Google Scholar]
- 36.Meuleman, P., L. Libbrecht, R. De Vos, B. de Hemptinne, K. Gevaert, J. Vandekerckhove, T. Roskams, and G. Leroux-Roels. 2005. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology 41847-856. [DOI] [PubMed] [Google Scholar]
- 37.Moradpour, D., F. Penin, and C. M. Rice. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5453-463. [DOI] [PubMed] [Google Scholar]
- 38.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 1037408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poynard, T., M. F. Yuen, V. Ratziu, and C. L. Lai. 2003. Viral hepatitis C. Lancet 3622095-2100. [DOI] [PubMed] [Google Scholar]
- 40.Rijnbrand, R. C., and S. M. Lemon. 2000. Internal ribosome entry site-mediated translation in hepatitis C virus replication. Curr. Top. Microbiol. Immunol. 24285-116. [DOI] [PubMed] [Google Scholar]
- 41.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2460. In D. M. Knipe and P. M. Howley (ed.), Fields violrogy, 4th ed. Lippincot, Williams and Wilkins, Philadelphia, PA.
- 42.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. T. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42962-973. [DOI] [PubMed] [Google Scholar]
- 43.Simmonds, P., A. Tuplin, and D. J. Evans. 2004. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: Implications for virus evolution and host persistence. RNA 101337-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, D. B., and P. Simmonds. 1997. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J. Mol. Evol. 45238-246. [DOI] [PubMed] [Google Scholar]
- 45.Steinmann, E., F. Penin, S. Kallis, A. H. Patel, R. Bartenschlager, and T. Pietschmann. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tellinghuisen, T. L., K. L. Foss, and J. Treadaway. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tellinghuisen, T. L., K. L. Foss, J. C. Treadaway, and C. M. Rice. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 821073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuplin, A., D. J. Evans, and P. Simmonds. 2004. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J. Gen. Virol. 853037-3047. [DOI] [PubMed] [Google Scholar]
- 49.Tuplin, A., J. Wood, D. J. Evans, A. H. Patel, and P. Simmonds. 2002. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 8824-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van den Hoff, M. J., V. M. Christoffels, W. T. Labruyere, A. F. Moorman, and W. H. Lamers. 1995. Electrotransfection with “intracellular” buffer. Methods Mol. Biol. 48185-197. [DOI] [PubMed] [Google Scholar]
- 51.Van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: the VIIth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA.
- 52.Varaklioti, A., N. Vassilaki, U. Georgopoulou, and P. Mavromara. 2002. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J. Biol. Chem. 27717713-17721. [DOI] [PubMed] [Google Scholar]
- 53.Vassilaki, N., H. Boleti, and P. Mavromara. 2008. Expression studies of the HCV-1a core+1 open reading frame in mammalian cells. Virus Res. 133123-135. [DOI] [PubMed] [Google Scholar]
- 54.Vassilaki, N., K. I. Kalliampakou, and P. Mavromara. 2008. Differences in the expression of the hepatitis C virus core+1 open reading frame between a nuclear and a cytoplasmic expression system. J. Gen. Virol. 89222-231. [DOI] [PubMed] [Google Scholar]
- 55.Vassilaki, N., and P. Mavromara. 2003. Two alternative translation mechanisms are responsible for the expression of the HCV ARFP/F/core+1 coding open reading frame. J. Biol. Chem. 27840503-40513. [DOI] [PubMed] [Google Scholar]
- 56.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walewski, J. L., T. R. Keller, D. D. Stump, and A. D. Branch. 2001. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 7710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walewski, J. L., D. D. Stump, T. R. Keller, and A. D. Branch. 1998. HCV patients have antibodies against a novel protein encoded in a second reading frame. Hepatology 28278A. [Google Scholar]
- 59.Wang, T. H., R. C. Rijnbrand, and S. M. Lemon. 2000. Core protein-coding sequence, but not core protein, modulates the efficiency of cap-independent translation directed by the internal ribosome entry site of hepatitis C virus. J. Virol. 7411347-11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J. Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 203840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.