Abstract
Live attenuated vaccines against measles have been developed through adaptation of clinical isolates of measles virus (MV) in various cultured cells. Analyses using recombinant MVs with chimeric genomes between wild-type and Edmonston vaccine strains indicated that viruses possessing the polymerase protein genes of the Edmonston strain exhibited attenuated viral gene expression and growth in cultured cells as well as in mice expressing an MV receptor, signaling lymphocyte activation molecule, regardless of whether the virus genome had the wild-type or vaccine-type promoter sequence. These data demonstrate that the polymerase protein genes of the Edmonston strain contribute to its attenuated phenotype.
Measles is a highly contagious disease associated with high morbidity and mortality. Measles virus (MV), the causative agent of the disease, is an enveloped virus with a nonsegmented negative-strand RNA genome of ∼16 kb that is classified in the genus Morbillivirus in the family Paramyxoviridae (12). The genome is encapsidated by the nucleocapsid (N) protein and associated with a viral RNA-dependent RNA polymerase composed of two subunits, phosphoprotein (P protein) and large (L) protein. The genome and these proteins form a helical ribonucleoprotein complex that acts as the active template for transcription and replication (12). The incidence and mortality of measles have been remarkably reduced in many countries as the vaccine coverage rate increases (12).
Live attenuated MV vaccines have been generated via many rounds of passages of clinical isolates of MV in various cultured cells (12, 29). The Edmonston strain, isolated in 1954 (9), was used as a seed strain to generate live attenuated vaccines (17, 29). Although some of the molecular mechanisms by which Edmonston lineage vaccines have adapted to grow in various cultured cells have been elucidated (32, 33, 40), the mechanisms of their attenuation are poorly understood. In the present study, we generated various recombinant MVs with chimeric genomes of the Edmonston vaccine and wild-type strains of MV, and we found that viruses possessing the polymerase protein genes of the Edmonston strain exhibited attenuated MV gene expression and growth in cultured cells as well as in mice expressing a cellular receptor for MV, human signaling lymphocyte activation molecule (hSLAM) (21). These data provide a molecular basis for the avirulence of the Edmonston strain in vivo.
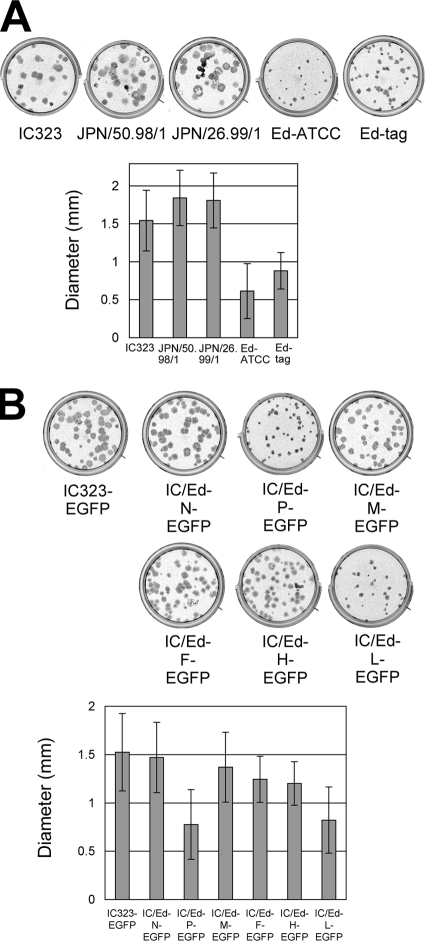
SLAM is a universal receptor for all MV strains, while CD46 functions as an additional receptor for vaccine strains but not for wild-type strains (41, 43). Thus, both wild-type and Edmonston strains replicate efficiently and produce high titers of progeny viruses in SLAM-expressing cells (10, 14). However, when plaque assays were performed, the Edmonston lineage strains produced smaller plaques than wild-type strains of MV (Fig. 1A). Wild-type strains of MV (JPN/50.98/1 and JPN/26.99/1) were isolated from patients with measles and passaged two or three times in B95a cells (15) to make stocks for experiments (kind gifts from H. Sakata). The Edmonston strain obtained from the American Type Culture Collection (ATCC) is designated the Ed-ATCC strain in this paper. Recombinant Edmonston tag (Ed-tag) (26) and wild-type IC323 (39) MV clones were recovered from cDNAs and grown in Vero/hSLAM cells (23). To clarify the genes responsible for the small-plaque-forming phenotype of the Edmonston strain, six recombinant MVs that each contained one of the Ed-tag strain genes in the backbone of the genome of the wild-type IC323 strain expressing enhanced green fluorescent protein (IC323-EGFP) (13) were prepared and designated IC/Ed-N-EGFP, IC/Ed-P-EGFP, IC/Ed-M-EGFP, IC/Ed-F-EGFP, IC/Ed-H-EGFP, and IC/Ed-L-EGFP. IC/Ed-N-EGFP, IC/Ed-P-EGFP, IC/Ed-F-EGFP, IC/Ed-H-EGFP, and IC/Ed-L-EGFP were reported previously and referred to as m2, m3, m7, m8, and m9, respectively (34). When the recombinant MVs were used to infect Vero/hSLAM cells, IC/Ed-P-EGFP and IC/Ed-L-EGFP produced smaller plaques than wild-type IC323-EGFP (Fig. 1B). These data indicate that the P and L genes of the Ed-tag strain contribute to the decreased plaque sizes produced by the Ed-tag strain. In addition to the P protein, the P gene encodes two accessory gene products, V and C (12). The V protein directly interferes with host interferon (IFN) induction and IFN signaling pathways, while the C protein modulates viral RNA synthesis to circumvent IFN induction (19). It should be noted that, unlike the V proteins of the Ed-ATCC and wild-type MV strains, the V protein of the Ed-tag strain is defective in counteracting IFN signaling pathways owing to tyrosine-to-histidine and cysteine-to-arginine substitutions at amino acid positions 110 and 272 (Y110H and C272R), respectively (5, 8, 11, 22). Substitutions at these positions are not unique to the Ed-tag strain and have been found in other vaccines and cultured cell-adapted MV strains, including the CAM-70 vaccine (4), the Changchun-47 vaccine (4), the chicken embryo fibroblast-adapted D-CEF strain (1), and the Vero cell-adapted 94YTV strain (containing the C272R substitution; our unpublished observation). These findings prompted us to carry out further experiments using the P gene of the Ed-tag strain.
FIG. 1.
Small plaque formed by the Edmonston strain and recombinant MVs with the P or L gene of the Edmonston strain on Vero/hSLAM cells. (A) Plaque assays were performed for various MV strains (three wild-type strains [IC323, JPN/50.98/1, and JPN/26.99/1] and two Edmonston lineage strains [Ed-ATCC and Ed-tag]). Monolayers of Vero/hSLAM cells on 12-well cluster plates were infected with 50 PFU of each virus and overlaid with Dulbecco's modified Eagle medium containing 2% fetal bovine serum and 1% methylcellulose. At 5 days p.i., the cells were stained with RTU Vectastain Elite ABC reagent (Vector Laboratories) using anti-MV H-protein monoclonal antibodies and a biotinylated secondary antibody. After high-resolution digital images were obtained, the sizes of all plaques were measured. The mean sizes ± standard deviations are shown in the bar graph. (B) Plaque assays were performed for wild-type IC323-EGFP and six recombinant MVs (each containing one of the Ed-tag strain genes in the backbone of the IC323-EGFP genome) on Vero/hSLAM cells, as described for panel A.
To quantify the expression levels of MV genes, four recombinant MVs expressing Renilla luciferase instead of EGFP were generated. The wild-type IC323 strain expressing Renilla luciferase (IC323-Luci) was reported previously (38). The P and/or L genes of the Ed-tag strain were introduced into the IC323-Luci genome, thereby generating IC/Ed-P-Luci, IC/Ed-L-Luci, and IC/Ed-PL-Luci. The Renilla luciferase activities induced by these recombinant MVs were analyzed in various cell types expressing SLAM (Vero/hSLAM [23], CV1/hSLAM, HeLa/hSLAM, A549/hSLAM [36], CHO/hSLAM [41], B95a, MT2, Raji, Ramos, BJAB-B95-8, and C91/PL) or an unidentified MV receptor on epithelial cells (NCI-H358) (38). CV-1/hSLAM and HeLa/hSLAM cells were generated by cotransfecting CV1 and HeLa cells with the eukaryotic expression vector pCA7 (36), a derivative of pCAGGS (20) encoding human SLAM, and the selection vector pCXN2 (20), encoding a neomycin resistance gene, followed by selection in the presence of Geneticin (G418; Nacalai Tesque). Among all these cell lines, IC/Ed-P-Luci and IC/Ed-L-Luci showed reduced Renilla luciferase expression levels compared with wild-type IC323-Luci (Fig. 2A and data not shown). The luciferase activities induced by IC/Ed-PL-Luci were even lower than those induced by IC/Ed-P-Luci and IC/Ed-L-Luci among all the cell lines examined (Fig. 2A and data not shown). These data indicate that introduction of the P and L genes of the Ed-tag strain into recombinant virus genomes reduces viral gene expression and growth, regardless of the cell type. The viral mRNA levels in Vero/hSLAM cells infected with the recombinant MVs were also quantified. Confluent monolayers of Vero/hSLAM cells in 6-cm culture plates were infected with 1.0 × 104 PFU of IC323-Luci, IC/Ed-P-Luci, IC/Ed-L-Luci, or IC/Ed-PL-Luci. Multiple rounds of infection were blocked by a fusion-blocking peptide (Peptide Institute) (28). At 18 h postinfection (p.i.), mRNAs were purified from the cells using TRIzol reagent (Invitrogen) and an Oligotex-dT30 mRNA purification kit (TaKaRa Bio Inc.), reverse transcribed into cDNAs with an iScript cDNA synthesis kit (Promega), and subjected to PCR using SYBR Premix Ex Taq II (TaKaRa Bio Inc.). The primers used to amplify the MV genes were described previously (36). β-Actin mRNA was also quantified and used as an internal control (Fig. 2B). The levels of the viral transcripts (N, P, M, F, H, and L mRNAs, which show a transcriptional gradient [6, 7, 25]) were reduced ∼10-fold in IC/Ed-P-Luci- and IC/Ed-L-Luci-infected cells, compared with the levels in IC323-Luci-infected cells (Fig. 2B). The viral transcript levels were even further reduced in IC/Ed-PL-Luci-infected cells, being about 100-fold lower than those in IC323-Luci-infected cells (Fig. 2B). These data show that the small-plaque-forming phenotype of the Edmonston strain is at least partly caused by reduced viral gene expression levels in cells. We also analyzed the activities of the P and L proteins using a minigenome assay (18, 19). All combinations of the P- and L-protein-expressing plasmids of the wild-type IC323 and Ed-tag strains produced equivalent Renilla luciferase activities (Fig. 2C). The discrepancy between the data obtained with the minigenome assays and the data obtained using recombinant viruses may be attributed to differences in the gene expression systems. In minigenome assays, the components required for viral RNA synthesis (N, P, and L proteins) are continuously supplied in trans from expression plasmids, whereas in virus infections, MV has to synthesize these proteins by itself to continue viral RNA synthesis. Furthermore, in virus infections, but not in minigenome assays, MV produces the C, V, and matrix (M) proteins, which modulate viral RNA synthesis (3, 24, 27, 31, 42).
FIG. 2.
Attenuated gene expression by recombinant MVs with the P and/or L genes of the Edmonston strain. (A) Confluent monolayers of various cell lines (Vero/hSLAM, CV1/hSLAM, HeLa/hSLAM, CHO/hSLAM, and B95a) and suspensions of nonadherent MT2 cells cultured in 24-well cluster plates were infected with 2.5 × 103 PFU of IC323-Luci (circles), IC/Ed-P-Luci (squares), IC/Ed-L-Luci (triangles), and IC/Ed-PL-Luci (diamonds). After various intervals, the Renilla luciferase activities were measured. Data are means ± standard deviations for triplicate samples. RLU, relative light units. (B) Confluent monolayers of Vero/hSLAM cells in 6-cm culture plates were infected with 1.0 × 104 PFU of IC323-Luci (light gray bars), IC/Ed-P-Luci (black bars), IC/Ed-L-Luci (dark gray bars), and IC/Ed-PL-Luci (white bars) and cultured in the presence of a fusion-blocking peptide. At 18 h p.i., mRNAs were purified from the cells, and the levels of N, P, M, F, H, and L mRNAs were determined by reverse transcription-quantitative PCR. Data are means ± standard deviations for triplicate samples. (C) Minigenome assays. The method was described in detail elsewhere (19). Monolayers of CHO/hSLAM cells cultured in Opti-MEM on 24-well plates were infected with vTF7-3 at a multiplicity of infection of 0.5 and then transfected with 0.2 μg of p18MGFLuc01-wt-Le, 0.2 μg of pCAG-T7-IC-N, 0.3 μg of a P-protein expression plasmid (pCAG-T7-IC-PΔC or -Ed-PΔC), and 0.2 μg of an L-protein expression plasmid (pGEMCR-IC-L or -Ed-L) using Lipofectamine 2000 (Invitrogen). At 6 h p.i., the culture media were replaced with RPMI 1640 medium supplemented with 7.5% fetal bovine serum. At 48 h p.i., the firefly luciferase activities were measured. (-), L-protein expression plasmid was omitted. Data are means ± standard deviations for triplicate samples.
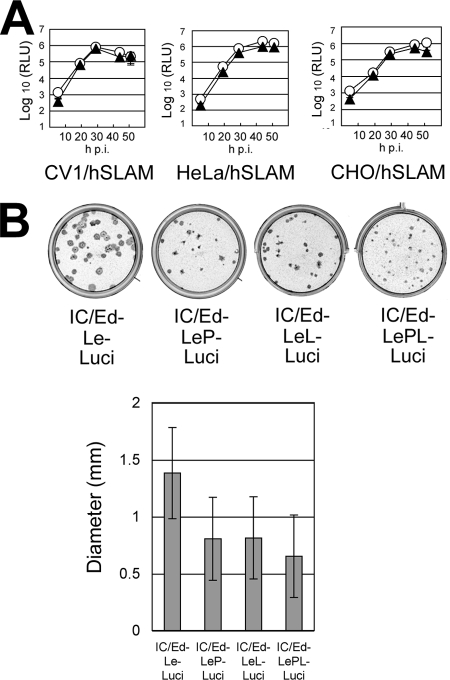
Compared with the genomes of wild-type MV strains, the genomes of the Edmonston vaccine strains have three nucleotide substitutions (uracil-to-adenine, uracil-to-guanine, and uracil-to-cytosine changes at nucleotide positions 26, 42, and 50, respectively) in the genomic promoter (leader [Le]) region (16, 37). Using minigenome assays, these substitutions have been shown to enhance the promoter activity (16). To analyze the effects of these substitutions on virus infection, they were introduced into the IC323-Luci genome, generating IC/Ed-Le-Luci. The substitutions were also introduced into the IC/Ed-P-Luci, IC/Ed-L-Luci, and IC/Ed-PL-Luci genomes, thereby generating IC/Ed-LeP-Luci, IC/Ed-LeL-Luci, and IC/Ed-LePL-Luci, respectively. The Renilla luciferase activities induced by IC323-Luci and IC/Ed-Le-Luci were compared in the 12 cell lines described above. No significant differences between the Renilla luciferase activities of IC323-Luci- and IC/Ed-Le-Luci-infected cells were observed for any of the 12 cell lines (Fig. 3A and data not shown). These data reveal that the three substitutions have neutral effects on the promoter function when introduced into the wild-type MV genome. Regardless of the presence or absence of these substitutions in the Le region, the P and L genes of the Ed-tag strain, especially when combined, reduced the plaque sizes of recombinant MVs (Fig. 3B).
FIG. 3.
Neutral effects of substitutions in the leader region of the Edmonston strain on MV gene expression levels and plaque sizes. (A) Confluent monolayers of CV1/hSLAM, HeLa/hSLAM, and CHO/hSLAM cells cultured in 24-well cluster plates were infected with 2.5 × 103 PFU of IC323-Luci (circles) and IC/Ed-Le-Luci (triangles). After various intervals, the Renilla luciferase activities were measured. Data are means ± standard deviations for triplicate samples. RLU, relative light units. (B) Plaque assays were performed for the recombinant MVs, as described for Fig. 1A. The IC/Ed-Le-Luci, IC/Ed-LeP-Luci, IC/Ed-LeL-Luci, and IC/Ed-LePL-Luci genomes contain Ed-tag genome regions encoding Le alone, Le and P, Le and L, or Le, P, and L, respectively, in the backbone of the IC323-Luci genome.
It is now clear that the P and L genes of the Ed-tag strain attenuate MV gene expression when introduced into recombinant virus genomes. To exclude possible growth-inhibitory effects caused by the inability of the Ed-tag V protein to block IFN signaling due to the Y110H and C272R substitutions (5, 8, 11, 22), the histidine and arginine residues at these positions were replaced with the corresponding residues of wild-type MV (tyrosine and cysteine, respectively), generating Ed-tag-VH110Y/R272C. An additional transcriptional unit for Renilla luciferase was also created in the Ed-tag strain genome, generating Ed-tag-VH110Y/R272C-Luci. Ed-tag-VH110Y/R272C-Luci still produced small plaques, with sizes equivalent to those of plaques produced by the original Ed-tag strain (Fig. 1A and 4A). The Renilla luciferase activities induced by Ed-tag-VH110Y/R272C-Luci were analyzed and compared with those induced by wild-type IC323-Luci in various cell lines described above (Fig. 4B and data not shown). The wild-type and Edmonston strains have been shown to enter SLAM-positive cells with similar efficiencies (41, 43). Nonetheless, Ed-tag-VH110Y/R272C-Luci induced 8- to 50-fold-lower Renilla luciferase activities in SLAM-positive cell lines than IC323-Luci (Fig. 4B). These data further confirmed the reduced gene expression levels induced by the Edmonston strain.
FIG. 4.
Attenuated gene expression by the Edmonston strain in cultured cells and SLAM-knock-in mice. (A) Plaque assays were performed for IC323-Luci and Ed-tag-VH110Y/R272C-Luci on Vero/hSLAM cells, as described for Fig. 1A. (B) Confluent monolayers of HeLa/hSLAM, A549/hSLAM, CV1/hSLAM, B95a, and CHO/hSLAM cultured in 12-well cluster plates were infected with 2.5 × 103 PFU of IC323-Luci (black bars) and Ed-tag-VH110Y/R272C-Luci (gray bars). At 30 h p.i., the Renilla luciferase activities were measured. Data are means ± standard deviations for triplicate samples. Mock-infected samples are shown by white bars. RLU, relative light units. (C) IC323-Luci, IC/Ed-L-Luci, and Ed-tag-VH110Y/R272C-Luci (1.5 × 105 PFU) were injected into the peritoneal cavities of IFNAR1−/− SLAM-knock-in mice. At 5 days p.i., the spleens were removed and analyzed for their Renilla luciferase activities. Each symbol indicates the sample from a single mouse, and bars indicate the median values. RLU, relative light units. *, P < 0.05; **, P < 0.001.
Aliquots (1.0 ml) containing 1.5 × 105 PFU of recombinant MVs (IC323-Luci, IC/Ed-L-Luci, and Ed-tag-VH110Y/R272C-Luci) were injected into the peritoneal cavities of SLAM-knock-in mice crossed with mice lacking type I IFN receptor subunit 1 (IFNAR1−/− SLAM-knock-in mice) (21). Eight mice were used for each recombinant MV. At 5 days p.i., the spleens were removed, homogenized using a BioMasher (Hi-Tech Inc.), and analyzed for their Renilla luciferase activities. The median Renilla luciferase activity in spleens from mice infected with Ed-tag-VH110Y/R272C-Luci was ∼10,000-fold lower than that in spleens from mice infected with wild-type IC323-Luci (P < 0.001; Fig. 4C). These data clearly indicate that the Edmonston strain is severely attenuated in IFNAR1−/− SLAM-knock-in mice. The median Renilla luciferase activity in spleens from IC/Ed-L-Luci-infected mice was ∼200-fold lower than that in spleens from mice infected with wild-type IC323-Luci (P < 0.001; Fig. 4C). These data indicate that the L gene of the Edmonston strain contributes to the in vivo attenuation of the Edmonston strain.
Many viruses can be adapted to various cultured cells by passaging in the cells, and it is empirically known that these adaptations often reduce virus virulence in natural host animals (15). Previous studies indicated that MV can adapt to grow in some cultured cells by acquiring specific substitutions in the receptor-binding hemagglutinin (H) and/or M proteins (43). Although some mechanisms by which these changes in the H and M proteins may cause attenuation have been proposed (30, 33), their contributions in vivo remain to be determined in animal models. Consistent with the present data, our previous study using a wild-type MV strain and its Vero cell-adapted strain suggested that substitutions introduced into the polymerase protein genes (L and P) during passages in Vero cells caused MV attenuation by reducing the transcriptional activities of viral polymerase (35). However, we observed attenuated gene expression levels by the Edmonston strain only in virus infection analyses, not in minigenome assays. Bankamp et al. (2) also reported that the polymerase proteins of MV vaccine strains show higher transcriptional activities than those of wild-type MV strains when analyzed by minigenome assays. The detailed mechanisms of the attenuated gene expression induced by the polymerase protein genes of the Edmonston strain remain to be elucidated.
In conclusion, the present study demonstrates that the polymerase protein genes of the Edmonston strain contribute to its attenuated phenotype. Our data further show that assays using infectious recombinant viruses are crucial for understanding the contribution of each viral gene to virus replication and virulence and that the SLAM-knock-in mouse is a useful animal model for elucidating the attenuation mechanisms of MV vaccines.
Acknowledgments
We thank M. A. Billeter and K. Komase for providing the p(+)MV2A plasmid and p18MGFLuc01. We also thank H. Sakata and T. A. Sato for providing the clinical isolates of MV and monoclonal antibodies against MV H protein.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labor and Welfare of Japan.
Footnotes
Published ahead of print on 17 September 2008.
REFERENCES
- 1.Bankamp, B., G. Hodge, M. B. McChesney, W. J. Bellini, and P. A. Rota. 2008. Genetic changes that affect the virulence of measles virus in a rhesus macaque model. Virology 37339-50. [DOI] [PubMed] [Google Scholar]
- 2.Bankamp, B., S. P. Kearney, X. Liu, W. J. Bellini, and P. A. Rota. 2002. Activity of polymerase proteins of vaccine and wild-type measles virus strains in a minigenome replication assay. J. Virol. 767073-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bankamp, B., J. Wilson, W. J. Bellini, and P. A. Rota. 2005. Identification of naturally occurring amino acid variations that affect the ability of the measles virus C protein to regulate genome replication and transcription. Virology 336120-129. [DOI] [PubMed] [Google Scholar]
- 4.Borges, M. B., E. Caride, A. V. Jabor, J. M. Malachias, M. S. Freire, A. Homma, and R. Galler. 2008. Study of the genetic stability of measles virus CAM-70 vaccine strain after serial passages in chicken embryo fibroblasts primary cultures. Virus Genes 3635-44. [DOI] [PubMed] [Google Scholar]
- 5.Caignard, G., M. Guerbois, J. L. Labernardiere, Y. Jacob, L. M. Jones, F. Wild, F. Tangy, and P. O. Vidalain. 2007. Measles virus V protein blocks Jak1-mediated phosphorylation of STAT1 to escape IFN-alpha/beta signaling. Virology 368351-362. [DOI] [PubMed] [Google Scholar]
- 6.Cattaneo, R., G. Rebmann, K. Baczko, V. ter Meulen, and M. A. Billeter. 1987. Altered ratios of measles virus transcripts in diseased human brains. Virology 160523-526. [DOI] [PubMed] [Google Scholar]
- 7.Cattaneo, R., G. Rebmann, A. Schmid, K. Baczko, V. ter Meulen, and M. A. Billeter. 1987. Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 6681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devaux, P., V. von Messling, W. Songsungthong, C. Springfeld, and R. Cattaneo. 2007. Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virology 36072-83. [DOI] [PubMed] [Google Scholar]
- 9.Enders, J. F., and T. C. Peebles. 1954. Propagation in tissue cultures of cytopathic agents from patients with measles. Proc. Soc. Exp. Biol. Med. 86277-286. [DOI] [PubMed] [Google Scholar]
- 10.Erlenhofer, C., W. Duprex, B. Rima, V. ter Meulen, and J. Schneider-Schaulies. 2002. Analysis of receptor (CD46, CD150) usage by measles virus. J. Gen. Virol. 831431-1436. [DOI] [PubMed] [Google Scholar]
- 11.Fontana, J. M., B. Bankamp, W. J. Bellini, and P. A. Rota. 2008. Regulation of interferon signaling by the C and V proteins from attenuated and wild-type strains of measles virus. Virology 37471-81. [DOI] [PubMed] [Google Scholar]
- 12.Griffin, D. E. 2007. Measles virus, p. 1551-1585. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 13.Hashimoto, K., N. Ono, H. Tatsuo, H. Minagawa, M. Takeda, K. Takeuchi, and Y. Yanagi. 2002. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J. Virol. 766743-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston, I. C. D., V. ter Meulen, J. Schneider-Schaulies, and S. Schneider-Schaulies. 1999. A recombinant measles vaccine virus expressing wild-type glycoproteins: consequences for viral spread and cell tropism. J. Virol. 736903-6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobune, F., H. Sakata, and A. Sugiura. 1990. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J. Virol. 64700-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, X., B. Bankamp, W. Xu, W. J. Bellini, and P. A. Rota. 2006. The genomic termini of wild-type and vaccine strains of measles virus. Virus Res. 12278-84. [DOI] [PubMed] [Google Scholar]
- 17.Moss, W. J., and D. E. Griffin. 2006. Global measles elimination. Nat. Rev. Microbiol. 4900-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakatsu, Y., M. Takeda, M. Kidokoro, M. Kohara, and Y. Yanagi. 2006. Rescue system for measles virus from cloned cDNA driven by vaccinia virus Lister vaccine strain. J. Virol. Methods 137152-155. [DOI] [PubMed] [Google Scholar]
- 19.Nakatsu, Y., M. Takeda, S. Ohno, Y. Shirogane, M. Iwasaki, and Y. Yanagi. 2008. Measles virus circumvents the host interferon response by different actions of the C and V proteins. J. Virol. 828296-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108193-199. [DOI] [PubMed] [Google Scholar]
- 21.Ohno, S., N. Ono, F. Seki, M. Takeda, S. Kura, T. Tsuzuki, and Y. Yanagi. 2007. Measles virus infection of SLAM (CD150) knockin mice reproduces tropism and immunosuppression in human infection. J. Virol. 811650-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno, S., N. Ono, M. Takeda, K. Takeuchi, and Y. Yanagi. 2004. Dissection of measles virus V protein in relation to its ability to block alpha/beta interferon signal transduction. J. Gen. Virol. 852991-2999. [DOI] [PubMed] [Google Scholar]
- 23.Ono, N., H. Tatsuo, Y. Hidaka, T. Aoki, H. Minagawa, and Y. Yanagi. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 754399-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parks, C. L., S. E. Witko, C. Kotash, S. L. Lin, M. S. Sidhu, and S. A. Udem. 2006. Role of V protein RNA binding in inhibition of measles virus minigenome replication. Virology 34896-106. [DOI] [PubMed] [Google Scholar]
- 25.Plumet, S., W. P. Duprex, and D. Gerlier. 2005. Dynamics of viral RNA synthesis during measles virus infection. J. Virol. 796900-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 145773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reutter, G. L., C. Cortese-Grogan, J. Wilson, and S. A. Moyer. 2001. Mutations in the measles virus C protein that up regulate viral RNA synthesis. Virology 285100-109. [DOI] [PubMed] [Google Scholar]
- 28.Richardson, C. D., A. Scheid, and P. W. Choppin. 1980. Specific inhibition of paramyxovirus and myxovirus replication by oligopeptides with amino acid sequences similar to those at the N-termini of the F1 or HA2 viral polypeptides. Virology 105205-222. [DOI] [PubMed] [Google Scholar]
- 29.Rota, J. S., Z. D. Wang, P. A. Rota, and W. J. Bellini. 1994. Comparison of sequences of the H, F, and N coding genes of measles virus vaccine strains. Virus Res. 31317-330. [DOI] [PubMed] [Google Scholar]
- 30.Schnorr, J., L. Dunster, R. Nanan, J. Schneider-Schaulies, S. Schneider-Schaulies, and V. ter Meulen. 1995. Measles virus-induced down-regulation of CD46 is associated with enhanced sensitivity to complement-mediated lysis of infected cells. Eur. J. Immunol. 25976-984. [DOI] [PubMed] [Google Scholar]
- 31.Suryanarayana, K., K. Baczko, V. ter Meulen, and R. R. Wagner. 1994. Transcription inhibition and other properties of matrix proteins expressed by M genes cloned from measles viruses and diseased human brain tissue. J. Virol. 681532-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tahara, M., M. Takeda, F. Seki, T. Hashiguchi, and Y. Yanagi. 2007. Multiple amino acid substitutions in hemagglutinin are necessary for wild-type measles virus to acquire the ability to use receptor CD46 efficiently. J. Virol. 812564-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tahara, M., M. Takeda, and Y. Yanagi. 2007. Altered interaction of the matrix protein with the cytoplasmic tail of hemagglutinin modulates measles virus growth by affecting virus assembly and cell-cell fusion. J. Virol. 816827-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tahara, M., M. Takeda, and Y. Yanagi. 2005. Contributions of matrix and large protein genes of the measles virus Edmonston strain to growth in cultured cells as revealed by recombinant viruses. J. Virol. 7915218-15225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeda, M., A. Kato, F. Kobune, H. Sakata, Y. Li, T. Shioda, Y. Sakai, M. Asakawa, and Y. Nagai. 1998. Measles virus attenuation associated with transcriptional impediment and a few amino acid changes in the polymerase and accessory proteins. J. Virol. 728690-8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda, M., S. Ohno, F. Seki, Y. Nakatsu, M. Tahara, and Y. Yanagi. 2005. Long untranslated regions of the measles virus M and F genes control virus replication and cytopathogenicity. J. Virol. 7914346-14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeda, M., T. Sakaguchi, Y. Li, F. Kobune, A. Kato, and Y. Nagai. 1999. The genome nucleotide sequence of a contemporary wild strain of measles virus and its comparison with the classical Edmonston strain genome. Virology 256340-350. [DOI] [PubMed] [Google Scholar]
- 38.Takeda, M., M. Tahara, T. Hashiguchi, T. A. Sato, F. Jinnouchi, S. Ueki, S. Ohno, and Y. Yanagi. 2007. A human lung carcinoma cell line supports efficient measles virus growth and syncytium formation via a SLAM- and CD46-independent mechanism. J. Virol. 8112091-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeda, M., K. Takeuchi, N. Miyajima, F. Kobune, Y. Ami, N. Nagata, Y. Suzaki, Y. Nagai, and M. Tashiro. 2000. Recovery of pathogenic measles virus from cloned cDNA. J. Virol. 746643-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatsuo, H., K. Okuma, K. Tanaka, N. Ono, H. Minagawa, A. Takade, Y. Matsuura, and Y. Yanagi. 2000. Virus entry is a major determinant of cell tropism of Edmonston and wild-type strains of measles virus as revealed by vesicular stomatitis virus pseudotypes bearing their envelope proteins. J. Virol. 744139-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tatsuo, H., N. Ono, K. Tanaka, and Y. Yanagi. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406893-897. [DOI] [PubMed] [Google Scholar]
- 42.Witko, S. E., C. Kotash, M. S. Sidhu, S. A. Udem, and C. L. Parks. 2006. Inhibition of measles virus minireplicon-encoded reporter gene expression by V protein. Virology 348107-119. [DOI] [PubMed] [Google Scholar]
- 43.Yanagi, Y., M. Takeda, and S. Ohno. 2006. Measles virus: cellular receptors, tropism and pathogenesis. J. Gen. Virol. 872767-2779. [DOI] [PubMed] [Google Scholar]