Abstract
Two members of the paramyxovirus family, Nipah virus (NiV) and Hendra virus (HeV), are recent additions to a growing number of agents of emergent diseases which use bats as a natural host. Identification of ephrin-B2 and ephrin-B3 as cellular receptors for these viruses has enabled the development of immunotherapeutic reagents which prevent virus attachment and subsequent fusion. Here we present the structural analysis of the protein and carbohydrate components of the unbound viral attachment glycoprotein of NiV glycoprotein (NiV-G) at a 2.2-Å resolution. Comparison with its ephrin-B2-bound form reveals that conformational changes within the envelope glycoprotein are required to achieve viral attachment. Structural differences are particularly pronounced in the 579-590 loop, a major component of the ephrin binding surface. In addition, the 236-245 loop is rather disordered in the unbound structure. We extend our structural characterization of NiV-G with mass spectrometric analysis of the carbohydrate moieties. We demonstrate that NiV-G is largely devoid of the oligomannose-type glycans that in viruses such as human immunodeficiency virus type 1 and Ebola virus influence viral tropism and the host immune response. Nevertheless, we find putative ligands for the endothelial cell lectin, LSECtin. Finally, by mapping structural conservation and glycosylation site positions from other members of the paramyxovirus family, we suggest the molecular surface involved in oligomerization. These results suggest possible pathways of virus-host interaction and strategies for the optimization of recombinant vaccines.
The emergence of highly virulent pathogens from previously undisturbed ecological niches is considered an increasing threat (58). This is exemplified by the recent emergence throughout Southeast Asia and Australia of Nipah virus (NiV) and Hendra virus (HeV), zoonotic paramyxoviruses characterized by high mortality rates. Outbreaks of these viruses originate from the fruit bat and are usually triggered by contamination of food and water or by direct contact with infected animals (39). NiV was first detected in 1998 in Malaysia, where it was transmitted from pigs to humans and resulted in the culling of over 1 million pigs to contain the outbreak (39). The first outbreak of HeV occurred in 1994 in the Brisbane suburb of Hendra, Australia, where infected horses transmitted the virus to humans (18, 52). Symptoms of infection for both of these viruses include acute encephalitis and respiratory illness, and the time from onset to death is usually 7 to 10 days. Due to their high mortality rates and rapid emergence, NiV and HeV have been designated biosafety level 4 pathogens by the Centers for Disease Control and Prevention (Atlanta, GA).
NiV and HeV are enveloped, single-stranded, negative-sense RNA viruses which constitute the Henipavirus (HNV) genus in the Paramoxyviridae family. Henipaviruses enter their host cell by a pH-independent mechanism utilizing two outer membrane proteins: HNV-G for cell attachment and HNV-F for fusion. NiV-G and HeV-G are oligomeric type II transmembrane glycoproteins composed of an N-terminal cytoplasmic tail (approximately 50 amino acids), a transmembrane domain (23 amino acids), a stalk region (110 amino acids), and a C-terminal globular head domain (approximately 420 amino acids). Analytical ultracentrifugation and size exclusion chromatography have demonstrated that the NiV-G ectodomain is predominantly tetrameric, although some lower-order species were also observed (7). On the virion surface, HNV-G associates with HNV-F through the stalk region, facilitating entry into the host (19).
Unlike the closely related parainfluenza viruses (PIVs) and Newcastle disease virus (NDV), which enter their hosts via sialic (neuraminic) acid-mediated attachment, henipaviruses use ephrin-B2 (EFNB2) and EFNB3 cell surface glycoproteins as high-affinity functional receptors (4, 44, 45). This correlates with the broad tissue tropism of these viruses, since ephrins are widely expressed in neurons, bone, stem cells, and epithelial cells and across the immune system and are used in many signaling processes underlying axon guidance, vascular development, and osteogenesis (31, 49). In addition, the ability of these viruses to infect a wide range of hosts is reflected in the broad species conservation of the ephrin ligands, which are expressed and well conserved (>95% sequence identity) in many vertebrates, including bats, horses, and pigs (6). Recent crystal structures of NiV-G and HeV-G in complex with EFNB2 revealed a highly conserved binding mode (7). Vaccination with recombinant HNV-G in animal models generates a neutralizing antibody response to protein surfaces utilized for henipavirus attachment (5, 22). These neutralizing antibodies can provide passive immunity to henipavirus (23, 62). The structural similarity of EFNB2 binding by NiV-G and HeV-G underscores the potential for the development for a single vaccine that protects against challenge from both viruses (5, 7). Encouragingly, recombinant HeV-G can protect against NiV infection in cats (42).
The use of proteins as functional receptors for host attachment within the Paramyxoviridae is not limited to henipaviruses. Morbillaviruses such as measles, canine distemper, and rinderpest viruses have recently been shown to require SLAM (CD151) for viral attachment (59). Structure-based phylogenetic analysis, however, shows that HNV-G is structurally more similar to attachment glycoproteins of sialic acid binding viruses such as NDV and PIVs (37, 60) than to that of measles virus (MV) (7, 12, 29), suggesting that the protein binding capacities of henipaviruses and morbilliviruses have evolved independently.
While the crystal structures of NiV-G and HeV-G in complex with EFNB2 reveal an interface dominated by protein-protein interactions, the carbohydrate moieties of many viral envelope glycoproteins can influence tissue tropism through interaction with host cell surface lectins (8), increasing the concentration of the virus at the cell surface and colocalizing it with cell attachment proteins. The β-propeller domain of NiV-G contains five N-linked glycosylation sequons (NXS/TX, where X is any amino acid except proline). N-linked glycosylation occurs in the endoplasmic reticulum and, through the calnexin/calreticulin folding pathways, can be critical for protein folding and expression (48). For mammalian glycoproteins, the immature oligomannose-type glycans are usually processed to complex-type structures in the Golgi apparatus (33). However, processing of viral glycosylation is not always complete, and many enveloped viruses contain immature oligomannose-type glycans. Oligomannose-type glycans are largely absent from the host cell surface (8), and their presence on pathogens can be a signal to the host immune system through recognition by serum and cell surface lectins (8). For example, in human immunodeficiency virus type 1 (HIV-1), a cluster of oligomannose-type glycans promotes infection through interaction with the C-type lectin DC-SIGN (8). Such oligomannose-type glycans can also upregulate the host immune response by activating the complement cascade through interaction with the serum mannose binding protein. Furthermore, oligomannose-type glycans can act as ligands for the macrophage mannose receptor, promoting antigen presentation. This mannose receptor targeting can be exploited in the optimization of vaccines (32).
To facilitate the development of antiviral drugs, we determined the crystal structure of the unbound globular domain of NiV-G. Furthermore, we extended our structural analysis of NiV-G with mass spectrometric (MS) analysis of the carbohydrate moiety. Together with mapping of the glycosylation sites from across the paramyxoviruses, our analyses suggest strategies for the optimization of recombinant vaccines.
MATERIALS AND METHODS
Protein expression and purification.
NiV-G (residues 183 to 602, GenBank accession number NC_002728; synthesized by GeneArt, Regensburg, Germany) was expressed and purified as previously described (7). Briefly, cDNA was cloned into the pHLsec vector (1) and transiently expressed in human embryonic kidney (HEK) 293T cells in the presence of the class I α-mannosidase inhibitor, kifunensine (Toronto Research Chemicals, Ontario, Canada) (9), using 2 mg DNA per liter cell culture. Protein was purified by immobilized metal affinity chromatography and then treated with endoglycosidase F1 (75 μg mg−1 protein, 12 h, 21°C) to release the N-linked glycans (see Fig. S1 in the supplemental material). Following partial deglycosylation, protein complexes were purified by size exclusion chromatography using a Superdex 200 10/30 column (Amersham, United Kingdom), in 150 mM NaCl-10 mM Tris (pH 8.0) buffer. Typical protein yields were about 2 mg of pure, deglycosylated NiV-G per liter cell culture.
Crystallization and structure determination.
Crystals were grown by sitting-drop vapor diffusion using 100 nl protein plus 100 nl precipitant as described previously (56). NiV-G crystals grew at 4°C (6.5 mg ml−1) in 20% (vol/vol) polyethylene glycol 6000, 0.1 M MES (morpholineethanesulfonic acid) (pH 6.0), 0.1 M LiCl, and 18% γ-butylactone after 3 days. Crystals were flash frozen by immersion in a cryoprotectant containing 25% (vol/vol) ethylene glycol followed by rapid transfer to a gaseous nitrogen stream. Data were collected at beamline ID23-2 at the European Synchotron Radiation Facility, Grenoble, France. Images were integrated and scaled using the programs DENZO and SCALEPACK (46). Details are presented in Table 1.
TABLE 1.
NiV-G crystallographic and structural data
| Parameter (unit) | Valuea |
|---|---|
| Data collection | |
| Resolution (Å) | 40.0-2.25 |
| Space group | P21 |
| Cell dimensions (Å) and angles (°) | a = 71.6, b = 86.9, c = 82.9, α = 90, β = 108.3, γ = 90 |
| No. of unique reflections | 45,810 (4,555) |
| Completeness (%) | 100 (100) |
| Rmerge (%)b | 8.6 (52.5) |
| I/σI | 18.4 (3.1) |
| Avg redundancy | 7.3 (5.8) |
| Refinement | |
| Resolution range (Å) | 40.0-2.25 |
| No. of reflections | 43,463 |
| Rfactor (%)c | 17.2 |
| Rfree (%)d | 22.1 |
| RMSDe | |
| Bonds (Å) | 0.008 |
| Angles (°) | 1.2 |
| Main-chain bond (Å2) | 0.6 |
| Side-chain bond (Å2) | 0.7 |
| Between noncrystallographic symmetry-related Cα atoms (Å) | 0.6 |
| Atoms per asymmetric unit (protein/water) | 6,510/579 |
| Avg B-factors (protein/water) (Å2) | 26.7/32.9 |
Numbers in parentheses refer to the relevant outer resolution shell.
Rmerge = Σhkl ΣiI(hkl;i) − <I(hkl)>/Σhkl ΣiI(hkl;i), where I(hkl;i) is the intensity of an individual measurement and <I(hkl)> is the average intensity from multiple observations.
Rfactor = Σhkl‖Fobs − kFcalc‖/Σhkl Fobs.
Rfree is Rfactor for 5% of the data not used at any stage of the structural refinement.
RMSD, RMS deviation from ideal geometry.
The structure of NiV-G was solved by molecular replacement using the program Phaser (41) with the NiV-G component of the NiV-G-EFNB2 complex (PDB accession number 2VSM) as the search model. Two molecules were identified in the asymmetric unit. Structure refinement was performed using Refmac 5 in the CCP4 suite and included iterative restrained refinement with TLS using medium noncrystallographic symmetry restraints between the two NiV-G molecules in the asymmetric unit (25, 43). The program COOT was used for manual rebuilding (17), and PROCHECK and Molprobity were used to validate the model (36, 38). Ramachandran analysis (36) of the final structure showed 84% of residues in the most favored region, with no residues in disallowed regions.
Molecular superimpositions were calculated using SHP (54). Sequence alignments were prepared with Multalign (13) and Espript (20). Figures were prepared using Adobe Photoshop and Pymol (http://pymol.sourceforge.net).
Enzymatic release of N-linked glycans for MS.
Glycans were released by the method of Küster et al. (35). Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis bands containing approximately 10 μg of target NiV-G were excised, washed with 20 mM NaHCO3 (pH 7.0), and dried in a vacuum centrifuge before rehydration with 30 μl of 30 mM NaHCO3 (pH 7.0) containing 100 U ml−1 of protein N-glycanase F (Prozyme, San Leandro, CA). After incubation for 12 h at 37°C, the enzymatically released N-linked glycans were eluted with water and the sample was passed through a 0.45-μl-pore-size filter (Millex-LH, hydrophobic polytetrafluoroethylene).
MALDI-TOF MS.
Positive-ion matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectra of glycans were generated according to previously described methods (28). Spectra were recorded with a Waters-Micromass TofSpec 2E reflectron-TOF mass spectrometer (Waters-MS Technologies, Manchester, United Kingdom) operated under the following conditions: accelerating voltage, 20 kV; pulse voltage, 3.0 kV; time lag focusing delay, 500 ns (setting 39); laser repetition rate, 10 Hz. Aqueous glycan samples (0.5 μl) were mixed on the MALDI target with the matrix (0.5 μl of a saturated solution of 2,5-dihydroxybenzoic acid in acetonitrile), allowed to dry under ambient conditions, and recrystallized from ethanol (0.2 μl).
Negative-ion nano-electrospray MS/MS.
Electrospray MS was performed with a Waters quadrupole-time-of-flight (Q-Tof) Ultima Global instrument in negative-ion mode according to previously described methods (26-28). Samples in 1:1 (vol/vol) methanol-water were infused through Proxeon nanospray capillaries (Proxeon Biosystems, Odense, Denmark). The ion source conditions were as follows: temperature, 120°C; nitrogen flow 50 liters h−1; infusion needle potential, 1.2 kV; cone voltage 100 V; RF-1 voltage, 150 V. Spectra (2-s scans) were acquired with a digitization rate of 4 GHz and accumulated until a satisfactory signal/noise ratio had been obtained. For MS/MS data acquisition, the parent ion was selected at low resolution (about 5 m/z mass window) to allow transmission of isotope peaks and fragmented with argon at a pressure of 50 Pa. The voltage on the collision cell was adjusted to give an even distribution of fragment ions across the mass scale. Typical values were 80 to 120 V. Other voltages were as recommended by the manufacturer. Instrument control, data acquisition, and processing were performed with MassLynx software version 4.0.
Protein structure accession number.
Coordinates and structure factors of NiV-G have been deposited in the Protein Data Bank (PDB accession number 2VWD).
RESULTS AND DISCUSSION
Structure of unbound NiV-G.
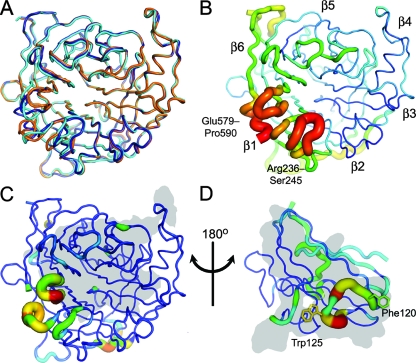
The globular six-bladed β-propeller domain (residues 183 to 602) of the NiV-G envelope glycoprotein was transiently expressed in HEK 293T cells in the presence of kifunensine and partially deglycosylated with endoglycosidase F1 (see Fig. S1 in the supplemental material). We have previously demonstrated by analytical ultracentrifugation that this recombinant form of NiV-G is monomeric (7). The domain was crystallized and the structure determined at 2.2-Å resolution by molecular replacement (Table 1 and Fig. 1). The two molecules in the crystallographic asymmetric unit, chains A and B, are, with the exception of the 579-590 loop (see below), mostly very similar to each other (root mean square [RMS] deviation in Cα position of 0.6 Å) and to the structure of the EFNB2-bound form (RMS deviation of 1.0 and 0.7 Å over 410 equivalent Cα atoms for chains A and B, respectively) (Fig. 1A and C). NiV-G is also quite similar to the hemagglutinins of PIV type 3 (PIV3-HN) (PDB accession number 1V2I [37]; RMS deviation of 2.3 Å over 380 equivalent Cα atoms), NDV (NDV-HN) (1E8T [10]; RMS deviation 2.4 Å over 372 equivalent Cα atoms), simian PIV5 structures (SV5-HN) (1Z4V [60]; RMS deviation of 2.4 Å over 368 equivalent Cα atoms), and HeV (HeV-G) (2VSK [7]; RMS deviation of 0.8 Å over 397 equivalent Cα atoms) but much less similar to the MV hemagglutinin (MV-H) (2RKC [12, 29]; RMS deviation of 3.3 Å over 320 equivalent Cα atoms). This is consistent with our previously published hypothesis that paramyxoviruses have adapted to attach to proteins rather than sugars in at least two separate evolutionary events (7).
FIG. 1.
Crystal structure of apo-NiV-G reveals an induced-fit mechanism for EFNB2 binding. (A) Cα trace of apo-NiV-G (orange, chain A; dark blue, chain B) superimposed with NiV-G of the NiV-G-EFNB2 complex (cyan; PDB accession number 2VSM [7]). (B) Relative B-factor values (ramped from blue to red) mapped onto the Cα trace structure of apo-NiV-G (chain B). Mobile regions have a thick radius and are colored red (high B-factor), while ordered regions have a thin radius and are colored blue (low B-factor). The β-propellers are numbered according to standard nomenclature (7, 14, 37, 60). (C) RMS displacement of equivalent residues between apo-NiV-G (average of chains A and B) and EFNB2-bound NiV-G mapped onto Cα trace structure of apo-NiV-G (chain B). The tube radius and color of the trace represent the RMS displacement (ramped from blue to red). Regions with high deviations between apo and EFNB2-bound forms are thick and colored red, while regions with low deviation are thin and colored blue. The gray region indicates the surface of the protein that interacts with EFNB2. (D) RMS displacement between NiV-G bound EFNB2 and the other reported EFNB2 structures mapped onto the Cα trace structure of NiV-G-bound EFNB2 (average of apo-EFNB2 [55], EPHB2-EFNB2 [30], and EPHB4-EFNB2 [11]; colored as in panel C). The gray region indicates the surface of the protein that interacts with NiV-G.
Specificity of NiV-G binding.
Both molecules in the crystallographic asymmetric unit of apo-NiV-G show structural differences from the EFNB2-bound form in the region of the 579-590 loop. In the NiV-G-EFNB2 and HeV-G-EFNB2 complexes (PDB accession numbers 2VSM and 2VSK, respectively), this loop is critical for complex formation (7). Specifically, Ile588NiV-G and Tyr581NiV-G participate in hydrophobic interactions and contribute to a binding pocket that accommodates Phe120EFNB2 (Fig. 2A). Consistent with these observations, mutagenesis experiments have indicated the importance of this residue in receptor binding (7). In the unbound form of NiV-G, the 579-590 loop is relatively flexible in both molecules of the crystallographic asymmetric unit, and 2Fo-Fc electron density for the side chains is poor. Nevertheless, continuous electron density is visible for the main chain, which is displaced by up to 6 Å (chain B) from its EFNB2-bound position. In both molecules in the crystallographic asymmetric unit, the 579-590 loop participates in crystal packing, perhaps explaining their different conformations. The loop structure in both of these molecules precludes EFNB2 binding with movement required to accommodate Phe120EFNB2 (Fig. 2A, B, and C). In addition, the 579-590 and 236-245 loops exhibit high B-factor values in the apo form (Fig. 1B and C). Interestingly, these loops account for only 22% of the total buried surface in the NiV-G-EFNB2 binding interface (34). The rest of the NiV-G surface is far more structurally conserved between the apo and complexed forms. We previously identified the Phe120EFNB2 binding pocket as a potential template for structure-based drug design (7). The structure of the apo form of NiV-G, reported here, further refines this potential drug target by highlighting the structural plasticity of this pocket.
FIG. 2.
Conformational plasticity of the EFNB2 binding pocket of NiV-G. (A) When bound to EFNB2, NiV-G residues Gln559, Glu579, Tyr581, and Ile588 (stick representation with nitrogen blue, oxygen red, and carbon white) form a pocket around Phe120EFNB2 (gray van der Waals surface; stick representation with nitrogen blue, oxygen red, and carbon yellow). (B and C) Superimposed Phe120EFNB2 is no longer accommodated in the apo structure of chain B (B) or chain A (C) in the asymmetric unit of NiV-G due to steric clashes resulting from 579-590 loop movements.
The flexibility of apo-NiV-G in the region of the 579-590 and 236-245 loops is mirrored in the complementary binding face of the EFNB2 structures (Fig. 1D) (7, 11, 30, 55). Comparison of the NiV-G-bound EFNB2 with apo-EFNB2 (PDB accession number 1IKO) and EFNB2 in complex with the cellular receptors EPHB2 (1NUK) and EPHB4 (2HLE) reveals that residues in the GH loop (Glu119 to Leu127) exhibit relatively high structural deviations (Fig. 1D). Thus, opposing loops of both NiV-G and EFNB2 undergo induced-fit conformational changes upon binding.
Monomeric NiV-G contains processed complex-type glycosylation.
Within the NiV-G crystal structure, electron density for N-acetylglucosamine (GlcNAc) β1-linked to Asn was observed at all five predicted N-linked glycosylation sites (Asn306, Asn378, Asn417, Asn481, and Asn529) (Fig. 3A; see Fig. S2 in the supplemental material). The presence of a single GlcNAc at each glycosylation site is consistent with full occupancy and effective cleavage of the kifunensine-induced oligomannose-type glycans by endoglycosidase F1 (9). Most GlcNAc residues project away from the protein surface with little carbohydrate-protein interaction, as illustrated for Asn378 in Fig. 3B. However, the GlcNAc at Asn306 packs against the aromatic side chain of Tyr309 (Fig. 3C). The hydrophobic faces of reducing-terminal GlcNAc residues are often found in contact with aromatic residues, effectively shielding hydrophobic patches on the protein surface (50).
FIG. 3.
NiV-G N-linked glycosylation sites. (A) Cartoon representation of NiV-G (chain B) with GlcNAcβ1-Asn structures shown in sticks. The carbons of GlcNAc are shown in green, while those of side chains are colored gray. Oxygen atoms are colored red, and nitrogen atoms are blue. A simulated annealing electron density omit map calculated in the absence of carbohydrate is displayed around the GlcNAc residues contoured at 1σ. (B and C) Enlarged views of the GlcNAc residue at Asn378 (B) and the GlcNAc residue at Asn306 (C); the omit map was calculated in the absence of carbohydrate and Asn378, Asn306, and Tyr309.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis reveals that recombinant NiV-G (residues 183 to 602, expressed in HEK 293T cells) comprises between 15 and 30% carbohydrate (see Fig. S1 in the supplemental material). The processing of glycosylation can be influenced by both tissue-specific factors, such as glycosyltransferase expression, and protein structure (53). Consequently, only limited conclusions can be drawn from the carbohydrate analysis of recombinant material. However, the occurrence of oligomannose-type glycans can indicate glycosylation sites that are sterically protected from processing (2, 8, 16); such glycans are often conserved between infectious virions and recombinant material. For example, the mannose epitope of a broadly neutralizing antibody to HIV-1 gp120 is independent of oligomerization state and is present in recombinant gp120 monomers expressed in CHO cells (8, 61). Furthermore, in direct comparison with our present study, we note that MS analysis of the surface glycoprotein of Ebola virus by Powlesland et al. also exploited recombinant expression in HEK 293T cells (51). Therefore, with the caveat that the envelope glycoprotein under investigation is monomeric, we investigated whether oligomannose-type glycans and other lectin targets were present in NiV-G. We determined the composition of the carbohydrate component of recombinant NiV-G (expressed in HEK 293T cells in the absence of kifunensine) by MALDI-TOF MS and assigned structural isomers by negative-ion nano-electrospray MS/MS (26-28) (data not shown) of purified glycans released by in-gel protein N-glycanase F. Glycan structures are shown in Fig. 4A and in Table S1 in the supplemental material.
FIG. 4.
MS analyses of NiV-G monomer, showing MALDI-TOF MS of N-linked glycans released by in-gel protein N-glycanase F digestion. (A) Spectra of [M + Na]+ ions of N-linked glycans from the NiV-G expressed in HEK 293T cells. (B) Spectra of [M + Na]+ ions of glycans from NiV-G expressed in HEK 293T cells in the presence of 5 μM kifunensine. Symbols used for the structural formulae: ⋄, Gal; ⧫, GalNAc; ▪, GlcNAc; ○, Man; ★, sialic acid; ·⃟, Fuc. The linkage position is shown by the angle of the lines linking the sugar residues (vertical line, 2-link; forward slash, 3-link; horizontal line, 4-link; back slash, 6-link). Anomericity is indicated by full lines for β-bonds and broken lines for α-bonds.
In contrast to the case for other enveloped viruses, such as HIV-1 (61) and Ebola virus (40, 51), NiV-G contains highly processed complex-type glycans with negligible levels of oligomannose-type glycans terminated with Manα1→2Man motifs (Fig. 4A). This is consistent with viral tropism mediated by protein-protein-type interaction with ephrin rather than binding via oligomannose-specific lectins such as DC-SIGNR (24). The processing to complex-type glycosylation is consistent with the accessibility and lack of clustering of the positions of the glycosylation sites in the crystal structure of the monomer. The GlcNAcβ1→2Man terminal structures in NiV-G are similar to those reported to exist on the Ebola virus surface glycoprotein. In Ebola virus, these structures mediate binding to the C-type lectin, LSECtin, expressed on sinusoidal endothelial cells of lymph nodes and liver (51). These carbohydrate motifs are common but not ubiquitous on viral surfaces. For example, LSECtin also binds to the S protein from severe acute respiratory syndrome coronavirus, whereas in contrast, hepatitis C virus pseudovirions and HEK 293T-derived HIV-1 virions do not interact with LSECtin (21). Therefore, despite the inherent limitations of recombinant systems, the effect of differential viral glycosylation on receptor binding can be revealed by comparison of material from matched expression systems. We suggest that NiV binding to host lectins, such as LSECtin, may influence viral infectivity and that based on our observations, an analysis of the glycosylation of native oligomeric NiV-G from primary isolates would be of significant interest.
We also determined the glycosylation status of the NiV-G expressed in the presence of kifunensine, generated for crystallization (9), revealing a series of endoglycosidase F1-sensitive oligomannose-type glycans ranging from Man9GlcNAc2 (m/z 1905.7) to a trace amount of Man5GlcNAc2 detectable at m/z 1257.2 (Fig. 4B), consistent with previous MALDI MS analysis of the effect of glycan processing by kifunensine (8, 9, 15). Comparison of the spectra in Fig. 4A and B supports our conclusion that NiV-G is largely devoid of oligomannose-type glycans. The paucity of these structures suggests that glycosylation engineering of vaccines based on recombinant NiV-G, to induce oligomannose-type glycosylation, may be a route by which to enhance immunogenicity, increasing the uptake of the vaccine by macrophages (32).
Mapping the putative oligomeric face of NiV-G.
Previously reported analysis of the full-length NiV-G ectodomain, by analytical ultracentrifugation, demonstrated that it was mainly tetrameric (7). Similarly, gel filtration of HeV-G revealed monomers, dimers, and tetramer (5). Oligomerization of NiV-G, by analogy with the NDV, is likely to be induced by interchain disulfide bonds within the stalk region, which is absent in our NiV-G crystal structure. However, the overall architecture of the oligomeric assembly remains unknown. We sought to infer the oligomeric architecture of NiV-G by comparing structural features of homologous proteins across the paramyxovirus family.
While the largest intermolecular interface in the NiV-G crystal occludes only 600 Å2 of surface (34), inconsistent with oligomerization, packing interactions of the β-propeller domains in the crystal structures of SV5-HN, NDV-HN, and human PIV3-HN revealed dimeric and tetrameric arrangements (14, 37, 60). The structure of human PIV3-HN demonstrates that the detection of oligomeric states within the crystal lattice is not necessarily precluded by the absence of the stalk domain. However, Yuan et al. suggested a putative oligomerization face of the envelope attachment glycoproteins of the Paramyxovirus family through the identification of regions of low structural diversity by superimposition of the available crystal structures of hemagglutinins from SV5, NDV, and PIV3 (60). We extend this analysis by including our NiV-G structure and find significant structural divergence along the face formed by the propeller blades β2 to β4 and structural similarity on the opposing face (Fig. 5A), consistent with the previous study (60), suggesting that the region of the propeller blades β1 and β6 of the NiV-G forms the putative oligomerization face. Interestingly, a further region of low structural diversity corresponds to the ephrin binding face of NiV-G (Fig. 5B).
FIG. 5.
Conservation of N-linked glycosylation sites on NiV-G across paramyxoviruses. (A) Cartoon representation (Cα trace) of the crystal structures of the globular β-propeller domains from envelope attachment glycoproteins of NiV-G (gray; PDB accession number 2VSM), NDV-HN (pink; 1E8T), PIV3-HN (green; 1V2I), and SV5-HN (blue; 1Z4Y). (B) Residues from the G-H loop of EFNB2 (sticks; residues 115 to 127) in complex with NiV-G (van der Waals surface), where sticks are colored yellow for carbon, blue for nitrogen, and red for oxygen. The surface is colored by a gradient of red (completely buried) to white (not buried) according to the fraction of buried surface area. (C) Superimposition of Cα trace ribbon of NiV-G with N-linked glycosylation sites of NiV-G, HeV-G, PIV1, PIV2, PIV3, Sendai virus, and NDV mapped as spheres at the Cα on equivalent residues. The position of the NiV-G glycans (at Asn306, Asn378, Asn417, Asn481, and Asn529) are indicated by orange spheres, and the positions of glycans from the other viruses are indicated by yellow spheres. (D) View of NiV-G formed by a 90° rotation of panel C. The putative oligomerization face is indicated by a blue bracket.
We further extend the analysis of the putative oligomerization face by considering the conservation of N-linked glycosylation sites across the Paramyxoviridae family. Our MS analysis demonstrated that over 2 kDa of carbohydrate may be attached to individual glycosylation sites on NiV-G (Fig. 4B). Such large and highly heterogeneous groups are generally not found on protein surfaces utilized for protein-protein interactions. To map the putative oligomeric face of NiV-G, we aligned sequences of the envelope glycoprotein from viruses across the paramyxovirus family and mapped the positions of the glycosylation sites onto the structure of apo-NiV-G (Fig. 5C and D). Inspection of Fig. 5C and D reveals significant diversity in the positions of the glycosylation sites. The low degree of glycosylation site conservation (see Fig. S4 in the supplemental material) suggests that there are no conserved structural roles for individual glycans, although the retention of glycosylation sites at nonspecific sites across the structure is consistent with a general role in chaperone-mediated protein folding. However, analysis reveals that the β1 and β6 propeller blades, which were identified above as a region of low structural divergence, are largely devoid of N-linked glycans. The small number of glycosylation sites within this region are located at the membrane-proximal and distal faces of the glycoprotein and are not anticipated to disrupt lateral interactions (Fig. 5D).
The suggestion that the surface of the β1 and β6 propeller blades forms the oligomerization face of NiV-G is further supported by the observation that this face mediates dimerization of SV5, PIV3, and NDV hemagglutinin neuraminidases (37, 60). The positions of the glycosylation sites on NiV-G are consistent with a model of the tetramer based on the crystal structure of the SV5 hemagglutinin (data not shown).
Structural guide to vaccine and antiviral drug design.
It is now evident that the use of ephrins as functional receptors defines the tropism of henipaviruses. We have demonstrated that both EFNB2 and NiV-G undergo conformational changes upon binding. These conformational changes underscore the importance of structural analysis of the apo and bound forms of an interaction pair for structure-based drug design. The NiV-G-EFNB2 interface is characterized by pockets that accommodate hydrophobic residues from EFNB2, most notably Trp125EFNB2 and Phe120EFNB2 (7). In light of recent advances in the identification of small-molecule antagonists which disrupt protein-protein interfaces exhibiting inherent structural plasticity (57), we anticipate that this deep hydrophobic groove of NiV-G, and in particular the structurally malleable Phe120EFNB2 binding pocket, is a tractable drug target.
Our analysis of the glycosylation status and N-linked glycosylation site conservation of recombinant NiV-G suggests that these carbohydrate structures provide a route for further optimization of recombinant vaccines against NiV. The HeV vaccine candidate, consisting of the full-length ectodomain of HeV-G, is a heterogeneous mixture of oligomers (5). The presentation of the oligomeric interface in a heterogeneous vaccine may lead to antibody responses toward the nonneutralizing epitopes. As oligomeric proteins generally have greater immunogenicity than monomeric constructs (3), structure-guided stabilization of the tetramer may be a route to vaccine optimization. Alternatively, the “immunofocusing” approach of Pantophlet and Burton (47), in which additional N-linked glycosylation sites are added to nonneutralizing epitopes, could be used to generate a homogenous monomer that directs a neutralizing antibody response toward the ephrin binding face. For such an approach, the positions of the additional glycosylation sites could be guided by the locations of sites in related paramyxoviruses (Fig. 5C and D; see Fig. S3 in the supplemental material) as well as by the putative oligomerization face.
Finally, our glycosylation analysis demonstrated putative ligands for the epithelial lectin, LSECtin, and revealed a paucity of oligomannose-type glycans on recombinant NiV-G. In contrast, our expression system for crystallographic analysis of glycoproteins using an α-mannosidase inhibitor exhibits uniformly oligomannose-type glycosylation. Such mannose-based expression systems may find utility in the development of recombinant henipavirus vaccines by stimulating mannose-dependent uptake by antigen-presenting cells (32).
Supplementary Material
Acknowledgments
We are grateful to W. Lu for help with tissue culture, to K. Harlos for data collection, and to the staff of beamline ID23.1 at the European Synchotron Radiation Facility for assistance.
This work was funded by the Wellcome Trust, Medical Research Council, Royal Society, Cancer Research UK, and Spine2 Complexes (FP6-RTD-031220).
Footnotes
Published ahead of print on 24 September 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aricescu, A. R., W. Lu, and E. Y. Jones. 2006. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D 621243-1250. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, J. N., C. M. Radcliffe, M. R. Wormald, L. Royle, D. J. Harvey, M. Crispin, R. A. Dwek, R. B. Sim, and P. M. Rudd. 2004. The glycosylation of human serum IgD and IgE and the accessibility of identified oligomannose structures for interaction with mannan-binding lectin. J. Immunol. 1736831-6840. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann, M. F., and R. M. Zinkernagel. 1997. Neutralizing antiviral B cell responses. Annu. Rev. Immunol. 15235-270. [DOI] [PubMed] [Google Scholar]
- 4.Bonaparte, M. I., A. S. Dimitrov, K. N. Bossart, G. Crameri, B. A. Mungall, K. A. Bishop, V. Choudhry, D. S. Dimitrov, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. USA 10210652-10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bossart, K. N., G. Crameri, A. S. Dimitrov, B. A. Mungall, Y. R. Feng, J. R. Patch, A. Choudhary, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J. Virol. 796690-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossart, K. N., M. Tachedjian, J. A. McEachern, G. Crameri, Z. Zhu, D. S. Dimitrov, C. C. Broder, and L. F. Wang. 2008. Functional studies of host-specific ephrin-B ligands as Henipavirus receptors. Virology 372357-371. [DOI] [PubMed] [Google Scholar]
- 7.Bowden, T. A., A. R. Aricescu, R. J. Gilbert, J. M. Grimes, E. Y. Jones, and D. I. Stuart. 2008. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 15567-572. [DOI] [PubMed] [Google Scholar]
- 8.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 3002065-2071. [DOI] [PubMed] [Google Scholar]
- 9.Chang, V. T., M. Crispin, A. R. Aricescu, D. J. Harvey, J. E. Nettleship, J. A. Fennelly, C. Yu, K. S. Boles, E. J. Evans, D. I. Stuart, R. A. Dwek, E. Y. Jones, R. J. Owens, and S. J. Davis. 2007. Glycoprotein structural genomics: solving the glycosylation problem. Structure 15267-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, L., P. M. Colman, L. J. Cosgrove, M. C. Lawrence, L. J. Lawrence, P. A. Tulloch, and J. J. Gorman. 2001. Cloning, expression, and crystallization of the fusion protein of Newcastle disease virus. Virology 290290-299. [DOI] [PubMed] [Google Scholar]
- 11.Chrencik, J. E., A. Brooun, M. L. Kraus, M. I. Recht, A. R. Kolatkar, G. W. Han, J. M. Seifert, H. Widmer, M. Auer, and P. Kuhn. 2006. Structural and biophysical characterization of the EphB4. EphrinB2 protein-protein interaction and receptor specificity. J. Biol. Chem. 28128185-28192. [DOI] [PubMed] [Google Scholar]
- 12.Colf, L. A., Z. S. Juo, and K. C. Garcia. 2007. Structure of the measles virus hemagglutinin. Nat. Struct. Mol. Biol. 141227-1228. [DOI] [PubMed] [Google Scholar]
- 13.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1610881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 71068-1074. [DOI] [PubMed] [Google Scholar]
- 15.Crispin, M., D. J. Harvey, V. T. Chang, C. Yu, A. R. Aricescu, E. Y. Jones, S. J. Davis, R. A. Dwek, and P. M. Rudd. 2006. Inhibition of hybrid and complex-type glycosylation reveals the presence of the GlcNAc transferase I-independent fucosylation pathway. Glycobiology 16748-756. [DOI] [PubMed] [Google Scholar]
- 16.Crispin, M. D. M., G. E. Ritchie, A. J. Critchley, B. P. Morgan, I. A. Wilson, R. A. Dwek, R. B. Sim, and P. M. Rudd. 2004. Monoglucosylated glycans in the secreted human complement component C3: implications for protein biosynthesis and structure. FEBS Lett. 566270-274. [DOI] [PubMed] [Google Scholar]
- 17.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 602126-2132. [DOI] [PubMed] [Google Scholar]
- 18.Field, H. E., P. C. Barratt, R. J. Hughes, J. Shield, and N. D. Sullivan. 2000. A fatal case of Hendra virus infection in a horse in north Queensland: clinical and epidemiological features. Aust. Vet. J. 78279-280. [DOI] [PubMed] [Google Scholar]
- 19.Gleeson, P. A., J. Feeney, and R. C. Hughes. 1985. Structures of N-glycans of a ricin-resistant mutant of baby hamster kidney cells. Synthesis of high-mannose and hybrid N-glycans. Biochemistry 24493-503. [DOI] [PubMed] [Google Scholar]
- 20.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15305-308. [DOI] [PubMed] [Google Scholar]
- 21.Gramberg, T., H. Hofmann, P. Moller, P. F. Lalor, A. Marzi, M. Geier, M. Krumbiegel, T. Winkler, F. Kirchhoff, D. H. Adams, S. Becker, J. Munch, and S. Pohlmann. 2005. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology 340224-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guillaume, V., H. Contamin, P. Loth, M. C. Georges-Courbot, A. Lefeuvre, P. Marianneau, K. B. Chua, S. K. Lam, R. Buckland, V. Deubel, and T. F. Wild. 2004. Nipah virus: vaccination and passive protection studies in a hamster model. J. Virol. 78834-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillaume, V., H. Contamin, P. Loth, I. Grosjean, M. C. Courbot, V. Deubel, R. Buckland, and T. F. Wild. 2006. Antibody prophylaxis and therapy against Nipah virus infection in hamsters. J. Virol. 801972-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo, Y., H. Feinberg, E. Conroy, D. A. Mitchell, R. Alvarez, O. Blixt, M. E. Taylor, W. I. Weis, and K. Drickamer. 2004. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 11591-598. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton, S. R., R. C. Davidson, N. Sethuraman, J. H. Nett, Y. Jiang, S. Rios, P. Bobrowicz, T. A. Stadheim, H. Li, B. K. Choi, D. Hopkins, H. Wischnewski, J. Roser, T. Mitchell, R. R. Strawbridge, J. Hoopes, S. Wildt, and T. U. Gerngross. 2006. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science 3131441-1443. [DOI] [PubMed] [Google Scholar]
- 26.Harvey, D. J. 2005. Fragmentation of negative ions from carbohydrates. 2. Fragmentation of high-mannose N-linked glycans. J. Am. Soc. Mass Spectrom. 16631-646. [DOI] [PubMed] [Google Scholar]
- 27.Harvey, D. J. 2005. Fragmentation of negative ions from carbohydrates. 3. Fragmentation of hybrid and complex N-linked glycans. J. Am. Soc. Mass Spectrom. 16647-659. [DOI] [PubMed] [Google Scholar]
- 28.Harvey, D. J., L. Royle, C. M. Radcliffe, P. M. Rudd, and R. A. Dwek. 2008. Structural and quantitative analysis of N-linked glycans by matrix-assisted laser desorption ionization and negative ion nanospray mass spectrometry. Anal. Biochem. 37644-60. [DOI] [PubMed] [Google Scholar]
- 29.Hashiguchi, T., M. Kajikawa, N. Maita, M. Takeda, K. Kuroki, K. Sasaki, D. Kohda, Y. Yanagi, and K. Maenaka. 2007. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. USA 10419535-19540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Himanen, J. P., K. R. Rajashankar, M. Lackmann, C. A. Cowan, M. Henkemeyer, and D. B. Nikolov. 2001. Crystal structure of an Eph receptor-ephrin complex. Nature 414933-938. [DOI] [PubMed] [Google Scholar]
- 31.Himanen, J. P., N. Saha, and D. B. Nikolov. 2007. Cell-cell signaling via Eph receptors and ephrins. Curr. Opin. Cell Biol. 19534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keler, T., V. Ramakrishna, and M. W. Fanger. 2004. Mannose receptor-targeted vaccines. Expert Opin. Biol. Ther. 41953-1962. [DOI] [PubMed] [Google Scholar]
- 33.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54631-664. [DOI] [PubMed] [Google Scholar]
- 34.Krissinel, E., and K. Henrick. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372774-792. [DOI] [PubMed] [Google Scholar]
- 35.Küster, B., S. F. Wheeler, A. P. Hunter, R. A. Dwek, and D. J. Harvey. 1997. Sequencing of N-linked oligosaccharides directly from protein gels: in-gel deglycosylation followed by matrix-assisted laser desorption/ionization mass spectrometry and normal-phase high-performance liquid chromatography. Anal. Biochem. 25082-101. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26283-291. [Google Scholar]
- 37.Lawrence, M. C., N. A. Borg, V. A. Streltsov, P. A. Pilling, V. C. Epa, J. N. Varghese, J. L. McKimm-Breschkin, and P. M. Colman. 2004. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 3351343-1357. [DOI] [PubMed] [Google Scholar]
- 38.Lovell, S. C., I. W. Davis, W. B. Arendall, 3rd, P. I. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson, and D. C. Richardson. 2003. Structure validation by Cα geometry: φ, ϕ and Cβ deviation. Proteins 50437-450. [DOI] [PubMed] [Google Scholar]
- 39.Luby, S. P., M. Rahman, M. J. Hossain, L. S. Blum, M. M. Husain, E. Gurley, R. Khan, B. N. Ahmed, S. Rahman, N. Nahar, E. Kenah, J. A. Comer, and T. G. Ksiazek. 2006. Foodborne transmission of Nipah virus, Bangladesh. Emerg. Infect. Dis. 121888-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marzi, A., A. Akhavan, G. Simmons, T. Gramberg, H. Hofmann, P. Bates, V. R. Lingappa, and S. Pohlmann. 2006. The signal peptide of the ebolavirus glycoprotein influences interaction with the cellular lectins DC-SIGN and DC-SIGNR. J. Virol. 806305-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCoy, A. J., R. W. Grosse-Kunstleve, L. C. Storoni, and R. J. Read. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D 61458-464. [DOI] [PubMed] [Google Scholar]
- 42.McEachern, J. A., J. Bingham, G. Crameri, D. J. Green, T. J. Hancock, D. Middleton, Y. R. Feng, C. C. Broder, L. F. Wang, and K. N. Bossart. 2008. A recombinant subunit vaccine formulation protects against lethal Nipah virus challenge in cats. Vaccine 263843-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 44.Negrete, O. A., E. L. Levroney, H. C. Aguilar, A. Bertolotti-Ciarlet, R. Nazarian, S. Tajyar, and B. Lee. 2005. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436401-405. [DOI] [PubMed] [Google Scholar]
- 45.Negrete, O. A., M. C. Wolf, H. C. Aguilar, S. Enterlein, W. Wang, E. Muhlberger, S. V. Su, A. Bertolotti-Ciarlet, R. Flick, and B. Lee. 2006. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski, A., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276307-326. [DOI] [PubMed] [Google Scholar]
- 47.Pantophlet, R., and D. R. Burton. 2003. Immunofocusing: antigen engineering to promote the induction of HIV-neutralizing antibodies. Trends Mol. Med. 9468-473. [DOI] [PubMed] [Google Scholar]
- 48.Parodi, A. J. 2000. Protein glucosylation and its role in protein folding. Annu. Rev. Biochem. 6969-93. [DOI] [PubMed] [Google Scholar]
- 49.Pasquale, E. B. 2008. Eph-ephrin bidirectional signaling in physiology and disease. Cell 13338-52. [DOI] [PubMed] [Google Scholar]
- 50.Petrescu, A.-J., A.-L. Milac, S. M. Petrescu, R. A. Dwek, and M. R. Wormald. 2004. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology 14103-114. [DOI] [PubMed] [Google Scholar]
- 51.Powlesland, A. S., T. Fisch, M. E. Taylor, D. F. Smith, B. Tissot, A. Dell, S. Pohlmann, and K. Drickamer. 2008. A novel mechanism for LSECtin binding to Ebola virus surface glycoprotein through truncated glycans. J. Biol. Chem. 283593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogers, R. J., I. C. Douglas, F. C. Baldock, R. J. Glanville, K. T. Seppanen, L. J. Gleeson, P. N. Selleck, and K. J. Dunn. 1996. Investigation of a second focus of equine morbillivirus infection in coastal Queensland. Aust. Vet. J. 74243-244. [DOI] [PubMed] [Google Scholar]
- 53.Rudd, P. M., and R. A. Dwek. 1997. Glycosylation: heterogeneity and the 3D structure of proteins. Crit. Rev. Biochem. Mol. Biol. 321-100. [DOI] [PubMed] [Google Scholar]
- 54.Stuart, D. I., M. Levine, H. Muirhead, and D. K. Stammers. 1979. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J. Mol. Biol. 134109-142. [DOI] [PubMed] [Google Scholar]
- 55.Toth, J., T. Cutforth, A. D. Gelinas, K. A. Bethoney, J. Bard, and C. J. Harrison. 2001. Crystal structure of an ephrin ectodomain. Dev. Cell 183-92. [DOI] [PubMed] [Google Scholar]
- 56.Walter, T. S., J. M. Diprose, C. J. Mayo, C. Siebold, M. G. Pickford, L. Carter, G. C. Sutton, N. S. Berrow, J. Brown, I. M. Berry, G. B. Stewart-Jones, J. M. Grimes, D. K. Stammers, R. M. Esnouf, E. Y. Jones, R. J. Owens, D. I. Stuart, and K. Harlos. 2005. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D 61651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wells, J. A., and C. L. McClendon. 2007. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 4501001-1009. [DOI] [PubMed] [Google Scholar]
- 58.Wild, T. F. 2008. Henipaviruses: a new family of emerging Paramyxoviruses. Pathol. Biol. (Paris) doi: 10.1016/j.patbio.2008.04.006. [DOI] [PubMed]
- 59.Yanagi, Y., M. Takeda, and S. Ohno. 2006. Measles virus: cellular receptors, tropism and pathogenesis. J. Gen. Virol. 872767-2779. [DOI] [PubMed] [Google Scholar]
- 60.Yuan, P., T. B. Thompson, B. A. Wurzburg, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2005. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13803-815. [DOI] [PubMed] [Google Scholar]
- 61.Zhu, X., C. Borchers, R. J. Bienstock, and K. B. Tomer. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 3911194-11204. [DOI] [PubMed] [Google Scholar]
- 62.Zhu, Z., A. S. Dimitrov, K. N. Bossart, G. Crameri, K. A. Bishop, V. Choudhry, B. A. Mungall, Y. R. Feng, A. Choudhary, M. Y. Zhang, Y. Feng, L. F. Wang, X. Xiao, B. T. Eaton, C. C. Broder, and D. S. Dimitrov. 2006. Potent neutralization of Hendra and Nipah viruses by human monoclonal antibodies. J. Virol. 80891-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.