Abstract
Homologous recombination is a high fidelity, template-dependent process that is used in repair of damaged DNA, recovery of broken replication forks, and disjunction of homologous chromosomes in meiosis. Much of what is known about recombination genes and mechanisms comes from studies on baker's yeast. Ustilago maydis, a basidiomycete fungus, is distant evolutionarily from baker's yeast and so offers the possibility of gaining insight into recombination from an alternative perspective. Here we have surveyed the genome of Ustilago maydis to determine the composition of its homologous recombination system. Compared to baker's yeast, there are fundamental differences in the function as well as in the repertoire of dedicated components. These include the use of a BRCA2 homolog and its modifier Dss1 rather than Rad52 as a mediator of Rad51, the presence of only a single Rad51 paralog, and the absence of Dmc1 and auxiliary meiotic proteins.
Keywords: homologous recombination, DNA repair, meiosis, BRCA2, Dss1, Rad51, Rad52, Dmc1, synaptonemal complex
1. Introduction
Genetic recombination is a nearly universal process in which information in the form of nucleotide sequence is exchanged or transferred between DNA molecules [for excellent recent reviews, see (Krogh and Symington, 2004; Neale and Keeney, 2006; Paques and Haber, 1999; Shrivastav et al., 2008; Sung and Klein, 2006; West, 2003; Wyman and Kanaar, 2006)]. In homologous recombination exchange takes place between sequences that are highly related, or nearly perfectly matching, spanning lengths of hundreds of base pairs. Homologous recombination plays important or even essential roles in the mitotic and meiotic cell cycles of most eukaryotes. In meiosis recombination functions to establish direct physical bonds between homologous chromosomes to insure their correct disjunction during reductional meiotic division. Crossing over during this process provides a means for rapid dissemination of new alleles through a population and serves as a mechanism for creating genetic diversity. In mitotic cells the primary function of homologous recombination is to repair DNA double-strand breaks (DSBs) and gaps resulting from replication fork collapse or damage by endogenous or exogenous genotoxins. Unknown mechanisms repress homologous recombination in G1 phase of the cell cycle during which nonhomologous end-joining is the favored mode of DSB repair.
Several models dating from 1964 that have been seminal in shaping thinking and experimentation about the molecular mechanism of recombination have tried to reconcile the relationship between crossing over and gene conversion in meiosis (Holliday, 1964; Meselson and Radding, 1975; Szostak et al., 1983). Crossing over between linked genetic markers is a reciprocal process that changes the configuration of the markers but does not alter their Mendelian ratio. Gene conversion is a non-reciprocal Mendelian segregation of markers resulting from transfer of information between homologous sequences to duplicate one allele but with the corresponding loss of the other. In meiosis there is a strong association between crossing over and gene conversion (Hurst et al., 1972). In mitotic cells crossing over with respect to conversion is minimized (Holliday, 1966; Hurst and Fogel, 1964; Roman and Jacob, 1958).
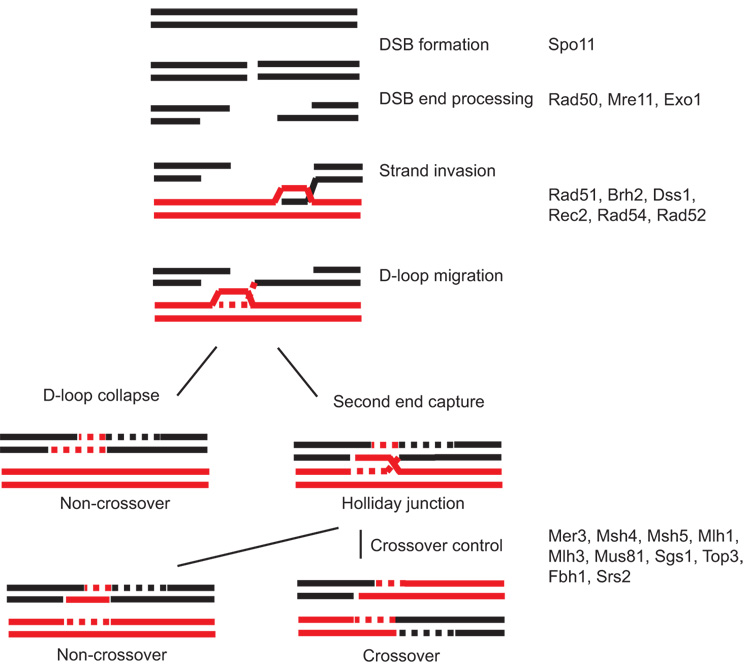
Variations of the DNA double-strand break repair (DSBR) model (Szostak et al., 1983) can be postulated to explain the association of crossing over with gene conversion during meiosis and the lack of associated crossing over during mitosis. In the migrating D-loop model (Ferguson and Holloman, 1996), which was formulated to explain recombinational repair in mitotic cells of the fungus Ustilago maydis, DNA strand invasion occurs to form a D-loop as envisioned in the DSBR model following resection of a double-strand break (DSB) end to reveal a protruding single-stranded tail (Figure 1). The invading single-strand primes DNA synthesis which then extends the invading strand and concomitantly drives migration of the D-loop. The freshly elongated invading strand can then be displaced from the D-loop in the homologous DNA template to pair with complementary sequence in the resected duplex on the other side of the DSB. By this mechanism there is repair exclusive of crossing over. Alternatively, the displaced strand of the D-loop can be captured by the second end of the DSB to pair with the complementary strand on that side of the break. With DNA synthesis primed from the non-invading end a Holliday junction intermediate will be formed. In this case alternative modes of resolving the Holliday junction open the possibility for crossing over. Balancing D-loop dissolution versus second end capture determines the outcome in terms of whether repair will occur with or without crossing over of the flanking regions on the two homologous duplexes.
Figure 1.
Migrating D-loop model of recombination. After DSB formation the ends are processed to reveal protruding 3′ single-stranded tails. One tail invades a homologous duplex to form a D-loop that migrates concomitant with DNA synthesis. With collapse of the D-loop as shown on the left side, the newly synthesized strand may anneal with the complementary protruding single-stranded tail on the other side of the break. Further fill-in synthesis will result in a non-crossover product. If the D-loop is captured by the protruding strand from the other side of the DSB, as shown on the right side, it may be processed to form an intermediate with one or possibly two Holliday junctions. Resolution of the Holliday junction by nicking the top and bottom strands will yield crossover products, while nicking the two inner strands will yield non-crossover products. The U. maydis gene products that serve in recombination are shown on the right side of the diagram at the approximate step in the pathway where they are thought to function.
Studies with the baker’s yeast Saccharomyces cerevisiae have illuminated many mechanistic aspect of homologous recombination in mitotic and meiotic cells. Genetic methods have led to the identification of most of the genes involved, and biochemical studies on the cloned, overexpressed gene products have provided a broad understanding of protein function. In addition, physical monitoring of chromosomal DNA dynamics in different mutant backgrounds has provided a powerful view of recombination gene action. A number of the yeast genes dedicated to recombination, i.e., the RAD52 epistasis group, were identified by their requirement for survival after damage by ionizing radiation (Symington, 2002). The genes can be categorized into two subgroups, those encoding the Mre11 complex, and those encoding Rad51 and associated proteins. Mre11, Rad50, and Xrs2 (Nbs1 in mammals) form a complex (MRX or MRN in mammals) that appears to sense DNA double-strand breaks and to function at an early step in processing the ends in preparation for subsequent repair events (Ivanov et al., 1994; Lisby et al., 2004). The central role of MRX complex in end recognition also impacts other processes leading to the non-homologous joining of DNA ends, maintenance of telomeres, and DNA damage checkpoint in mitotic cells. Rad51 and associated proteins are dedicated to the actual mechanics of homologous sequence recognition and DNA strand exchange.
Additional genes that function in recombination or that appear to provide auxiliary functions in the homologous pairing process have been identified from screens and methods focusing on mitotic recombination, meiotic defects, interactions with the Rad51 group of components or related proteins, or genes that contribute to the control of recombination [e.g., (Bai and Symington, 1996; Bishop et al., 1992; Interthal and Heyer, 2000; Malone et al., 1991; Menees and Roeder, 1989; Rong et al., 1991)]. These investigations have led to the discovery of the Spo11 complex that provides the initiating event for homologous recombination in meiosis, of proteins associated with meiotic homologous pairing including Dmc1 and auxiliary factors, and proteins involved in regulating or processing recombination intermediates.
Given the important role of recombination in maintaining genomic stability and its near universal occurrence, it would be expected that proteins involved are highly conserved. Indeed this expectation is broadly borne out and there are striking examples of conservation of both structure and function of recombination proteins in all domains of life, for instance the RecA/Rad51 protein (Lin et al., 2006). Here we were interested in surveying the genome of the basidiomycete U. maydis (Kamper et al., 2006) for its repertoire of recombination genes. U. maydis is a well-established experimental system for studying recombination and repair. However, since it is evolutionarily distant from the ascomycete S. cerevisiae (James et al., 2006), we were curious to learn how far the paradigm of homologous recombination proteins identified in S. cerevisiae might extend. We performed BLAST analyses with sequences of recombination proteins primarily from yeast (but also from human on a limited basis) as queries to identify candidates for the orthologous proteins in the U. maydis proteome and used information in the literature on genetics, protein domain structure, and/or configuration of key residues when available for verification. Sequence identity was computed over the total length of the shorter protein to account for non-overlap. Unless supporting information from genetic studies or additional information on domain structure or arrangement of critical amino acid residues was available to suggest otherwise, a threshold value of about 20% identity over protein length was taken as an indicator of orthology or to be more accurate, isofunctional homology (Gerlt and Babbitt, 2000; Jensen, 2001). Analysis suggests that U. maydis uses a simplified set of functions and that some components are in common with the yeast recombination system while others are more related to the recombination system of human (Table 1).
Table 1.
Homologous recombination proteins in U. maydis
| Protein | Organism | Probable function | U.maydis gene model | E-Valuea | Identitiesb | length of protein (Sc or Hs)c | length of protein (Um)c | identity * overlap / lengthd |
|---|---|---|---|---|---|---|---|---|
| End processing | ||||||||
| Rad50 | S. cerevisiae | DSB end sensing, processing | um01085 | 3e-167 | 419/1335 (31%) | 1312 | 1309 | 32% |
| Mre11 | S. cerevisiae | DSB end sensing, processing | um04704 | 1e-78 | 129/408 (31%) | 692 | 883 | 19% |
| Xrs2 | S. cerevisiae | DSB end sensing, processing | none | n.a. | n.a. | 854 | n.a. | n.a. |
| Sae2 | S. cerevisiae | DSB end processing | none | n.a. | n.a. | 345 | n.a. | n.a. |
| Exo1 | S. cerevisiae | DSB end processing | um03141 | 2e-71 | 134/300 (44%) | 702 | 828 | 19% |
| DNA strand exchange | ||||||||
| Rad51 | S. cerevisiae | Homologous pairing | um03290 | 1e-125 | 229/339 (67%) | 400 | 339 | 67% |
| BRCA2 | H. sapiens | Rad51 regulator | um03200, brh2 | 2e-32 | 88/254 (34%) | 3418 | 1075 | 8% |
| DSS1 | H. sapiens | BRCA2 regulator | um10918, dss1 | 5e-16 | 33/50 (66%) | 70 | 119 | 47% |
| Rad52 | S. cerevisiae | Rad51 mediator | um04989, rad52 | 4e-40 | 79/157 (50%) | 471 | 722 | 17% |
| Rad59 | S. cerevisiae | Homologous recombination, Rad52 related | none | n.a. | n.a. | 238 | n.a. | n.a. |
| Rad55 | S. cerevisiae | Homologous pairing, Rad51 paralog | none | n.a. | n.a. | 406 | n.a. | n.a. |
| Rad57 | S. cerevisiae | Homologous pairing, Rad51 paralog | um03095, rec2 | 4e-4 | 16/39 (41%) | 460 | 781 | 3% |
| Rad54 | S. cerevisiae | Chromatin remodelling ATPase | um02083 | 2e-209 | 363/573 (63%) | 898 | 870 | 42% |
| Rdh54 | S. cerevisiae | Chromatin remodelling ATPase | um03676 | 2e-111 | 307/904 (33%) | 924 | 1060 | 33% |
| Meiotic recombination | ||||||||
| Spo11 | S. cerevisiae | Meiotic double-strand break formation | um10420 | 3e-5 | 29/81 (35%) | 398 | 393 | 7% |
| Rec102 | S. cerevisiae | Meiotic double-strand break formation | none | n.a. | n.a. | 264 | n.a. | n.a. |
| Rec104 | S. cerevisiae | Meiotic double-strand break formation | none | n.a. | n.a. | 182 | n.a. | n.a. |
| Rec114 | S. cerevisiae | Meiotic double-strand break formation | none | n.a. | n.a. | 428 | n.a. | n.a. |
| Mei4 | S. cerevisiae | Meiotic double-strand break formation | none | n.a. | n.a. | 408 | n.a. | n.a. |
| Mer2 | S. cerevisiae | Meiotic double-strand break formation | none | n.a. | n.a. | 314 | n.a. | n.a. |
| Ski8 | S. cerevisiae | Meiotic double-strand break formation | none | n.a. | n.a. | 397 | n.a. | n.a. |
| Dmc1 | S. cerevisiae | Meiotic homologous pairing | none | n.a. | n.a. | 334 | n.a. | n.a. |
| Mnd1 | S. cerevisiae | Meiotic homologous pairing | none | n.a. | n.a. | 219 | n.a. | n.a. |
| Hop2 | S. cerevisiae | Meiotic homologous pairing | none | n.a. | n.a. | 218 | n.a. | n.a. |
| Mei5 | S. cerevisiae | Meiotic homologous pairing | none | n.a. | n.a. | 222 | n.a. | n.a. |
| Sae3 | S. cerevisiae | Meiotic homologous pairing | none | n.a. | n.a. | 91 | n.a. | n.a. |
| Hed1 | S. cerevisiae | Meiotic homologous pairing | none | n.a. | n.a. | 162 | n.a. | n.a. |
| Hop1 | S. cerevisiae | Synaptonemal complex | none | n.a. | n.a. | 605 | n.a. | n.a. |
| Red1 | S. cerevisiae | Synaptonemal complex | none | n.a. | n.a. | 827 | n.a. | n.a. |
| Zip1 | S. cerevisiae | Synaptonemal complex | none | n.a. | n.a. | 875 | n.a. | n.a. |
| Zip2 | S. cerevisiae | Synaptonemal complex | none | n.a. | n.a. | 704 | n.a. | n.a. |
| Zip3 | S. cerevisiae | Synaptonemal complex | none | n.a. | n.a. | 482 | n.a. | n.a. |
| Mer3 | S. cerevisiae | Meiotic crossing over | um11008 | 1e-126 | 181/528 (34%) | 1187 | 1265 | 15% |
| Msh4 | S. cerevisiae | Holliday junction clamp | um12336 | 8e-54 | 147/484 (30%) | 878 | 1046 | 17% |
| Msh5 | S. cerevisiae | Holliday junction clamp | um12155 | 8e-57 | 141/564 (25%) | 901 | 963 | 16% |
| Mlh1 | S. cerevisiae | Meiotic crossing over | um05208 | 3e-91 | 174/354 (49%) | 768 | 831 | 23% |
| Mlh3 | S. cerevisiae | Meiotic crossing over | um03481 | 2e-22 | 83/247 (33%) | 715 | 828 | 12% |
| Mus81 | S. cerevisiae | Structure-specific endonuclease | um04630 | 3e-42 | 92/310 (29%) | 632 | 605 | 15% |
| Recombination regulators and modifiers | ||||||||
| Sgs1 | S. cerevisiae | Recombination regulation | um02874 | 2e-134 | 284/678 (41%) | 1447 | 1291 | 22% |
| Top3 | S. cerevisiae | Recombination regulation | um11929 | 2e-135 | 187/440 (42%) | 656 | 986 | 28% |
| Rmi1 | S. cerevisiae | Recombination regulation | none | n.a. | n.a. | 241 | n.a. | n.a. |
| Srs2 | S. cerevisiae | Recombination regulation | um01691 | 8e-74 | 167/471 (35%) | 1174 | 1176 | 14% |
| FBH1 | H. sapiens | Recombination regulation | um03756 | 2e-26 | 71/219 (32%) | 1094 | 1166 | 6% |
| Shu1 | S. cerevisiae | Recombination regulation | none | n.a. | n.a. | 150 | n.a. | n.a. |
| Psy3 | S. cerevisiae | Recombination regulation | none | n.a. | n.a. | 242 | n.a. | n.a. |
| Shu2 | S. cerevisiae | Recombination regulation | none | n.a. | n.a. | 223 | n.a. | n.a. |
| Csm3 | S. cerevisiae | Recombination regulation | none | n.a. | n.a. | 317 | n.a. | n.a. |
E value <1e-3, "n.a.": not applicable.
As determined by BLASTP analysis of the protein sequence specified in column 1 to the predicted proteins of U. maydis (MUMDB, 01/2008) using the BLAST server at http://mips.gsf.de/genre/proj/ustilago/.
Sc, S. cerevisiae; Hs, H. sapiens; Um, U. maydis.
The length of the shorter protein was used.
2. DNA end processing
The Mre11-Rad50-Xrs2 (MRX) complex (MRN in mammals) is among the earliest responders to DNA double-strand breaks enabling structural, enzymatic, and signaling functions (Borde, 2007; Lee and Paull, 2007). The architecture of the complex is organized by Mre11, which binds Rad50, Xrs2 (Nbs1 in mammals) and DNA (Symington, 2002). Mre11 plays a critical role in appropriate processing of DNA ends that is a prelude required for repair by homologous recombination. Rad50 is related to the SMC (structural maintenance of chromosomes) coiled-coil domain family proteins and utilizes an ATP binding cassette, zinc hook, and coiled-coil domain to bridge DSBs and to facilitate DNA end processing by Mre11 (Hopfner et al., 2002). Contributing to a MRX regulatory role, Xrs2 harbors an N-terminal phosphopeptide interacting forkhead-associated domain (FHA) and C-terminal Mre11-interaction domain. In mammals the functional homolog of Xrs2 appears to be Nbs1 (mutated in the cancer predisposition disorder Nijmegan breakage syndrome), although similarity is restricted to the N-terminal region.
Sae2, which has distant isofunctional homologs Ctp1 and CtIP in Schizosaccharomyces pombe and human (Limbo et al., 2007), respectively, collaborates with the MRX(N) complex and contributes to processing DSBs by an inherent nuclease activity (Clerici et al., 2005; Lengsfeld et al., 2007). Cell cycle regulation of Sae2/Ctp1/CtIP levels could be a key controlling factor in limiting homologous recombination to the S/G2 phase of the cell cycle. CtIP, the isofunctional homolog of Sae2 in mammals has a number of interacting partners including the human breast cancer tumor suppressor BRCA1. The emerging evidence suggests that MRX(N) together with Sae2/Ctp1/CtIP has critical roles in DSB sensing, stabilization by scaffold formation, processing and DNA damage response signaling by recruitment of the ATM (ataxia-telangiectasia mutated) protein kinase (Takeda et al., 2007). Exo1, a 5′–3′ directed exonuclease also is likely to act in concert with Sae2 and the MRX(N) complex to play a role in end processing (Clerici et al., 2005; Fiorentini et al., 1997). Mre11, Rad50, and Exo1 are highly conserved and homologs are evident in the U. maydis genome. However, Xrs2 and Sae2 are poorly conserved and no obvious homolog to Xrs2 or Sae2 is recognizable by BLAST analysis.
3. DNA strand exchange
The defining step in recombinational repair is recognition of sequence homology coupled with DNA strand invasion (Folta-Stogniew et al., 2004). Rad51 promotes this process to achieve strand exchange over hundreds of base pairs (Sung, 1994). Rad51 forms a nucleoprotein filament by polymerizing on single-stranded DNA, which becomes a molecular machine that catalyzes DNA strand exchange in an ATP dependent manner. The filament is conserved in structure and function across the domains of life exemplifying the universal importance of its role in recombination. Once assembled the Rad51 filament is capable of interacting with a second DNA molecule to search for sequence homology and to initiate strand exchange. Self-polymerization leading to filament formation is an intrinsic property of Rad51, but assembly and disassembly of the filament is regulated by a number of other proteins (West, 2003).
The single-strand DNA binding protein RPA when bound to single-stranded DNA blocks Rad51 from polymerization. Therefore a key step in Rad51 filament assembly is overcoming the barrier to filament formation imposed by RPA. In yeast Rad52 enables loading of Rad51 on RPA-coated single-stranded DNA and thus provides a mediator function (New et al., 1998; Shinohara and Ogawa, 1998; Sung, 1997a). Upon binding to RPA-coated single-stranded DNA, Rad52 creates a seeding site for Rad51 to assemble and displace RPA. Rad52 also has a powerful DNA annealing activity (Mortensen et al., 1996) that could be important in pairing complementary strands on the second side of a DSB to complete synapsis of proximal and distal ends. Mutants lacking Rad52 in yeast are severely defective in all aspects of recombination and DSB repair and show the most profound defects of all the rad52 group mutants (Symington, 2002).
Rad51 is highly conserved in U. maydis with 75% sequence identity to human Rad51 and 67% identity to yeast Rad51. As in yeast loss of Rad51 results in profound recombination deficiency, meiotic failure, and extreme sensitivity to ionizing radiation and to other genotoxins such as DNA damaging chemicals and UV light (Ferguson et al., 1997; Kojic et al., 2002). Based on sequence alignment with residues known from the crystal structure to be involved in DNA binding and protein-protein association (Kagawa et al., 2002; Singleton et al., 2002), a Rad52 homolog is also present in U. maydis. The purified U. maydis protein exhibits powerful DNA annealing activity (Mazloum et al., 2007), but unlike the situation in yeast, disruption of the structural gene has little consequence in recombinational repair or meiosis in U. maydis (Kojic et al., 2008). A second related protein with overlapping function might serve as an explanation for this lack of phenotype. Rad59 in yeast is related to Rad52 in sequence across the DNA-binding, self-association domain. However, no clear Rad59 or other Rad52-related protein appears present in U. maydis.
Rad51 mediator activity in U. maydis appears to be performed by Brh2, a homolog of BRCA2, the product of a hereditary breast cancer predisposition gene in human (Kojic et al., 2002; Yang et al., 2002). Like Rad52, Brh2 has an associated DNA annealing activity (Mazloum et al., 2007) in addition to the ability to nucleate Rad51 filament formation on RPA-coated single-stranded DNA (Yang et al., 2005). In contrast to Rad52, loss of Brh2 results in profound radiation sensitivity and recombination deficiency (Kojic et al., 2002). Brh2 is structurally divergent from its counterpart in animals and plants, at one-third the size, with only a single BRC (a Rad51-interacting element) compared to four in Arabidopsis thailana and eight in vertebrates (Warren et al., 2002), and only two tandem OB (oligonucleotide/oligosaccharide binding) folds compared to three in vertebrates (Kojic et al., 2002). There is no apparent Brh2 equivalent in S. cerevisiae, but related proteins are clearly recognizable in fungi other than ascomycetes.
Dss1 is a small acidic protein that was originally identified in a two-hybrid screen for human proteins interacting with BRCA2 and was found in structural studies to co-crystallize with the BRCA2 DNA-binding domain (Yang et al., 2002). It is highly conserved in eukaryotes and an homolog is present in U. maydis (Kojic et al., 2003) as recognized by the two domains of conserved acidic residues with spaced aromatics that are known from the crystal structure of BRCA2 to intertwine with the helix-rich domain and OB (oligonucleotide/oligosaccharide binding) folds (Yang et al., 2002). Dss1 forms a strong complex with Brh2 and serves as a cofactor essential for its activity (Kojic et al., 2003; Kojic et al., 2005), similar to what has been observed with mammalian BRCA2 and DSS1 (Gudmundsdottir et al., 2004). Deletion of the Dss1 structural gene results in a phenocopy the brh2 mutant in radiation sensitivity and recombination deficiency (Kojic et al., 2003). Dss1 appears to serve in additional cellular processes. Besides its function in recombination, Dss1 has been identified as a non-essential component of the proteasome 19S regulatory particle (Krogan et al., 2004; Sone et al., 2004) and as a member of a nuclear mRNA processing complex (Thakurta et al., 2005). Sem1, a Dss1 homolog is present in S. cerevisiae, which appears to have no Brh2-related protein. However, deletion of the gene encoding Sem1 does not confer sensitivity to DNA damage or recombination deficiency (Marston et al., 1999).
In yeast the Rad51 paralogs Rad55 and Rad57 contribute to Rad51-filament formation and to assembling recombination complexes (Gasior et al., 1998; Lisby et al., 2004; Sung, 1997b). These proteins are related in primary sequence to Rad51 and are important for proficiency in recombination and repair. In vitro they stimulate Rad51 pairing reactions, but do not exhibit any autonomous capacity for DNA strand exchange. The proteins appear to form a heterodimer complex and to interact with Rad51 through the Rad55 component. In vertebrates there is an even greater expansion in Rad51 paralogs that includes Rad51B, Rad51C, Rad51D, Xrcc2 and Xrcc3 (Thacker, 2005). As in the case of yeast, the human paralogs form specific complexes among themselves (Schild et al., 2000; Yonetani et al., 2005) that stimulate Rad51 reactions (Liu et al., 2002), but by themselves lack the intrinsic capacity for ATP-dependent DNA strand exchange (Lio et al., 2003). In U. maydis Rec2 appears to be the only Rad51 paralog present in the genome (Rubin et al., 1994). Rec2 has a DNA-dependent ATPase activity, but unlike the Rad51 paralogs of yeast and human, it has an inherent ATP-dependent homologous pairing activity (Bennett and Holloman, 2001). The radiation sensitivity of rec2 can be suppressed by overexpressing Brh2, suggesting that by making more Rad51 available DNA repair can be executed in the absence of Rec2 (Kojic et al., 2006). Rad52 appears to overlap with Rec2 in DNA repair in some as yet unknown way because the rec2 rad52 double mutant exhibits a synthetic fitness phenotype with respect to radiation sensitivity that is even more severe than rad51 (Kojic et al., 2008).
From a historical perspective, the rec2 mutant defining the structural gene was the first recombinational repair mutant identified (originally named uvs-2) not only in U. maydis, but indeed in any eukaryote (Holliday, 1967). In early studies during the time of development of systems for exploring the biochemistry of recombination in eukaryotes, a protein preparation, which subsequently was shown to contain a proteolytic form of Rec2, was purified by conventional methods from extracts of U. maydis and was shown to promote homologous pairing and strand invasion in vitro (Bennett and Holloman, 2001).
Rad54 and the Rad54-related protein Rdh54 are members of the Swi2/Snf2 DNA-dependent ATPase family that serve in remodeling DNA and protein/DNA complexes. The proteins are viewed as motors that translocate on duplex DNA (Amitani et al., 2006) and respond to interactions with other proteins through domains outside of the core motor assembly (Thoma et al., 2005). Rad54 functions in all phases of Rad51-promoted DNA strand exchange including stimulating synapsis, heteroduplex extension, and disassembly of recombination complexes (Heyer et al., 2006). In yeast loss of Rad54 function results in extreme sensitivity to ionizing radiation, alkylating chemicals, DNA crosslinkers, and other agents causing DSB formation. Rad54 is partially redundant with the highly related Rdh54. The overlapping function is most notable during meiosis in which ascospore viability of rad54 rdh54 double mutants is reduced to the level of the rad51 mutant. Rad54 is thought to play a more dominant role in sister chromatid interaction while Rdh54 may be more dedicated to interactions between homologues. Clearly identifiable Rad54 and Rdh54 homologs are evident in U. maydis as well as in higher eukaryotes. Genetic studies on Rad54 in mouse and chicken have confirmed its importance in recombinational repair (Bezzubova et al., 1997; Essers et al., 1997), however, no comparable studies have yet been performed in U. maydis.
4. Meiotic recombination
Meiotic recombination is initiated by formation of DSBs through the action of Spo11, which cleaves DNA through a toposiomerase-like transesterification reaction breaking the phosphodiester backbone concomitant with generation of a covalent protein-DNA intermediate [reviewed in (Keeney and Neale, 2006)]. In yeast, at least nine other proteins are required for DSB formation. These include Mer2, Mei4, Rec102, Rec104, Rec114, and Ski8 as well as the MRX complex that is required for end-processing and removal of the Spo11-DNA adduct (Keeney, 2001). Except for MRX and Ski8, which contains multiple repeats of WD motifs that are thought to mediate protein-protein interactions, there is no clue to the function of any of the others. From genetics, immuno-cytochemistry, and chromatin immunoprecipitation studies the model that has emerged is that, rather than making up a single holoenzyme of defined stoichiometry, these proteins form subcomplexes that collaborate through a network of interactions to promote Spo11-dependent DSB formation (Maleki et al., 2007). In light of the highly sophisticated choreography implied by these interactions, it would be no surprise that Spo11-mediated DSB formation should be tightly regulated, and the components highly conserved. However, this is not the case. With the exception of Mre11 and Rad50 of the end-processing MRX(N) subcomplex as noted above and the WD-repeat protein Ski8, there are no proteins in U. maydis recognizable by BLAST analysis that correspond to Mer2, Mei4, Rec102, Rec104, or Rec114. Similar observations have been reported in S. pombe (Young et al., 2004). Ski8 is known to interact directly with Spo11 in yeast, but is all the more mysterious because it is also expressed mitotically and seems to have a role in regulating translation of nonpolyadenylated RNA (Maleki et al., 2007).
Proper disjunction of chromosomes during the reductional division of meiosis requires crossing over of homologues to make certain that the spindle apparatus can mediate segregation symmetrically [reviewed in (Baudat and de Massy, 2007; Bishop and Zickler, 2004; Neale and Keeney, 2006)]. Central to this process is the Holliday junction, which is formed as a key intermediate in the pathway leading to crossing over, and which provides the physical link connecting the homologues during the course of recombinational repair of Spo11-induced DSBs (Cromie et al., 2006; Schwacha and Kleckner, 1995). Many of the unique features of meiosis are thought to be devoted to insuring that obligatory crossover events take place between homologues rather than their being entirely associated with sister chromatids. In yeast, the Rad51-related protein Dmc1 and associated mediators Hop2 and Mnd1 are expressed in meiosis, promote DNA strand exchange in vitro, and may be part of the constellation of proteins that helps establish homologous pairing between homologues (Petukhova et al., 2005; Tsubouchi and Roeder, 2003). Dmc1, Hop2, and Mnd1 are highly conserved in eukaryotes from yeast to vertebrates and flowering plants. Loss of Dmc1 reduces sporulation, spore viability, and crossing over in yeast. The Sae3 and Mei5 proteins appear to act together with Dmc1 to function as accessory factors (Tsubouchi and Roeder, 2004). Mutation in these genes results in the same phenotype as the dmc1 mutant. When Dmc1 function is impaired Hed1 serves as a regulator to attenuate Rad51 activity (Tsubouchi and Roeder, 2006). Remarkably, there are no apparent homologs of any of these proteins in U. maydis. Drosophila, Caenorhabditis, and Neurospora have also been reported to lack Dmc1, Mnd1 and Hop2 (Ramesh et al., 2005).
In many systems meiotic cells develop specialized higher order chromosome structures culminating in formation of the synaptonemal complex (SC), the tripartite pairing structure, that eventually extends the length of each chromosome pair (Zickler, 2006). The pathways leading to recombination and SC formation interdigitate at a number of points. In S. cerevisiae, Spo11 is required for normal SC formation (Henderson and Keeney, 2004) as are several of the other gene products involved in Spo11-mediated DSB formation [for review see (Keeney, 2001)]. Red1 and Hop1 are thought to be structural components of meiotic chromosomes and are both required for SC formation (Hollingsworth et al., 1990; Rockmill and Roeder, 1988). Inactivation of the genes causes defects in DSB formation and meiotic recombination implying that DSB formation is coordinated with SC development. Zip1 is a structural component of the central region of the SC (Sym et al., 1993) while Zip2 and Zip3 are proteins required for polymerization of Zip1 along the homologues and initiating assembly of the SC central region (Fung et al., 2004). All three of these proteins contribute to crossover efficiency. Likewise, a DNA helicase Mer3 (Mazina et al., 2004), and Msh4 (Ross-Macdonald and Roeder, 1994) and Msh5 (Hollingsworth et al., 1995), which are both related to the mismatch repair MutS protein of E. coli, but are not involved in mismatch repair, are required for normal levels of meiotic crossing over in S. cerevisiae. These proteins, collectively known as ZMM (Zip/Mer/Msh), plus the Msh4-associated proteins Mlh1 and Mlh3 (Wang et al., 1999) contribute to a crossover pathway in yeast (Lynn et al., 2007). However, crossovers also proceed to a lesser extent independent of ZMM through the action of Mus81, which forms a heterodimeric structure-specific endonuclease with a partner protein, able to cleave Holliday junctions and other branched structures (Hollingsworth and Brill, 2004). U. maydis has recognizable homologs of Mer3, Msh4, Msh5, Mlh1, Mlh3, and Mus81, although the meiotic phenotype has not yet been determined. However, there are no candidates related to the SC structural or assembly proteins Red1, Hop1, Zip1, Zip2, or Zip3. There is a candidate protein in U. maydis that is related to mammalian SYCP1, a coiled coil protein thought to be a distant functional homologue of Zip1 (Costa and Cooke, 2007), but no protein related to other mammalian SC proteins such as SYCP2 or SYCP3.
In U. maydis compatible haploid strains fuse on maize plant surfaces to generate a dikaryon, which undergoes a morphological change to a pathogenic hyphal form that branches and spreads through maize tissue. Tumors induced in the plant as a result of the infection provide an environment for hyphal differentiation leading to karyogamy and development of teliospores that are arrested in what has been supposed to be a premeiotic state. It has been thought that teliospore germination is coupled with meiosis. However, in ultrastructural studies of germinating U. maydis teliospores no evidence for synaptonemal complexes was found (Fletcher, 1981; O'Donnell and McLaughlin, 1984). It is possible that meiosis in U. maydis proceeds without SC formation as is the case in certain other fungi such as S. pombe (Bahler et al., 1993). If this were the case, then the absence of recognizable structural and assembly proteins involved in SC formation would be understandable. Nevertheless, it remains a possibility that SC formation precedes teliospore development in U. maydis.
5. Recombination regulators and modifiers
Avoiding crossovers is a key feature of mitotic recombination as such events have the potential to generate genomic rearrangements and loss of heterozygosity. The BLM helicase (mutated in the human chromosome instability disorder Bloom’s syndrome), which is related to Sgs1 in yeast, can dissolve D-loops and Holliday junction recombination intermediates (van Brabant et al., 2000; Wu and Hickson, 2003). Dissociating D-loops after the invading strand has been extended by DNA synthesis, followed by its reannealing with the second end of a DSB, as in the migrating D-loop model (Figure 1), results in a repair event unaccompanied by crossing over. In association with the type I topoisomerase Top3. BLM can promote migration of two Holliday junctions towards each other and enable removal of the hemi-catenane that topologically links the two DNA duplexes (Plank et al., 2006). This dissolution mechanism avoids a crossover outcome as well. Thus, BLM and Top3 serve as important regulators of recombination. A third protein, Rmi1 (BLAP75 in human), associates with BLM and Top3 and promotes the dissolution activity (Wu et al., 2006). Homologs of BLM/Sgs1 and Top3 are present in U. maydis but no Rmi1 is recognizable.
Elevated recombination can have a destabilizing effect on the genome. Since the Rad51-single strand DNA nucleoprotein filament is the active element in initiating recombination events by strand invasion, controlling stability of the filament represents an important means for governing recombination. In addition, removal of Rad51 from DNA following recombination would appear to be an important step in cleansing the repaired DNA and bringing the process to a close. The Srs2 helicase in S. cerevisiae has the capacity to strip Rad51 from single-stranded DNA and genetic experiments provide evidence that it can serve as a negative regulator of recombination (Krejci et al., 2003; Veaute et al., 2003). Srs2 is conserved in other fungi including U. maydis, but no orthologs have been identified in vertebrates. On the other hand, a related helicase Fbh1 has been found in mouse, chickens, human, and S. pombe, but not in S. cerevisiae (Kim et al., 2002; Kohzaki et al., 2007; Morishita et al., 2005; Osman et al., 2005)}. Fbh1 helicases possess an F-box motif that forms part of a Skp/Cullin/F-box ubiquitin ligase complex. Studies with the human Fbh1 have shown that it can substitute for Srs2 in yeast in regulating recombination (Chiolo et al., 2007). Notably, in U. maydis there is also an Fbh1 related protein. Thus, there appears that U. maydis has an overlapping or redundant system in place possibly to insure that recombination is held in check. A similar situation exists in S. pombe where genetic studies suggest that the fbh1 srs2 double mutant is lethal due to unrestrained recombination (Morishita et al., 2005; Osman et al., 2005).
Interference with replication fork progression, or unrepaired single-strand breaks induced by endogenous or exogenous sources can result in fork collapse and breakage producing a one-sided DSB. Homologous recombination supports replication by contributing to repair of such lesions. This mode of repair in the context of replication appears to require some additional components that are specialized for fork support. The Shu complex consisting of Shu1, Psy3, Shu2, and Csm2, appears to constitute part of this system. The structural genes for these proteins were identified in a genetic screen in S. cerevisiae for suppressors that could alleviate the slow growth phenotype of top3 mutants and the sensitivity of sgs1 mutants to the replication inhibitor hydroxyurea (Shor et al., 2002). Shu2 and Psy3 are related to mammalian Rad51 paralogs (Martin et al., 2006) and are thought to have a specialized role in Rad51 filament formation dedicated to replication fork support (Mankouri et al., 2007), but none of the Shu complex components are evident in U. maydis.
6. Conclusions
There are two primary conclusions from this investigation. First, in mitotic cells the repertoire of gene products devoted to recombinational repair seems to comprise a simplified set of functional components many of which are shared in yeast. But in addition there are several components in common with the human system that appear to have been lost from yeast. The U. maydis system utilizes Brh2, a functional homolog of the human breast cancer tumor suppressor BRCA2, rather than Rad52 as the major controller of Rad51-promoted recombination, and relies on Dss1 as a regulator of Brh2. No BRCA2-related protein is present in yeast and there is no role for its Dss1-related protein in recombination. Unlike yeast or human U. maydis employs only a single Rad51 paralog, but has representatives from both yeast and human, namely, Srs2 and Fbh1, as negative regulators of the Rad51 filament.
The second major conclusion is that a number of genes involved in meiotic recombination appear to be absent in U. maydis. Some of these genes comprise subsets whose encoded proteins appear to function together and are widely represented throughout the domain of eukaryotes. Striking is the shared absence of Dmc1 and auxiliary factors dedicated to meiotic recombination and the apparent absence of components of the attendant meiotic chromosome structure, the synaptonemal complex. U. maydis has twenty-three chromosome pairs to distribute during meiosis, but has a chromosome missegregation frequency of only one to two percent [based on the appearance of disomes of chromosome 1 in meiotic products (Kojic et al., 2005)]. Assuming this frequency to be representative of the entire complement, such a degree of precision implies that pairing of homologues in meiosis, and by extension crossing over mediated by the homologous recombination system, is actively operational, appropriately regulated, and well-executed. Therefore, it would appear that Brh2, Rad51, and the single Rad51 paralog Rec2 suffice to supply the fundamental machinery for homologous pairing during meiosis. Whatever activities or subtleties in homologue versus sister recognition preference that are provided by Dmc1 and its auxiliary proteins in S. cerevisiae are not evident in U. maydis. It is interesting to consider that Rec2 by itself has the ability to promote Dmc1-like DNA strand exchange reactions in vitro, unlike other paralogs that have been investigated. Is it possible that Rec2 evolved not only for its Rad51-paralog function but also as a substitute for Dmc1?
A remarkable feature of U. maydis is its extreme resistance to killing by both UV and ionizing radiation (Holloman et al., 2007). The resistance is due in large part to its homologous recombination system. This conclusion can be immediately appreciated from the DNA repair phenotype of mutants defective in homologous recombination, i.e., they are extremely sensitive to UV as well as to ionizing radiation. On the other hand, in S. cerevisiae homologous recombination mutants are, for the most part, sensitive to ionizing radiation but not UV (Game and Mortimer, 1974). One interpretation of these observations is that U. maydis prefers to use recombination as a means for cleansing its genome of lesions that might ordinarily be channeled into more specialized repair systems in S. cerevisiae. This is not to imply that specialized repair systems are lacking in U. maydis; they most clearly are, as analysis has revealed a fairly standard collection of genes for nucleotide and base excision repair, translesion synthesis polymerases, postreplication repair, and non-homologous end-joining. Reliance on recombination as a primary line of defense against DNA damage attests to the efficacy of this system in maintaining genome stability in U. maydis.
Acknowledgments
We thank Lorraine Symington (Columbia University) and Joe Heitman (Duke University) for reading the manuscript. WKH gratefully acknowledges receiving alternative viewpoints from Milorad Kojic and financial support from National Institutes of Health grants GM42482 and GM79859.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Bahler J, Wyler T, Loidl J, Kohli J. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J Cell Biol. 1993;121:241–256. doi: 10.1083/jcb.121.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Symington LS. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007;15:565–577. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- Bennett RL, Holloman WK. A RecA homologue in Ustilago maydis that is distinct and evolutionarily distant from Rad51 actively promotes DNA pairing reactions in the absence of auxiliary factors. Biochemistry. 2001;40:2942–2953. doi: 10.1021/bi002494i. [DOI] [PubMed] [Google Scholar]
- Bezzubova O, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, Buerstedde JM. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Zickler D. Early decision: meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 2007;15:551–563. doi: 10.1007/s10577-007-1147-9. [DOI] [PubMed] [Google Scholar]
- Chiolo I, Saponaro M, Baryshnikova A, Kim JH, Seo YS, Liberi G. The human F-Box DNA helicase FBH1 faces Saccharomyces cerevisiae Srs2 and postreplication repair pathway roles. Mol Cell Biol. 2007;27:7439–7450. doi: 10.1128/MCB.00963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- Costa Y, Cooke HJ. Dissecting the mammalian synaptonemal complex using targeted mutations. Chromosome Res. 2007;15:579–589. doi: 10.1007/s10577-007-1142-1. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, Hendriks RW, Swagemakers SM, Troelstra C, de Wit J, Bootsma D, Hoeijmakers JH, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- Ferguson DO, Holloman WK. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci U S A. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DO, Rice MC, Rendi MH, Kotani H, Kmiec EB, Holloman WK. Interaction between Ustilago maydis REC2 and RAD51 genes in DNA repair and mitotic recombination. Genetics. 1997;145:243–251. doi: 10.1093/genetics/145.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher HL. A search for synaptonemal complexes in Ustilago maydis. J Cell Sci. 1981;50:171–180. doi: 10.1242/jcs.50.1.171. [DOI] [PubMed] [Google Scholar]
- Folta-Stogniew E, O'Malley S, Gupta R, Anderson KS, Radding CM. Exchange of DNA base pairs that coincides with recognition of homology promoted by E. coli RecA protein. Mol Cell. 2004;15:965–975. doi: 10.1016/j.molcel.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Fung JC, Rockmill B, Odell M, Roeder GS. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell. 2004;116:795–802. doi: 10.1016/s0092-8674(04)00249-1. [DOI] [PubMed] [Google Scholar]
- Game JC, Mortimer RK. A genetic study of x-ray sensitive mutants in yeast. Mutat Res. 1974;24:281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC. Can sequence determine function? Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-5-reviews0005. reviews0005.1–0005.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsdottir K, Lord CJ, Witt E, Tutt AN, Ashworth A. DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep. 2004;5:989–993. doi: 10.1038/sj.embor.7400255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KA, Keeney S. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc Natl Acad Sci U S A. 2004;101:4519–4524. doi: 10.1073/pnas.0400843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- Holliday R. Studies on mitotic gene conversion in Ustilago. Genet. Res. 1966;8:323–337. doi: 10.1017/s0016672300010181. [DOI] [PubMed] [Google Scholar]
- Holliday R. Altered recombination frequencies in radiation sensitivie strains of Ustilago. Mutation Research. 1967;4:275–288. doi: 10.1016/0027-5107(67)90022-x. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Goetsch L, Byers B. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990;61:73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- Holloman WK, Schirawski J, Holliday R. Towards understanding the extreme radiation resistance of Ustilago maydis. Trends Microbiol. 2007;15:525–529. doi: 10.1016/j.tim.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JH, Tainer JA. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Hurst DD, Fogel S. Mitotic recombination and heteroallelic repair in Saccharomyces cerevisiae. Genetics. 1964;50:435–458. doi: 10.1093/genetics/50.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst DD, Fogel S, Mortimer RK. Conversion-associated recombination in yeast (hybrids-meiosis-tetrads-marker loci-models) Proc Natl Acad Sci U S A. 1972;69:101–105. doi: 10.1073/pnas.69.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interthal H, Heyer WD. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O'Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schussler A, Longcore JE, O'Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lucking R, Budel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Jensen RA. Orthologs and paralogs - we need to get it right. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-interactions1002. Interactions1002.1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa W, Kurumizaka H, Ishitani R, Fukai S, Nureki O, Shibata T, Yokoyama S. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol Cell. 2002;10:359–371. doi: 10.1016/s1097-2765(02)00587-7. [DOI] [PubMed] [Google Scholar]
- Kamper J, Kahmann R, Bolker M, Ma LJ, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Muller O, Perlin MH, Wosten HA, de Vries R, Ruiz-Herrera J, Reynaga-Pena CG, Snetselaar K, McCann M, Perez-Martin J, Feldbrugge M, Basse CW, Steinberg G, Ibeas JI, Holloman WK, Guzman P, Farman M, Stajich JE, Sentandreu R, Gonzalez-Prieto JM, Kennell JC, Molina L, Schirawski J, Mendoza-Mendoza A, Greilinger D, Munch K, Rossel N, Scherer M, Vranes M, Ladendorf O, Vincon V, Fuchs U, Sandrock B, Meng S, Ho EC, Cahill MJ, Boyce KJ, Klose J, Klosterman SJ, Deelstra HJ, Ortiz-Castellanos L, Li W, Sanchez-Alonso P, Schreier PH, Hauser-Hahn I, Vaupel M, Koopmann E, Friedrich G, Voss H, Schluter T, Margolis J, Platt D, Swimmer C, Gnirke A, Chen F, Vysotskaia V, Mannhaupt G, Guldener U, Munsterkotter M, Haase D, Oesterheld M, Mewes HW, Mauceli EW, DeCaprio D, Wade CM, Butler J, Young S, Jaffe DB, Calvo S, Nusbaum C, Galagan J, Birren BW. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444:97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans. 2006;34:523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim JH, Lee SH, Kim DH, Kang HY, Bae SH, Pan ZQ, Seo YS. The novel human DNA helicase hFBH1 is an F-box protein. J Biol Chem. 2002;277:24530–24537. doi: 10.1074/jbc.M201612200. [DOI] [PubMed] [Google Scholar]
- Kohzaki M, Hatanaka A, Sonoda E, Yamazoe M, Kikuchi K, Vu Trung N, Szuts D, Sale JE, Shinagawa H, Watanabe M, Takeda S. Cooperative roles of vertebrate Fbh1 and Blm DNA helicases in avoidance of crossovers during recombination initiated by replication fork collapse. Mol Cell Biol. 2007;27:2812–2820. doi: 10.1128/MCB.02043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic M, Kostrub CF, Buchman AR, Holloman WK. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell. 2002;10:683–691. doi: 10.1016/s1097-2765(02)00632-9. [DOI] [PubMed] [Google Scholar]
- Kojic M, Mao N, Zhou Q, Lisby M, Holloman WK. Compensatory role for Rad52 during recombinational repair in Ustilago maydis. Mol Microbiol. 2008;67:1156–1168. doi: 10.1111/j.1365-2958.2008.06116.x. [DOI] [PubMed] [Google Scholar]
- Kojic M, Yang H, Kostrub CF, Pavletich NP, Holloman WK. The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2003;12:1043–1049. doi: 10.1016/s1097-2765(03)00367-8. [DOI] [PubMed] [Google Scholar]
- Kojic M, Zhou Q, Lisby M, Holloman WK. Brh2-Dss1 interplay enables properly controlled recombination in Ustilago maydis. Mol Cell Biol. 2005;25:2547–2557. doi: 10.1128/MCB.25.7.2547-2557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic M, Zhou Q, Lisby M, Holloman WK. Rec2 interplay with both Brh2 and Rad51 balances recombinational repair in Ustilago maydis. Mol Cell Biol. 2006;26:678–688. doi: 10.1128/MCB.26.2.678-688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, Greenblatt JF. Proteasome involvement in the repair of DNA double-strand breaks. Molecular Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Kong H, Nei M, Ma H. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci U S A. 2006;103:10328–10333. doi: 10.1073/pnas.0604232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio YC, Mazin AV, Kowalczykowski SC, Chen DJ. Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J. Biol. Chem. 2003;278:2469–2478. doi: 10.1074/jbc.M211038200. [DOI] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Liu N, Schild D, Thelen MP, Thompson LH. Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res. 2002;30:1009–1015. doi: 10.1093/nar/30.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn A, Soucek R, Borner GV. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- Maleki S, Neale MJ, Arora C, Henderson KA, Keeney S. Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma. 2007;116:471–486. doi: 10.1007/s00412-007-0111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone RE, Bullard S, Hermiston M, Rieger R, Cool M, Galbraith A. Isolation of mutants defective in early steps of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics. 1991;128:79–88. doi: 10.1093/genetics/128.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Ngo HP, Hickson ID. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol Biol Cell. 2007;18:4062–4073. doi: 10.1091/mbc.E07-05-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol. 1999;19:4633–4642. doi: 10.1128/mcb.19.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR, 3rd, McGowan CH, Russell P. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. Embo J. 2006;25:2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina OM, Mazin AV, Nakagawa T, Kolodner RD, Kowalczykowski SC. Saccharomyces cerevisiae Mer3 helicase stimulates 3′–5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell. 2004;117:47–56. doi: 10.1016/s0092-8674(04)00294-6. [DOI] [PubMed] [Google Scholar]
- Mazloum N, Zhou Q, Holloman WK. DNA binding, annealing, and strand exchange activities of Brh2 protein from Ustilago maydis. Biochemistry. 2007;46:7163–7173. doi: 10.1021/bi700399m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menees TM, Roeder GS. MEI4, a yeast gene required for meiotic recombination. Genetics. 1989;123:675–682. doi: 10.1093/genetics/123.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson MS, Radding CM. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975;72:358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T, Furukawa F, Sakaguchi C, Toda T, Carr AM, Iwasaki H, Shinagawa H. Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates. Mol Cell Biol. 2005;25:8074–8083. doi: 10.1128/MCB.25.18.8074-8083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci U S A. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- O'Donnell KL, McLaughlin DJ. Ultrastructure of meiosis in Ustilago maydis. Mycologia. 1984;76:465–485. [Google Scholar]
- Osman F, Dixon J, Barr AR, Whitby MC. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol Cell Biol. 2005;25:8084–8096. doi: 10.1128/MCB.25.18.8084-8096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova GV, Pezza RJ, Vanevski F, Ploquin M, Masson JY, Camerini-Otero RD. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat Struct Mol Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- Plank JL, Wu J, Hsieh TS. Topoisomerase IIIalpha and Bloom's helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci U S A. 2006;103:11118–11123. doi: 10.1073/pnas.0604873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh MA, Malik SB, Logsdon JM., Jr A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. RED1: a yeast gene required for the segregation of chromosomes during the reductional division of meiosis. Proc Natl Acad Sci U S A. 1988;85:6057–6061. doi: 10.1073/pnas.85.16.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman HL, Jacob F. A comparison of spontaneous and ultraviolet-induced allelic recombination with reference to the recombination of outside markers. Cold Spring Harbor Symp. Quant. Biol. 1958;23:155–160. doi: 10.1101/sqb.1958.023.01.019. [DOI] [PubMed] [Google Scholar]
- Rong L, Palladino F, Aguilera A, Klein HL. The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics. 1991;127:75–85. doi: 10.1093/genetics/127.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P, Roeder GS. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 1994;79:1069–1080. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Rubin BP, Ferguson DO, Holloman WK. Structure of REC2, a recombinational repair gene of Ustilago maydis, and its function in homologous recombination between plasmid and chromosomal sequences. Mol Cell Biol. 1994;14:6287–6296. doi: 10.1128/mcb.14.9.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D, Lio YC, Collins DW, Tsomondo T, Chen DJ. Evidence for simultaneous protein interactions between human Rad51 paralogs. J Biol Chem. 2000;275:16443–16449. doi: 10.1074/jbc.M001473200. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Shor E, Gangloff S, Wagner M, Weinstein J, Price G, Rothstein R. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics. 2002;162:647–662. doi: 10.1093/genetics/162.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB. Structure of the single-strand annealing domain of human RAD52 protein. Proc Natl Acad Sci U S A. 2002;99:13492–13497. doi: 10.1073/pnas.212449899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone T, Saeki Y, Toh-e A, Yokosawa H. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:28807–28816. doi: 10.1074/jbc.M403165200. [DOI] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 1997a;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997b;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- Sym M, Engebrecht JA, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Takeda S, Nakamura K, Taniguchi Y, Paull TT. Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell. 2007;28:351–352. doi: 10.1016/j.molcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–135. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Thakurta AG, Gopal G, Yoon JH, Kozar L, Dhar R. Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast. EMBO J. 2005;24:2512–2523. doi: 10.1038/sj.emboj.7600713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS. The budding yeast mei5 and sae3 proteins act together with dmc1 during meiotic recombination. Genetics. 2004;168:1219–1230. doi: 10.1534/genetics.103.025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 2006;20:1766–1775. doi: 10.1101/gad.1422506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Warren M, Smith A, Partridge N, Masabanda J, Griffin D, Ashworth A. Structural analysis of the chicken BRCA2 gene facilitates identification of functional domains and disease causing mutations. Hum Mol Genet. 2002;11:841–851. doi: 10.1093/hmg/11.7.841. [DOI] [PubMed] [Google Scholar]
- West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- Yonetani Y, Hochegger H, Sonoda E, Shinya S, Yoshikawa H, Takeda S, Yamazoe M. Differential and collaborative actions of Rad51 paralog proteins in cellular response to DNA damage. Nucleic Acids Res. 2005;33:4544–4552. doi: 10.1093/nar/gki766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Hyppa RW, Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]