Abstract
The insecticidal nature of Cry δ-endotoxins produced by Bacillus thuringiensis is generally believed to be caused by their ability to form lytic pores in the midgut cell membrane of susceptible insect larvae. Here we have analyzed membrane-associated structures of the 65-kDa dipteran-active Cry4Ba toxin by electron crystallography. The membrane-associated toxin complex was crystallized in the presence of DMPC bilayers via detergent dialysis. Depending upon the charge of the adsorbed surface, 2D crystals of the oligomeric toxin complex have been captured in two distinct conformations. The projection maps of those crystals have been generated at 17 Å resolution. Both complexes appeared to be trimeric; as in one crystal form, its projection structure revealed a symmetrical pinwheel-like shape with virtually no depression in the middle of the complex. The other form revealed a propeller-like conformation displaying an obvious hole in the center region, presumably representing the toxin-induced pore. These crystallographic data thus demonstrate for the first time that the 65-kDa activated Cry4Ba toxin in association with lipid membranes could exist in at least two different trimeric conformations, conceivably implying the closed and open states of the pore.
Keywords: Bacillus thuringiensis, Cry δ-endotoxins, Electron crystallography, Membrane-associated toxin complex, Trimeric structure
Introduction
Cry δ-endotoxins made by Bacillus thuringiensis (Bt) are cytolytic pore-forming toxins that have been shown to be highly active against a wide variety of insect larvae [1]. For example, the 130-kDa Cry4Ba toxin produced from Bt subsp. israelensis is specifically toxic to mosquito-larvae of the genus Aedes and Anopheles [1]. These mosquitoes currently continue to be the most important vectors of certain serious human diseases such as dengue haemorrhagic fever and malaria [2].
The transformation of Bt Cry toxins from inactive protoxins, found within inclusion bodies, to cytolytic pore-forming toxins is a multistep process [3]. After being ingested by susceptible insect larvae, and dissolution in the midgut lumens, the solubilized protoxins are activated by larval gut proteases to yield toxic fragments of ∼65 kDa. The active toxins subsequently bind to specific receptors in the luminal plasma membrane of midgut epithelial cells. It is believed that active toxin subunits oligomerlize to form structures capable of inserting into the membrane to generate ion-leakage pores. These pores cause a net influx of ions and water, resulting in cell swelling, lysis, and the eventual death of the target larvae [3]. Although the molecular mechanistic view of how these toxins work has increased substantially over the last decade [4,5], the actual underlying mechanism of this toxicity process remains to be explored. Particularly, elucidation the pore-forming structure within the lipid membrane could lead to a more critical understanding of structural basis for membrane-pore formation of these toxins, and facilitating the design of more potent toxins.
To date, X-ray crystal structures of almost all the major specificity classes of Cry δ-endotoxins have been solved [6-10]. Regardless of target insect specificities and amino acid sequence identity, all the known Cry toxin structures reveal a high degree of overall structural similarity with three-distinct domains. The N-terminal domain is a group of seven helices in which the most hydrophobic helix α5 is encircled by six other amphipathic helices. Structurally, it is immediately apparent that domain I is likely to be the transmembrane pore-forming apparatus (see Fig. 1C). In addition, it has been experimentally evident that this helical domain is able to form functional pores, albeit in the absence of specific receptors [11,12]. The middle domain comprises three anti-parallel β-sheets aligned in β-prism motifs with surface-exposed loops that have been demonstrated to be involved in receptor binding, and hence determines specificity [4,5]. The C-terminal domain, whose function is still unclear, is a β-sandwich made up of two anti-parallel β-sheets arranged in a jelly-roll-like topology [5].
Fig. 1.
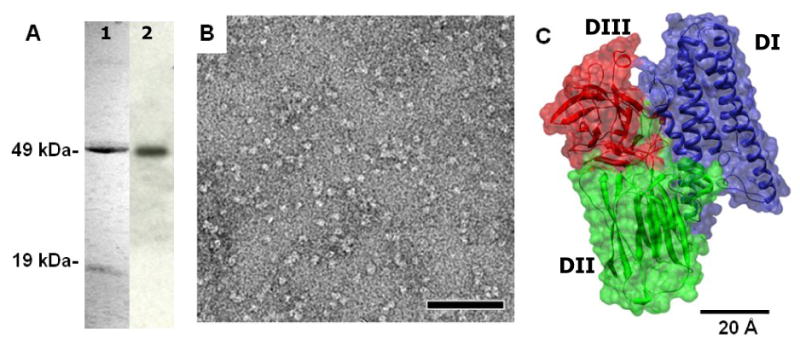
Purification of the 65-kDa activated Cry4Ba toxin. (A) SDS-PAGE (Coomassie brilliant blue-stained 12% gel) analysis of the FPLC-purified toxin (lane 1) and Western blotting, probed with the anti Cry4Ba-domain III MAb (lane 2). (B) Images of negatively stained Cry4Ba toxins, showing homogeneous population of particles at the size corresponding to the single Cry4Ba particle. The scale bar is 50 nm. (C) Ribbon and surface representation of the three-domain Cry4Ba toxin organization; a bundle of α-helices (DI), a three-β-sheet domain (DII), and a β-sandwich (DIII). The measured dimension of the 65-kDa activated Cry4Ba X-ray structure is 55×66×76 Å.
Despite the fact that several pore models have been proposed [13-16], substantial evidence has preferentially supported an “umbrella-like” model for describing the membrane-bound state of the Cry toxins [13]. This model involves an insertion of α4-α5 transmembrane hairpins from domain I into the lipid bilayers, with the remaining helices spreading over the membrane surface. According to the pore size estimated, ranging from 10-26 Å [17,18], a single molecule Cry protein would not be sufficient to constitute the pore [17]. Thus, toxin oligomerization would obviously have to take place. Indeed, various approaches have been used to enlighten the oligomerization process of the Cry toxins [14,19,20]. Albeit the tetrameric building is widely believed to be a real oligomericity of the Cry toxins [19-21], several arguments could still be made (see Table 1). In other words, the structural information obtained from the soluble monomeric Cry toxin has not been able to provide detailed insights into the structural basis of the toxin-pore formation. To address this question, visualizing the pore structure and oligomeric state of the membrane-associated form would be mandatory.
Table 1.
Proposed oligomericity of Cry toxins
| Oligomericity | Toxin | Experimental approach | Dimensions | Reference |
|---|---|---|---|---|
| Dimer | Cry1Ab | Incubating toxins with susceptible larval BBMVs1, analyzed by modified SDS-PAGE and Western blotting | ∼ 130 kDa | [35] |
| Dimer & Trimer | Cry1Ac | ∼ 130 kDa & ∼ 210 kDa | ||
| Dimer & Trimer | Cry1Ac | Using dynamics light scattering in buffer with different ionic strengths | Molecular mass of each oligomer was derived from hydrodynamic radius of proteins from dynamics light scattering | [36] |
| Trimer | Cry1Aa,
Cry1Ab Cry1Ac |
Incubating toxins with susceptible larval BBMVs1, analyzed by modified SDS-PAGE and Western blotting | ∼ 200 kDa | [37,38] |
| Trimer | Cry4Ba | Incubating toxins with liposomes, analyzed by modified SDS-PAGE and Western blotting | ∼ 200 kDa | [30] |
| Tetramer | Cry1Aa | AFM2 of toxin inserted in bilayers in liquid cell | Diameter 5 nm, each subunit 1.4 nm, depression 1.5 nm | [21] |
| Tetramer | Cry4Ba | AFM2 of toxin inserted in bilayers in liquid cell | Diameter 20-30 nm, height 2-4 nm | [20] |
| Tetramer | Cry1Ab | ScFV73 cross-linked toxin3, binding with ANS 4 | ∼ 250 kDa | [19] |
| Large multimer | Cry1Aa
Cry1Ac Cry1Ca Cry1D Cry3Aa |
Analyzed by native and modified SDS-PAGE, and size-exclusion chromatography | >600 kDa | [39] |
BBMVs = Brush-border membrane vesicles
AFM = Atomic force microscopy
ScFV73 = A single chain antibody mimicking Bt-R1 receptor
ANS = 8-Anilino-1-naphthalenesulfonate
In this study, we have employed the 2D crystallization technique available for membrane proteins [22,23] to gain structural insights into the membrane-bound states of the Cry4Ba toxin. Interestingly, two different forms of well-ordered 2D crystals were observed on DMPC bilayers. Despite the fact that they all appeared to be trimeric, the projection density maps of both kinds of 2D crystals calculated at 17 and 20 Å revealed significant structural discrepancy between these two trimeric states.
Materials and methods
Expression and preparation of toxin inclusions
Escherichia coli cells strain JM109 harboring the recombinant plasmid (pMU388) which encodes the 130-kDa Cry4Ba toxin [24] were grown at 37°C in Luria-Bertani broth supplemented with 100 μg/ml ampicillin. When the culture reached OD600 0.4-0.5, IPTG was added to a final concentration of 0.1 mM, and incubation was continued for 4 hrs. Cells expressing the toxin as inclusion bodies were harvested by centrifugation, resuspended in distilled water and then disrupted in a French Pressure Cell at 12,000 psi. After being collected by centrifugation, toxin inclusions were washed 3 times with cold distilled water. Protein concentrations of the partially purified inclusions were determined by using the bicinchoninic acid assay (Pierce), with bovine serum albumin fraction V (Sigma) as a standard.
Purification and characterization of the activated toxin
Protoxin inclusions were incubated in 50mM Na2CO3, pH 10.5 at 37°C for 1 hr as previously described [25]. The solubilized protoxins were then converted to active toxins by digesting with trypsin (TPCK-treated, Sigma) at toxin/enzyme ratio of 20/1 (w/w) in 50mM Na2CO3, pH 10.5, at 37°C for 16 hrs, and enzyme activity was inhibited by adding PMSF to give a final concentration of 1 mM. Purification of the 65-kDa trypsin-activated toxin was accomplished using a size-exclusion FPLC system (Superdex 200, 10/300 GL Amersham Pharmacia Bioscience) eluted with carbonate buffer (50 mM Na2CO3, pH 10.5) at a flow rate of 0.4 ml/min. Gel filtration standards (Bio-Rad) were used to calibrate the column. Immonoblotting of the purified toxin was performed with the 2F-1H2 MAb which is specific to the Cry4Ba-domain III fragment as previously described [26]. The homogeneity of the purified Cry4Ba toxin was finally evaluated by visualizing under negative stained electron microscopy.
2D crystallization of the 65-kDa Cry4Ba toxin
Purified Cry4Ba proteins were diluted to a final concentration of 0.5-1 mg/ml in 12.5 mM Tris-HCl, pH 8.0, 37.5 mM NaCl, 37.5 mM Na2CO3 and 10% OG in a total volume of 100 μl. DMPC, solubilized in 10% OG, was added at 25°C to give a final lipid-to-protein mass ratio of 1.0. Highly ordered arrays of the membrane associated toxin were produced by transferring the solution to home-made dialysis buttons (14-kDa cutoff) and dialyzing against 50 mM Na2CO3, pH 10.5 for 24-48 hrs at 25°C.
Negative stain electron microscopy and image analysis
2D crystals were placed either on air glow discharged (negatively charged surface) or ten day-old desiccated (hydrophobic) carbon coated Cu/Rh grids. Excess sample was removed by blotting with filter paper before staining with three washes of 1% uranyl acetate. Samples were analyzed using a Tecnai T12 electron microscope equipped with a LaB6-filament, operated at 120 kV and ×30,000 magnification. Electron microscopic images were recorded with a GATAN 794, 1,024 × 1,024-pixel charge-coupled device camera using a dose of approximately 25 electrons/Å2 and underfocus values that placed the first zero of the contrast transfer function past the resolution cutoff of the reconstructions. Images were corrected for lattice distortion using the MRC software package [27] and projection density maps were calculated in CCP4.
Results and discussion
It is has been proposed that the Cry toxins lyse target midgut epithelial cells by forming an ion-leakage pore, size ranging from 10-26 Å in diameter [17,18], which rapidly dissipates the membrane potential. Despite their high specificity, various electrophysiological experiments have shown that the Cry toxins were able to form lytic pores even in the absence of their receptors [11,28,29]. Previous studies have also shown that the Cry toxin-induced pores are cation selective [11,29]. Nonetheless, these leakage pores do not seem to be very discriminating about ion species since several large solutes, e.g. sucrose [28], raffinose [28], calcein [30] and polyetheleneglycol [18], have been shown to permeate through these pores. Thus far, the number of subunits that constitute a functional pore complex and whether the oligomarization occurs in the membrane-bound state has been still under debate (see Table I). In this study, we have therefore employed electron crystallography to gain more critical insights into the lipid-associated structure and oligomeric state of the Cry4Ba toxin.
As previously demonstrated, the 130-kDa Cry4Ba protoxin was cleaved by trypsin into two protease-resistant polypeptides of ∼49 and ∼19 kDa, in addition to the removal of the C-terminal half of the protoxin [25]. These two-main products were produced by a cleavage at Arg203 located in the exposed loop linking helices 5 and 6 within the pore-forming domain [25]. Upon purification on a size-exclusion FPLC column, the trypsin-treated Cry4Ba fragments (49 and 19 kDa) were found associated with each other under the non-denaturing condition used (50 mM Na2CO3, pH 10.5) and eluted from the column in a single peak whose retention time corresponded to that of the 66-kDa BSA marker (data not shown). This indicated that the 65-kDa purified Cry4Ba toxin exists in a monomeric form.
When freshly purified Cry4Ba proteins were evaluated by Western blotting with the anti-Cry4Ba-domain III MAb [26], a single band was detected at ∼49 kDa, corresponding to α6-loop-α7 linked with domains II and III of the 65-kDa activated toxin (Fig. 1A). Subsequently, electron microscopy of uranylacetate stained samples revealed a homogeneous population of particles with a size corresponding to the monomeric toxin (Fig. 1B, 1C).
To determine the structure and oligomeric state of the Cry4Ba toxin-induced pore in the lipid bilayers, we have crystallized the 65-kDa purified toxins in the presence of DMPC lipid by detergent dialysis technique. Well-ordered 2D crystal patches of up to 1×1 μm were diffracted to ∼15 Å after correction of the lattice for translational disorder (Fig 2A, 2B). To our surprise, the properties of the carbon support film (hydrophobic or glow discharge) affected both the unit cell parameters and apparent conformational state of the toxin. While generally obeying p3 symmetry, the cell of 2D crystal deposited on the hydrophobic carbon film was a = b = 112 Å, γ = 120° while the crystal had unit cell dimensions of a = b = 107 Å, γ = 120° when deposited on a hydrophilic carbon substrate. Moreover, the calculated projection structures displayed different conformation of the three-domain Cry4Ba protein. The propeller-like structure was obtained from the charged surface (Fig 3A, 3C) while the hydrophobic-bound trimeric structure has a pinwheel-like appearance (Fig 3B, 3D), demonstrating that this 65-kDa three-domain toxin had undergone some significant rearrangement.
Fig. 2.
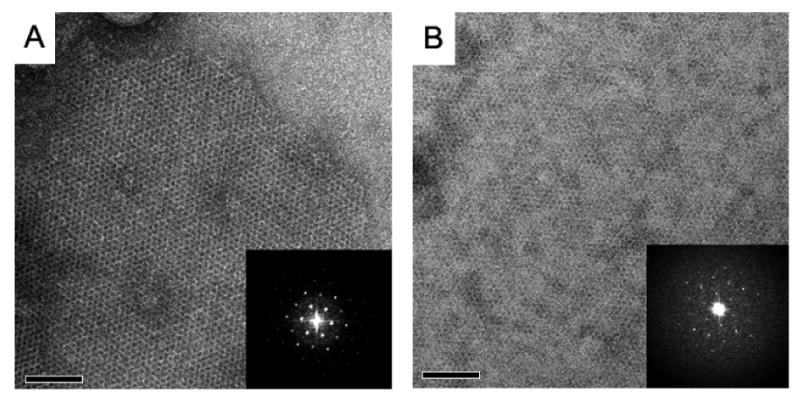
Two types of 2D crystals imaged in negative stain. (A) The 65-kDa Cry4Ba toxin crystal grown in DMPC on the air glow discharged carbon surface, and (B) the toxin crystal in DMPC attached on the hydrophobic carbon surface. Crystals were up to 1×1 μm in size. Insets show Fourier transforms of these crystalline areas.
Fig. 3.
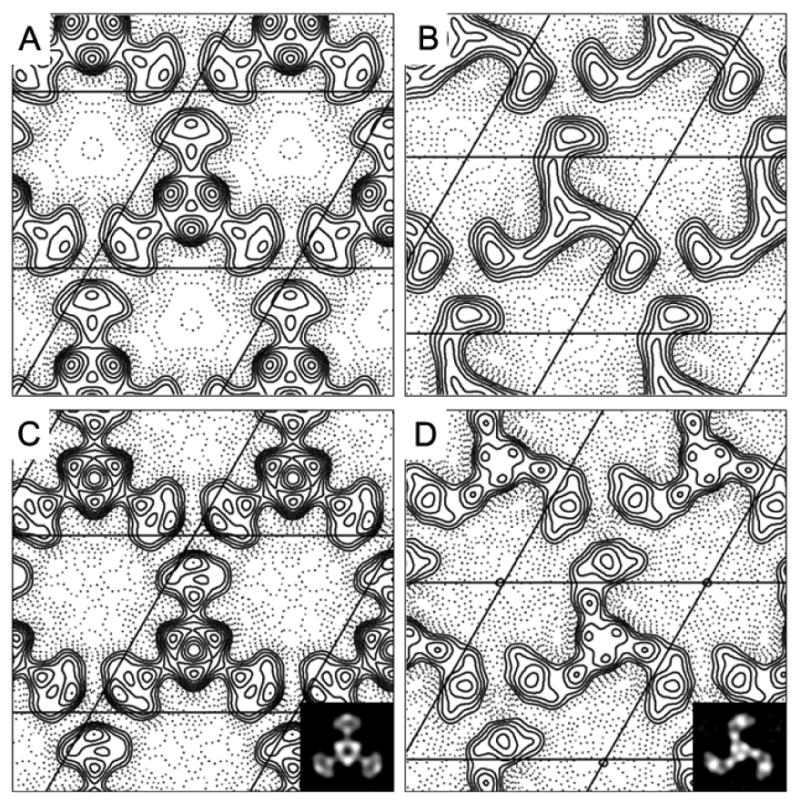
Projection maps of propeller and pinwheel crystal forms. The maps belong to p3 plane group. The contours of the propeller-like and pinwheel-like crystals were plotted at 20 Å (A) and (B), and at 15 Å resolution (C) and (D). Insets show projection density maps of the propeller-like (C) and pinwheel-like structures (D).
Like other ion channels [23,31], the Cry toxin-induced pores or channels would possibly undergo a conformational change upon gating of the protein. Even at this stage the gating mechanism of the pore-forming Cry toxins is still unrevealed, several planar lipid bilayer experiments suggested that the channels induced by Cry toxins exhibit at least two different functional states: closed and open [11,32]. Could these two trimeric conformations observed here be the closed and open states of the Cry4Ba toxin? Indeed, they could likely be each functional state since some major structural differences were seen at the pore mouth and peripheral region for both trimers (see Fig. 3).
It is noteworthy that the pinwheel-like structure exhibited a very clear clockwise handedness in that the L-shape blades which could represent the domains II and III of the Cry4Ba toxin pointing towards the same direction, whilst the central region which is likely to embody the pore-forming domain I is relatively small (Fig. 3B, 3D). Surprisingly, the projection map of this pinwheel structure showed virtually no depression in this region even when the map was calculated at 17 Å resolution (Fig. 3D). Notwithstanding the lack of detailed functional characterization, this pinwheel-like structure might perhaps represent the closed state of the Cry4Ba-induced pore in the membrane.
On the other hand, the central region of the propeller-like conformation is significantly larger with the stain excluded region in the center of the map (Fig 3A, 3C), which could possibly reflect a cavity of an open pore. It should be noted that this pore configuration can be seen even when the map was calculated at 20 Å resolution (Fig. 3A). This observation is in agreement with the estimated pore size from permeation experiments for Cry1Ab [17] and the blockade of various sizes of polyethyleneglycols for Cry1Ca [18] in that the pore size should be in the range of 10-26 Å. In addition, a major conformational change observed in the blade region of the trimer leads us to an assumption that the toxin molecules are associated peripherally with the lipid membrane and would insert only small part of the protein, i.e. the α4-loop-α5 hairpin, into the lipid layers as proposed earlier [13].
In conclusions, the two symmetrical trimeric conformations of the membrane-associated Cry4Ba toxin complex were observed in the 2D crystals on the different charge surfaces. Although the crystallographic resolution was still insufficient to give such critical insights into the structural details of the toxin-induced pore architecture, we have here provided pivotal evidence for the first time that the 65-kDa activated Cry4Ba toxin could have at least two conformational states in association with the membrane, implying the closed and open states of the pore. Further investigation for more details of the toxin-induced pore complex within the membrane is of great interest since this would shed light on precise toxicity mechanism of the insecticidal proteins in the Cry family.
Acknowledgments
Support for this work was funded by National Institute of Health Grant No. NS21501 (to F.J.S.), and Thailand Research Fund (TRF) in cooperation with Commission of Higher Education (to C.A.). Work in Unger laboratory is supported through NIH (GM66145.GM071590). The Royal Golden Jubilee PhD scholarship from TRF (to P.O.) is gratefully acknowledged.
Abbreviations
- Bt
Bacillus thuringiensis
- BSA
bovine serum albumin
- Cry
crystal
- 2D
two-dimensional
- DMPC
1, 2 dimyristoyl-sn-glycero-3-phosphocholine
- FPLC
fast-performance liquid chromatography
- IPTG
isopropyl-β-D-thiogalactopyranoside
- MAb
monoclonal antibody
- OG
octylglucoside
- PMSF
phenylmethane sulfonyl fluoride
- SDS-PAGE
sodium dodecyl sulphate-polyacrylamide gel electrophoresis
- TPCK
L-1-tosylamide-2-phenylethyl chloromethyl ketone
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.De Maagd RA, Bravo A, Berry C, Crickmore N, Schnepf E. Structure, diversity, and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu Rev Genet. 2003;37:409–433. doi: 10.1146/annurev.genet.37.110801.143042. [DOI] [PubMed] [Google Scholar]
- 2.Becker N, Margalit J. Use of Bacillus thuringiensis subsp. israelensis against mosquitoes and blackflies. In: Entwistle PF, Cory JS, Bailey MJ, Higgs S, editors. Bacillus thuringiensis, an Environmental Biopesticide: Theory and Practice. John Wiley and Sons; 1993. pp. 147–170. [Google Scholar]
- 3.Whalon ME, Wingerd BA. Bt: mode of action and use. Arch Insect Biochem Physiol. 2003;54:200–211. doi: 10.1002/arch.10117. [DOI] [PubMed] [Google Scholar]
- 4.Pigott CR, Ellar DJ. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol Mol Biol Rev. 2007;71:255–281. doi: 10.1128/MMBR.00034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gómez I, Pardo-López L, Muñoz-Garay C, Fernandez LE, Pérez C, Sánchez J, Soberón M, Bravo A. Role of receptor interaction in the mode of action of insecticidal Cry and Cyt toxins produced by Bacillus thuringiensis. Peptides. 2007;71:169–173. doi: 10.1016/j.peptides.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Li J, Carroll J, Ellar DJ. Crystal structure of insecticidal δ-endotoxin from Bacillus thuringiensis at 2.5 Å resolution. Nature. 1991;353:815–821. doi: 10.1038/353815a0. [DOI] [PubMed] [Google Scholar]
- 7.Grochulski P, Masson L, Borisova S, Pusztai-Carey M, Schwartz JL, Brousseau R, Cygler M. Bacillus thuringiensis Cry1A(a) insecticidal toxin crystal structure and channel formation. J Mol Biol. 1995;254:447–464. doi: 10.1006/jmbi.1995.0630. [DOI] [PubMed] [Google Scholar]
- 8.Morse RJ, Yamamoto T, Stroud RM. Structure of Cry2Aa suggests an unexpected receptor binding epitope. Structure (Camb) 2001;9:409–417. doi: 10.1016/s0969-2126(01)00601-3. [DOI] [PubMed] [Google Scholar]
- 9.Boonserm P, Davis P, Ellar DJ, Li J. Crystal structure of the mosquito-larvicidal toxin Cry4Ba and its biological implications. J Mol Biol. 2005;348:363–382. doi: 10.1016/j.jmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Boonserm P, Min M, Angsuthanasombat C, Lescar J. Structure of the functional form of the mosquito-larvicidal Cry4Aa toxin from Bacillus thuringiensis at a 2.8-angstorm resolution. J Bacteriol. 2006;188:3391–3401. doi: 10.1128/JB.188.9.3391-3401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puntheeranurak T, Uawithya P, Potvin L, Angsuthanasombat C, Schwartz JL. Ion channels formed in planar lipid bilayers by the diptheran-specific Cry4Ba Bacillus thuringiensis and its α1-α5 fragment. Mol Membr Biol. 2004;21:67–74. doi: 10.1080/09687680310001625792. [DOI] [PubMed] [Google Scholar]
- 12.Rausell C, Pardo-López L, Sánchez J, Muñoz-Garay C, Morera C, Soberón M, Bravo A. Unfolding events in the water-soluble monomeric Cry1Ab toxin during transition to oligomeric pre-pore and membrane-inserted pore channel. J Biol Chem. 2004;274:31996–32000. doi: 10.1074/jbc.M406279200. [DOI] [PubMed] [Google Scholar]
- 13.Gazit E, la Rocca P, Samson MS, Shai Y. The structure and organization within the membrane of the helices composing the pore-forming domain of Bacillus thuringiensis delta-endotoxin are consistent with an “umbrella-like” structure of the pore. Proc Natl Acad Sci USA. 1998;95:12289–12294. doi: 10.1073/pnas.95.21.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loseva OI, Tiktopulo EI, Vasiliev VD, Nikulin AD, Dobritsa AP, Potekhin SA. Structure of Cry3A delta-endotoxin within phospholipid membranes. Biochemistry. 2001;40:14143–14151. doi: 10.1021/bi010171w. [DOI] [PubMed] [Google Scholar]
- 15.Alzate O, You T, Claybon M, Osorio C, Curtiss A, Dean DH. Effects of disulfide bridges in domain I of Bacillus thuringiensis Cry1Aa delta-endotoxin on ion-channel formation in biological membranes. Biochemistry. 2006;45:13597–605. doi: 10.1021/bi061474z. [DOI] [PubMed] [Google Scholar]
- 16.Tomimoto K, Hayakawa T, Hori H. Pronase digestion of brush border membrane-bound Cry1Aa shows that almost the whole activated Cry1Aa molecule penetrates into the membrane. Comp Biochem Physiol B. 2006;144:413–422. doi: 10.1016/j.cbpb.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Soberón M, Pérez RV, Nunez-Valdez ME, Lorence A, Gómez I, Sánchez J, Bravo A. Evidence for intermolecular interaction as a necessary step for pore-formation activity and toxicity of Bacillus thuringiensis Cry1Ab toxin. FEMS Microbiol Lett. 2000;191:221–225. doi: 10.1111/j.1574-6968.2000.tb09343.x. [DOI] [PubMed] [Google Scholar]
- 18.Peyronnet O, Nieman B, Genereux F, Vachon V, Laprade R, Schwartz JL. Estimation of the radius of the pores formed by the Bacillus thuringiensis Cry1C delta-endotoxin in planar lipid bilayers. Biochim Biophys Acta. 2002;1567:113–122. doi: 10.1016/s0005-2736(02)00605-3. [DOI] [PubMed] [Google Scholar]
- 19.Gómez I, Sánchez J, Miranda R, Bravo A, Soberón M. Cadherin-like receptor binding facilitates proteolytic cleavage of helix alpha-1 in domain I and oligomer pre-pore formation of Bacillus thuringiensis Cry1Ab toxin. FEBS Lett. 2002;513:242–246. doi: 10.1016/s0014-5793(02)02321-9. [DOI] [PubMed] [Google Scholar]
- 20.Puntheeranurak T, Stroh C, Zhu R, Angsuthanasombat C, Hinterdorfer P. Structure and distribution of the Bacillus thuringiensis Cry4Ba toxin in lipid membranes. Ultramicroscopy. 2005;105:115–124. doi: 10.1016/j.ultramic.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 21.Vie V, Van Mau N, Pomarede P, Dance C, Schwartz JL, Laprade R, Frutos R, Rang C, Masson L, Heitz F, Le Grimellec C. Lipid-induced pore formation of the Bacillus thuringiensis Cry1Aa insecticidal toxin. J Membr Biol. 2001;180:195–203. doi: 10.1007/s002320010070. [DOI] [PubMed] [Google Scholar]
- 22.Unger VM, Kumar NM, Gilula NB, Yeager M. Projection structure of a gap junction membrane channel at 7 Å resolution. Nature Struct Biol. 1997;4:39–43. doi: 10.1038/nsb0197-39. [DOI] [PubMed] [Google Scholar]
- 23.Vinothkumar KR, Smits SH, Kuhlbrandt W. pH-induced structural change in a sodium/proton antiporter from Methanococcus jannaschii. EMBO J. 2005;24:2720–9. doi: 10.1038/sj.emboj.7600727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angsuthanasombat C, Chungjatupornchai W, Kertbundit S, Luxananil P, Settasatian C, Wilairat P, Panyim S. Cloning and expression of 130-kd mosquito-larvicidal delta-endotoxin gene of Bacillus thuringiensis var. israelensis in Escherichia coli. Mol Gen Genet. 1987;208:384–389. doi: 10.1007/BF00328128. [DOI] [PubMed] [Google Scholar]
- 25.Angsuthanasombat C, Uawithya P, Leetachewa S, Pornwiroon W, Ounjai P, Kerdcharoen T, Katzenmeier G, Panyim S. Bacillus thuringiensis Cry4A and Cry4B mosquito-larvicidal proteins: homology-based 3D model and implications for toxin activity. J Biochem Mol Biol. 2004;37:304–313. doi: 10.5483/bmbrep.2004.37.3.304. [DOI] [PubMed] [Google Scholar]
- 26.Moonsom S, Chaisri U, Kasinrerk W, Angsuthanasombat C. Binding characteristics to mosquito-larval midgut proteins of the cloned domain II-III fragment from the Bacillus thuringiensis Cry4Ba toxin. J Biochem Mol Biol. 2007 doi: 10.5483/bmbrep.2007.40.5.783. in press. [DOI] [PubMed] [Google Scholar]
- 27.Crowther RA, Henderson R, Smith JM. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 28.Vachon V, Prefontaine G, Rang C, Coux F, Juteau M, Schwartz JL, Brousseau R, Frutos R, Laprade R, Masson L. Helix 4 mutants of the Bacillus thuringiensis insecticidal toxin Cry1Aa display altered pore-forming abilities. Appl Environ Microbiol. 2004;70:6123–30. doi: 10.1128/AEM.70.10.6123-6130.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slatin SL, Abrams CK, English L. Delta-endotoxins form cation-selective channels in planar lipid bilayers. Biochem Biophys Res Commun. 1990;169:765–772. doi: 10.1016/0006-291x(90)90397-6. [DOI] [PubMed] [Google Scholar]
- 30.Likitvivatanavong S, Katzenmeier G, Angsuthanasombat C. Asn183 in alpha 5 is essential for oligomerisation and toxicity of the Bacillus thuringiensis Cry4Ba toxin. Arch Biochem Biophys. 2006;445:46–55. doi: 10.1016/j.abb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Kuo A, Domene C, Johnson LN, Doyle DA, Venien-Bryan C. Two different conformational states of the KirBac3.1 potassium channel revealed by electron crystallography. Structure. 2005;13:1463–72. doi: 10.1016/j.str.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Masson L, Tabashnik BE, Mazza A, Prefontaine G, Potvin L, Brousseau R, Schwartz JL. Mutagenic analysis of a conserved region of domain III in the Cry1Ac toxin of Bacillus thuringiensis. Appl Environ Microbiol. 2002;68:194–200. doi: 10.1128/AEM.68.1.194-200.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–22. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 34.Sigworth FJ. Voltage gating of ion channels. Q Rev Biophys. 1994;27:1–40. doi: 10.1017/s0033583500002894. [DOI] [PubMed] [Google Scholar]
- 35.Tigue NJ, Jacoby J, Ellar DJ. The alpha-helix 4 residue, Asn135, is involved in the oligomerization of Cry1Ac1 and Cry1Ab5 Bacillus thuringiensis toxins. Appl Environ Microbiol. 2001;67:5715–5720. doi: 10.1128/AEM.67.12.5715-5720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masson L, Mazza A, Sangadala S, Adang MJ, Brousseau R. Polydispersity of Bacillus thuringiensis Cry1 toxins in solution and its effect on receptor binding kinetics. Biochim Biophys Acta. 2002;1594:266–275. doi: 10.1016/s0167-4838(01)00312-0. [DOI] [PubMed] [Google Scholar]
- 37.Aronson AI, Geng C, Wu L. Aggregation of Bacillus thuringiensis Cry1A toxins upon binding to target insect larval midgut vesicles. Appl Environ Microbiol. 1999;65:2503–2507. doi: 10.1128/aem.65.6.2503-2507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar AS, Aronson AI. Analysis of mutations in the pore-forming region essential for insecticidal activity of a Bacillus thuringiensis delta-endotoxin. J Bacteriol. 1999;181:6103–6107. doi: 10.1128/jb.181.19.6103-6107.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guereca L, Bravo A. The oligomeric state of Bacillus thuringiensis Cry toxins in solution. Biochim Biophys Acta. 1999;1429:342–350. doi: 10.1016/s0167-4838(98)00241-6. [DOI] [PubMed] [Google Scholar]


