Abstract
BACKGROUND
Human homologs (FEM1A, FEM1B, FEM1C) of nematode sex determination genes are candidate genes for polycystic ovary syndrome (PCOS). We previously identified a FEM1A mutation (H500Y) in a woman with PCOS; FEM1B has been implicated in insulin secretion.
METHODS
Women with and without PCOS (287 cases, 187 controls) were genotyped for H500Y and six FEM1A single nucleotide polymorphisms (SNPs), five FEM1B SNPs and five FEM1C SNPs. SNPs and haplotypes were determined and tested for association with PCOS and component phenotypes.
RESULTS
No subject carried the FEM1A H500Y mutation. FEM1A SNPs rs8111933 (P = 0.001) and rs12460989 (P = 0.046) were associated with an increased likelihood of PCOS whereas FEM1A SNP rs1044386 was associated with a reduced probability of PCOS (P = 0.013). FEM1B SNP rs10152450 and a linked SNP were associated with a reduced likelihood of PCOS (P = 0.005), and lower homeostasis model assessment (HOMA) for beta-cell function (HOMA-%B, P = 0.011) and lower HOMA for insulin resistance (HOMA-IR, P = 0.018). FEM1B SNP rs12909277 was associated with lower HOMA-%B (P = 0.008) and lower HOMA-IR (P = 0.037). Haplotype associations were consistent with SNP results, and also revealed association of FEM1B haplotype TGAGG with increased HOMA-%B (P = 0.007) and HOMA-IR (P = 0.024). FEM1C variants were not associated with PCOS.
CONCLUSIONS
This study presents evidence suggesting a role for FEM1A and FEM1B in the pathogenesis of PCOS. Only FEM1B variants were associated with insulin-related traits in PCOS women, consistent with prior evidence linking this gene to insulin secretion. Replication of these associations and mechanistic studies will be necessary to establish the role of these genes in PCOS.
Keywords: polycystic ovary syndrome, insulin secretion, FEM1A, FEM1B, FEM1C
Introduction
Susceptibility to polycystic ovary syndrome (PCOS) appears to run in families. First degree female relatives of women with PCOS have a 3- to 4-fold increased prevalence of PCOS compared with the general population (Legro et al., 1998; Kahsar-Miller et al., 2001), and a recent twin study documented a high degree of heritability of PCOS (Vink et al., 2006). Over the past decade, many investigations into possible candidate genes for PCOS susceptibility have been conducted; however, no genes are universally agreed upon as important in the development or expression of PCOS (Goodarzi and Azziz, 2006). In this study, we examine the possible role of human homologs (FEM1A, FEM1B, FEM1C) of the nematode Caenorhabditis elegans feminizing (fem) genes. In C. elegans, gender is determined by the X chromosome to autosome (X:a) ratio; XX worms develop as hermaphrodites, XO worms as males. Fem-1 mutant worms develop as females, regardless of X:a ratio (Doniach and Hodgkin, 1984). Fem-1 thus appears to play a role in the formation of the male body and spermatogenesis. Given the role of fem-1 in reproductive development in C. elegans, we hypothesize that genetic variants in the human homologs may be risk factors for PCOS and modulate the PCOS phenotype.
There are at least three human homologs of fem-1 (FEM1A, 19p13.3; FEM1B, 15q22; and FEM1C, 5q22). The function of the human genes is unknown, although they are predicted to encode for anykrin repeat proteins. We first considered FEM1A, given its expression in the ovaries (Maher et al., 2005) and chromosomal location near the D19S884 microsatellite (19p13.2) that has been implicated as a genetic factor for PCOS risk (Stewart et al., 2006). In a pilot study, we identified a heterozygous H500Y mutation in one of 35 individuals with PCOS (Maher et al., 2005). We now also considered FEM1B as a PCOS candidate gene, given that mice with knockout of the fem1b gene displayed abnormal glucose tolerance, impaired acute phase insulin secretion, and possibly insulin resistance (Lu et al., 2005).
Herein, we present the first genetic association study of the FEM1A, FEM1B and FEM1C genes in PCOS, using single nucleotide polymorphisms (SNPs) as well as haplotypes to capture common variation across genomic regions. We report that variants in both FEM1A and FEM1B appear to be risk factors for PCOS; additionally, variants in FEM1B are associated with altered insulin secretion and insulin sensitivity in affected women.
Materials and Methods
Subjects
A total of 287 consecutive White patients with PCOS, aged 13–47 years, and 187 healthy White control women, aged 14–60, were recruited from Birmingham, AL, USA. The clinical characteristics of these subjects are presented in Table I. PCOS subjects were recruited consecutively from the reproductive endocrine practice of one of the investigators (RA) at the University of Alabama at Birmingham (UAB). Participation in research studies was offered to patients meeting inclusion criteria (premenopausal, non-pregnant, on no hormonal therapy for at least 3 months, and meeting diagnostic criteria for PCOS). Presence of PCOS was defined by the 1990 National Institutes of Health consensus criteria (Zawadzki and Dunaif, 1992), including (i) clinical hyperandrogenism and/or hyperandrogenemia, (ii) oligo-ovulation and (iii) the exclusion of related disorders, including androgen-producing tumors, non-classic 21-hydroxylase-deficient adrenal hyperplasia, hyperprolactinemia, active thyroid disease, or Cushing’s syndrome. The specific parameters for defining hirsutism, hyperandrogenemia, ovulatory dysfunction and exclusion of related disorders were previously reported (Azziz et al., 2004).
Table I.
Clinical characteristics of the study cohort.
| Control (n = 187) | PCOS (n = 287) | |
|---|---|---|
| Age (year) | 33.0 (25.0–42.0) | 27.5 (21.7–33.2)* |
| BMI (kg/m2) | 24.1 (21.9–28.3) | 34.7 (27.6–41.1)* |
| WHR | 0.78 (0.75–0.83) | 0.83 (0.79–0.89)* |
| mFG score | 0 (0) | 7.0 (5.0–10.0)* |
| Hirsute (%) | 0 | 73.9* |
| Total testosterone (nmol/l) | 1.42 (1.00–1.94) | 2.77 (2.25–3.33)* |
| Free testosterone (pmol/l) | 12.1 (8.0–17.0) | 29.1 (22.9–39.2)* |
| DHEAS (μmol/l) | 2.58 (1.61–3.66) | 5.66 (3.55–8.17)* |
| SHBG (nmol/l) | 220.0 (160.0–280.0) | 150.0 (130.0–200.0)* |
| Insulin (pmol/l) | 49.5 (33.2–79.5) | 129.15 (71.8–200.9)* |
| Glucose (mmol/l) | 4.77 (4.55–5.11) | 4.77 (4.50–5.22) |
| HOMA-IR | 0.92 (0.63–1.47) | 2.29 (1.30–3.24)* |
| HOMA-%B | 103.9 (80.0–135.6) | 175.3 (117.6–218.1)* |
Data are median (interquartile range). *P < 0.001 compared with control group, by unpaired T-tests or chi-square tests as appropriate; quantitative data were transformed to approximate normality. PCOS, polycystic ovary syndrome; WHR, waist-to-hip ratio; mFG, modified Ferriman–Gallwey; DHEAS, dehydroepiandrosterone sulfate; SHBG, sex hormone-binding globulin; HOMA-IR, homeostasis model assessment of insulin resistance; HOMA-%B, homeostasis model assessment of beta-cell function (insulin secretion).
Controls were healthy women, with regular menstrual cycles or a history of regular menstrual cycles before menopause, and without family history of hirsutism. These women had no evidence of hirsutism, acne, or alopecia, or endocrine dysfunction. Controls were recruited by word of mouth and advertisements in the Birmingham, Alabama area, through a call for ‘healthy women’ without detailing further the nature of the studies. All controls underwent a brief history and physical exam (see below) to ensure that all control women included were non-hirsute and eumenorrheic.
No subject (PCOS or control) had used hormonal preparations, including oral contraceptives, for at least 3 months preceding the study, and none was pregnant. All subjects also gave written informed consent, and the study was performed according to the guidelines of the Institutional Review Boards of UAB and Cedars-Sinai Medical Center.
Phenotyping
Subjects underwent a brief physical examination, including hirsutism scoring using a modification of the Ferriman–Gallwey method (mFG) (Hatch et al., 1981), and underwent blood sampling. Subjects were deemed hirsute if their mFG score was six or greater (Knochenhauer et al., 1998); all hirsutism scoring was performed by the same investigator (RA). Hormonal measures, including total and free testosterone, dehydroepiandrosterone sulfate (DHEAS), 17α-hydroxyprogesterone (17-HP) and sex hormone-binding globulin (SHBG), were obtained between Days 3 and 8 (follicular phase) following a spontaneous menstrual cycle or progesterone-induced withdrawal bleed, per our research protocol (Azziz et al., 2004). Total testosterone was measured after serum extraction by an in-house radioimmunoassay (RIA) method, SHBG activity was measured by competitive-binding analysis, using Sephadex G-25 (Sigma–Aldrich Corp., St Louis, MO, USA) and [3H] testosterone as the ligand, and the free testosterone was calculated as previously described (Pearlman et al., 1967; Boots et al., 1998). The SHBG method gives values of ∼100–300 nmol/L in normal adult women. DHEAS and 17-HP were measured by direct RIA using commercially available kits (from Diagnostic Products Corp., Los Angeles, CA, USA). The intra- and interassay variations for the hormonal assays have been previously reported (Knochenhauer et al., 1998). The same laboratory assays were employed for all subjects. For these hirsutism and androgen-related traits measured in the women with PCOS, completeness of data was over 98%. The total and free testosterone values of three cases were statistical outliers; therefore, these values were deleted from analysis.
Fasting glucose and insulin were also obtained in a subset of the cohort (∼70%). The glucose assays were performed with Ektachem DT slides (Johnson & Johnson Clinical Diagnostics, Rochester, NY, USA). Serum insulin was measured by RIA (Diagnostic Systems Laboratories, Webster, TX, USA). The computer-based homeostasis model assessment (HOMA, www.dtu.ox.ac.uk/homa) was used to calculate indices of insulin resistance (HOMA-IR) and insulin secretion (i.e. percent beta-cell function or HOMA-%B) utilizing the fasting glucose and insulin levels (Levy et al., 1998; Wallace et al., 2004). An ideal, normal-weight person less than 35 years of age has a HOMA-IR = 1 and HOMA-%B = 100% (Matthews et al., 1985).
Genotyping and haplotype determination
SNPs were selected using frequency and validation data from the International HapMap database (The International HapMap Consortium, 2003) with the aim of exploiting linkage disequilibrium (LD) for the study of each gene. Six SNPs (rs12460989, rs3087692, rs11085099, rs1044386, rs8111933 and rs4806998) across the 3.8 kb FEM1A gene (19p13.3) were genotyped; these SNPs are predicted to tag the haplotypes across the entire gene (+7 kb upstream and 4 kb downstream) in the Caucasian population of the HapMap database. We also genotyped the H500Y mutation in FEM1A. Five SNPs (rs10152450, rs11636081, rs7172340, rs12909277 and rs6494730) across the 13.5 kb FEM1B gene (15q22) were also genotyped; these SNPs are predicted to tag the haplotypes across the entire gene (+5 kb upstream and 5 kb downstream) in the Caucasian population of the HapMap database. Finally, five SNPs (rs12187973, rs372702, rs256940, rs17472710 and rs256958) across the 24 kb FEM1C gene (5q22) were genotyped to encompass the common haplotypes spanning the gene (+10 kb upstream and 10 kb downstream), again using HapMap. H500Y and six SNPs were genotyped in the 474 subjects using the 5′-exonuclease assay (TaqMan MGB, Applied Biosystems, Foster City, CA, USA) described previously (Livak, 1999; Goodarzi et al., 2003); duplicate genotyping of 96 samples yielded 100% concordance. The PCR primers and TaqMan MGB probes are presented in Table II. The genotyping success rate was 93.4%. The remaining 10 SNPs were genotyped using the oligo ligation assay on an Illumina (San Diego, CA, USA) bead station (Gunderson et al., 2006; Steemers et al., 2006). The genotyping success rate was 94%.
Table II.
Primers and probe sequences used in the 5′-exonuclease assay.
| Variant | PCR primers | TaqMan MGB probes |
|---|---|---|
| FEM1A | ||
| H500Y | CGGAGACTCAGCCCAGTTCA | CATCCTCcACCTGCT |
| GGTAGACGGTCTGGTGCTTCA | TCATCCTCtACCTGCTC | |
| rs11085099 | CTGAAGCCATTGCGTTCCTT | TTGAGGACgGTCTGC |
| TTCAGCGGCTGGAGTTATCTG | TTGAGGACtGTCTGCC | |
| rs1044386 | GGTTCTTGGCCAGCGTAGAC | CATTCTCACTgCATCCT |
| GCCTGGCCGTGGCTACT | ATTCTCACTaCATCCTC | |
| rs8111933 | AAAAGCCCCAGTATGTAACACAGAA | CTGTTCACTCAgACCAA |
| TCGGAGGTGGGTGAGTCAGT | TCTGTTCACTCAcACCAA | |
| FEM1B | ||
| rs10152450 | TGCTGCTGGTTCTAGGTGCTT | TTGACATTAAaGCCCAGC |
| TTCAAGTGAGTGGTTCCGTAAAAC | TTGACATTAAcGCCCAGC | |
| rs11636081 | CCTTTTCACCTTCCACCATGT | AACTGAAcCTACTGGCA |
| GAAGCTGGAATGTTCAAGATCAA | CAACTGAAtCTACTGGCA | |
| rs7172340 | CCACTAACCAAAAGACCTGATAAGTAGA | TTGCTGGAGAtAGTCTA |
| AACACTTCCAAATGTACTTATTGCTTCT | TTGCTGGAGAcAGTC | |
Primers for PCR are listed 5′–3′ and were synthesized by Invitrogen (Carlsbad, CA, USA). Probes are listed 5′ to 3′ and were synthesized by Applied Biosystems (Foster City, CA, USA). The probes are labeled at the 5′ end with 6FAM or VIC (laser-activated fluorescent dyes), and at the 3′ end with a quencher/minor groove binder. In the probe sequences, variant nucleotides are indicated in lower case.
Haploview 3 (Barrett et al., 2005) was used to determine haplotypes as well as haplotype blocks. Haploview constructs haplotypes using an accelerated expectation maximization algorithm similar to the partition/ligation method (Qin et al., 2002). Haploview was used to calculate LD (the D’ statistic and r2) between each pairwise combination of all the SNPs within each gene. Haplotype blocks were determined using the solid spine of LD algorithm in Haploview (Barrett et al., 2005). Haplotypes within each block were assigned to individual subjects only when the assignment could be made with a greater than 95% certainty.
Statistical analysis
For all analyses, quantitative trait values were log- or square root-transformed as appropriate to reduce non-normality. Unpaired T-tests and chi-square tests were used to compare clinical characteristics between women with and without PCOS. Quantitative data are presented as median (interquartile range). Interquartile range is given as first and third quartiles. Statistical significance was set at P < 0.05.
The primary phenotypes for genetic association analysis differed for each gene, and were selected based on hypotheses derived from our prior work (Lu et al., 2005; Maher et al., 2005). For FEM1A, the primary phenotypes were the presence or absence of PCOS and testosterone levels; for FEM1B, the primary phenotypes were presence or absence of PCOS, HOMA-IR and HOMA-%B. For FEM1C, the primary phenotype was PCOS status. Quantitative traits were considered in PCOS subjects and premenopausal controls separately. For each gene, all other available phenotypes were considered secondary. To minimize multiple testing, secondary traits were analyzed for association only with variants that showed association with primary traits. For the insulin-related traits only, subjects with diabetes (n = 6) were excluded because the hyperglycemia of diabetes may induce secondary changes in insulin-related traits that reduce their utility for genetic analyses.
SNPs and haplotypes were the genotypic units utilized in association analyses. Haplotypes were assigned prior to association analyses. Association of haplotypes with PCOS and presence/absence of hirsutism was evaluated using logistic regression. Association with androgens, insulin-related traits and mFG score was evaluated using analysis of covariance (ANCOVA). Logistic regression and ANCOVA analyses were adjusted for age and BMI by including them as independent variables in every analysis. Potential gene–gene interaction was tested by including an interaction term (between pairs of SNPs from different genes) as an additional independent variable in selected analyses. Analyses were carried out using Statview 5.0 (SAS Institute, Cary, NC, USA).
To confirm any significant associations, we estimated empirical P-values by permutation analysis. For each significant association, the samples were permuted by shuffling genotypic data 1000 times, and subsequent association analyses were carried out to obtain the distribution of the test statistic under the null hypothesis of no association. The empiric P-values were obtained as the proportion of the 1000 replicates that had a P-value less than or equal to the nominal ones obtained from the actual (unshuffled) data. These empiric P-values are reported in the Results.
Results
FEM1A
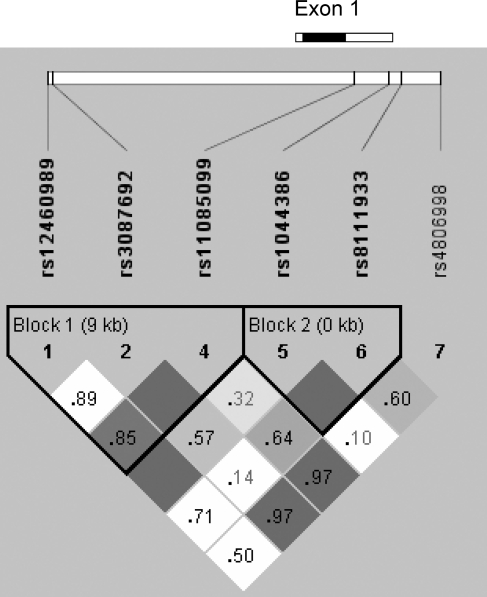
None of the 474 genotyped subjects carried out the H500Y mutation. We genotyped six SNPs in the FEM1A region (Table III, Fig. 1). All markers were in Hardy–Weinberg equilibrium. The SNPs were organized in two haplotype blocks and a single SNP (Fig. 1). Table IV displays the FEM1A haplotypes and their frequencies.
Table III.
Frequency and location information on FEM1A and FEM1B variants.
| Variant designation | Alleles | Location | Overall MAF | PCOS MAF | Control MAF |
|---|---|---|---|---|---|
| FEM1A | |||||
| rs12460989 | T/G | 5′ to gene | 0.052 | 0.062 | 0.037 |
| rs3087692 | C/T | 5′ to gene | 0.181 | 0.186 | 0.174 |
| H500Y | C/T | Exon 1 | 0 | 0 | 0 |
| rs11085099 | G/T | Exon 1 | 0.313 | 0.287 | 0.357 |
| rs1044386 | G/A | Exon 1 | 0.270 | 0.214 | 0.362 |
| rs8111933 | C/G | 3′ to gene | 0.259 | 0.296 | 0.199 |
| rs4806998 | G/A | 3′ to gene | 0.451 | 0.432 | 0.481 |
| FEM1B | |||||
| rs10152450 | T/G | Intron 1 | 0.316 | 0.283 | 0.370 |
| rs11636081 | G/A | Intron 1 | 0.029 | 0.033 | 0.024 |
| rs7172340 | A/G | Intron 1 | 0.034 | 0.039 | 0.027 |
| rs12909277 | G/A | Intron 1 | 0.415 | 0.396 | 0.445 |
| rs6494730 | G/T | 3′ to gene | 0.317 | 0.289 | 0.362 |
MAF, minor allele frequency.
Figure 1:
Gene structure and linkage disequilibrium plot for FEM1A.
The gene structure of FEM1A is shown at top; the gene has one exon and no introns. The coding region is indicated by the black box; untranslated regions by white boxes. The locations of the genotyped SNPs relative to the gene are indicated. H500Y was not present in this cohort. The linkage disequilibrium plot at the bottom displays D’ values for each pair of SNPs in the box at the intersection of the diagonals from each SNP. The solid blocks indicate D’ = 1 for the corresponding pair of variants. The haplotype block 1 is composed of three SNPs, and haplotype block 2 of two SNPs, as indicated.
Table IV.
FEM1A and FEM1B haplotypes and haplotype frequencies.
| Haplotype | Overall freq | PCOS freq | PCOS count* | Control freq | Control count |
|---|---|---|---|---|---|
| FEM1A | |||||
| Block 1 (rs12460989-rs3087692-rs11085099) | |||||
| TCG | 0.453 | 0.465 | 254 | 0.433 | 143 |
| TCT | 0.311 | 0.285 | 156 | 0.355 | 117 |
| TTG | 0.183 | 0.188 | 103 | 0.174 | 58 |
| GCG | 0.051 | 0.059 | 33 | 0.037 | 12 |
| Block 2 (rs1044386-rs8111933) | |||||
| GC | 0.471 | 0.490 | 268 | 0.439 | 144 |
| AC | 0.269 | 0.214 | 117 | 0.361 | 118 |
| GG | 0.260 | 0.296 | 161 | 0.200 | 66 |
| FEM1B | |||||
| (rs10152450-rs11636081-rs7172340-rs12909277-rs6494730) | |||||
| TGAGG | 0.576 | 0.599 | 332 | 0.539 | 182 |
| GGAAT | 0.314 | 0.285 | 158 | 0.361 | 122 |
| TGAAG | 0.072 | 0.078 | 43 | 0.061 | 21 |
| TAGAG | 0.029 | 0.032 | 18 | 0.024 | 8 |
*Count represents number of chromosomes assigned a particular haplotype by the expectation maximization algorithm.
SNPs, rs8111933, rs12460989 and rs1044386, were associated with PCOS. Carriers of the minor allele of rs8111933 had an increased frequency of PCOS (odds ratio (OR) 2.12, 95% confidence interval (CI) 1.26–3.57, P = 0.001), as did minor allele carriers of rs12460989 (OR 2.45, 95% CI 1.01–5.97, P = 0.046). Carriers of the minor allele of rs1044386 had a reduced frequency of PCOS (OR 0.53, 95% CI 0.32–0.88, P = 0.013). Genotype frequencies of the SNPs associated with PCOS are given in Supplementary Table I. No SNPs were associated with free or total testosterone in women with PCOS; however, in controls rs3087692 was associated with increased total testosterone (minor allele carrier 1.73 (1.14–2.38) nmol/l, non-carrier 1.35 (0.93–1.91) nmol/l; P = 0.015) as was rs8111933 (minor allele carrier 1.77 (1.14–2.37) nmol/l, non-carrier 1.35 (0.90–1.89) nmol/l; P = 0.022).
Haplotype association analysis was consistent with the SNP association analysis. Carriers of the block 1 haplotype (GCG) carrying the minor allele of rs12460989 exhibited increased PCOS frequency (OR 2.50, 95% CI 1.02–6.11, P = 0.042). The block 2 haplotype (AC) carrying the minor allele of rs1044386 was associated with PCOS with a similar magnitude of effect (OR 0.53, 95% CI 0.32–0.88, P = 0.013). Carriers of the third most common block 2 haplotype GG, that bearing the minor allele of rs8111933, had an increased frequency of PCOS (OR 2.18, 95% CI 1.28–3.72, P = 0.001). No haplotypes were associated with free or total testosterone in women with PCOS. In controls, the block 1 haplotype (TTG) carrying the minor allele of rs3087692 and the block 2 haplotype (GG) carrying the minor allele of rs8111933 were also associated with increased total testosterone, with similar significance and magnitude of effect as those SNPs.
Given the association of the above SNPs and haplotypes with PCOS (and total testosterone in controls), secondary analyses were carried out to evaluate whether these same variants were associated with BMI, DHEAS, SHBG, mFG score, hirsutism, fasting glucose or fasting insulin. No associations with these secondary traits were detected.
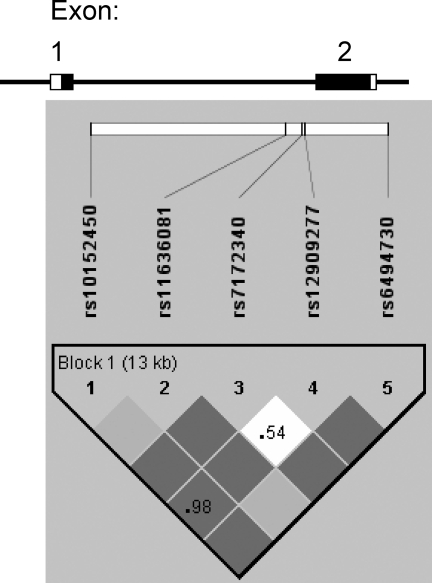
FEM1B
The five SNPs spanning the FEM1B gene were in Hardy–Weinberg equilibrium (Table III). The SNPs formed a haplotype block across the entire region (Fig. 2). Table IV displays the FEM1B haplotypes and frequencies.
Figure 2:
Gene structure and linkage disequilibrium plot for FEM1B.
The gene structure of FEM1B is shown at top; the gene has two exons and one intron. The coding regions are indicated by the black boxes; untranslated regions by white boxes. The locations of the genotyped SNPs relative to the exons are indicated. The linkage disequilibrium plot at the bottom displays D’ values for each pair of SNPs in the box at the intersection of the diagonals from each SNP. The darker solid blocks indicate D’ = 1 for the corresponding pair of variants; lighter solid blocks also indicate D’ = 1, but with a wide confidence interval. The haplotype block comprising all five SNPs is indicated.
SNP rs10152450 was associated with PCOS. Carriers of the minor allele of rs10152450 had a reduced frequency of PCOS (OR 0.49, 95% CI 0.30–0.82, P = 0.005). Among women with PCOS, minor allele carriers of this SNP also had lower HOMA-%B (P = 0.011) and HOMA-IR (P = 0.018). Fig. 3 displays the median level of insulin-related traits according to genotype at rs10152450; an apparent dose–response effect is seen with increasing number of minor alleles. SNP rs6494730 was in high LD (r2 = 0.98) with rs10152450 and therefore displayed the same associations with PCOS, HOMA-%B and HOMA-IR. Genotype frequencies of the SNPs associated with PCOS are given in Supplementary Table I. SNP rs12909277, while not associated with PCOS, was associated with decreased HOMA-%B (minor allele carriers 158.8 (105.9–215.1), non-carriers 186.2 (133.3–239.5); P = 0.008) and decreased HOMA-IR (minor allele carriers 2.08 (1.25–3.00), non-carriers 2.34 (1.53–3.48); P = 0.037) in affected women.
Figure 3:
Median levels of insulin resistance and insulin secretion indices by FEM1B rs10152450 genotype.
The black bars indicate homeostasis model assessment of insulin resistance (HOMA-IR), the white bars homeostasis model assessment of beta-cell function (HOMA-%B). The HOMA-IR values have been multiplied by 100. The levels are significantly different under both dominant (HOMA-%B, P = 0.011; HOMA-IR, P = 0.018) and additive models (HOMA-%B, P = 0.020; HOMA-IR, P = 0.011).
Haplotype association results were consistent with the SNP association analysis and revealed additional associations. The haplotype (GGAAT) carrying the minor alleles of rs10152450 and rs6494730 was associated with PCOS with a similar magnitude of effect (OR 0.49, 95% CI 0.29–0.83, P = 0.004). PCOS women who were haplotype GGAAT carriers also had lower HOMA-%B (P = 0.003) and lower HOMA-IR (P = 0.002), with magnitude of effects as predicted by the SNP analysis. In addition, the most common haplotype (TGAGG) was associated with higher HOMA-%B (carriers 177.9 (126.5–221.6), non-carriers 138.6 (83.8–186.6); P = 0.007) and higher HOMA-IR (carriers 2.29 (1.46–3.25), non-carriers 1.40 (0.66–3.17); P = 0.024) in women with PCOS.
Secondary analyses of association of FEM1B variants with BMI, free and total testosterone, DHEAS, SHBG, mFG, hirsutism and fasting glucose were negative. In PCOS women, SNP rs6494730 was associated with decreased fasting insulin (minor allele carrier 118.4 (61.5–180.8) pmol/l, non-carrier 132.7 (85.4–206.5) pmol/l; P = 0.015) as was haplotype GGAAT (carrier 118.4 (64.6–183.7) pmol/l, non-carrier 129.2 (84.7–203.4) pmol/l; P = 0.017).
FEM1B variants were not associated with quantitative traits in control subjects.
FEM1C
All five FEM1C SNPs were in Hardy–Weinberg equilibrium, and formed a haplotype block spanning the entire gene (Supplementary Figure 1 and Supplementary Tables II and III). No variants were associated with PCOS. Therefore, SNPs and haplotypes in FEM1C were not examined further.
Gene–gene interaction analyses
Given the observed associations of FEM1A and FEM1B variants with PCOS and quantitative traits, we tested for interaction between the associated FEM1A and FEM1B SNPs in association with PCOS diagnosis, HOMA-%B, HOMA-IR, fasting glucose and fasting insulin. There was no evidence of gene–gene interaction in any of these analyses.
Discussion
This is the first genetic association study evaluating whether variants in the genes for the three human homologs of fem-1, FEM1A, FEM1B and FEM1C are associated with PCOS and component phenotypes within PCOS. Specific FEM1A and FEM1B SNPs were associated with PCOS frequency (increased or decreased, depending on the variant). Specific FEM1A variants were associated with total testosterone in control subjects. In FEM1B only, several variants were also associated with HOMA-%B and HOMA-IR values in women with PCOS. Gene–gene interaction analyses suggested these loci act independently of each other.
One basis for considering FEM1A a candidate gene for PCOS is its homology to the fem1 gene of C. elegans, which functions in masculinization of the nematode (Doniach and Hodgkin, 1984). The hyperandrogenism of PCOS may be considered a form of mild masculinization. The Calpain-10 and -5 genes (Mugita et al., 1997; Horikawa et al., 2000), both homologs of the tra-3 gene of C. elegans, which functions as an upstream regulator of fem-1 in nematode sex determination, have been implicated in PCOS (Ehrmann et al., 2002; Escobar-Morreale et al., 2002; Gonzalez et al., 2002, 2006; Haddad et al., 2002; Vollmert et al., 2007). However, the function of FEM1A in mammals is unknown, including whether or not it may operate in a signaling pathway with Calpain-10 or -5. In mice, the expression of FEM1A in the ovaries increases at the time of puberty, particularly in androgen-producing secondary interstitial cells (Maher et al., 2005). This location and timing of expression is consistent with the ovarian hyperandrogenism that occurs at the time of puberty in PCOS. Therefore, the primary traits we analyzed for association with FEM1A variants were PCOS diagnosis and free and total testosterone levels. Although we found association with PCOS diagnosis, association with total testosterone was seen only in controls, which may reflect the challenge of measuring testosterone in women (Rosner et al., 2007). Alternatively, FEM1A may be a minor effector of testosterone production in PCOS women or may participate in other processes in the pathogenesis of PCOS.
The FEM1A protein may play a role in anti-inflammatory signaling by liganded prostaglandin E2 subtype E4 receptors (Takayama et al., 2006). Thus, if FEM1A participates in the pathogenesis of PCOS, it may do so via modulation of chronic inflammation; recent studies have found elevated markers of inflammation in PCOS (Diamanti-Kandarakis et al., 2006; Luque-Ramirez et al., 2006). Reduction of FEM1A function by small interfering RNA treatment decreased prostaglandin E2-mediated suppression of tumor necrosis factor alpha (TNFα) expression in human macrophages (Takayama et al., 2006). Circulating TNFα levels have been found to be elevated in PCOS (Gonzalez et al., 1999; Araya et al., 2002; Amato et al., 2003; Sayin et al., 2003; Yang et al., 2006). Also, TNFα levels have been found to be increased in the follicular fluid of polycystic ovaries (Amato et al., 2003); it is intriguing to hypothesize that inherited reduction in ovarian FEM1A expression or function is responsible. Whether inflammation is a cause or consequence of PCOS is currently unknown; it appears to be related to the central adiposity often present (Puder et al., 2005).
Another fact supporting FEM1A as a candidate gene for PCOS is its location on chromosome 19p13.3. At least four independent association studies (both case/control and family based) have confirmed the microsatellite D19S884, on chromosome 19p13.2, as a PCOS susceptibility locus (Urbanek et al., 1999, 2005; Tucci et al., 2001; Stewart et al., 2006). There are numerous potential PCOS candidate genes near this microsatellite (e.g. insulin receptor, resistin, FEM1A), raising the possibility that this locus is in a control region that modulates expression of multiple genes that, when dysregulated, increase risk of PCOS. Alternatively, the location of D19S884 in the gene for fibrillin-3 raises the possibility that fibrillin-3 itself is a PCOS susceptibility locus coincidentally located on the same chromosome as FEM1A (Urbanek et al., 2007). We do not believe that the FEM1A associations observed are a result of LD with D19S884, as they are separated by 3.2 MB.
The FEM1A H500Y mutation, previously identified in one out of 35 studied women with PCOS (Maher et al., 2005), was not found in any subjects in this study, suggesting that other variants must be responsible for the associations observed herein. Because the associated SNPs rs1044386 and rs8111933 lie in or near the 3' untranslated region of the gene, they may represent functional variants that alter FEM1A expression. Alternatively, they may be in LD with as yet unidentified functional variants. We plan to conduct further investigations, including sequencing analysis, to identify functional variation in FEM1A that may influence risk of PCOS.
Fem1b knockout mice displayed abnormal glucose tolerance; while the major effect appeared to be impaired acute phase insulin secretion, a minor effect on insulin sensitivity/resistance could not be ruled out (Lu et al., 2005). Thus, the primary traits we tested for association with FEM1B variants were PCOS diagnosis and insulin secretion and insulin resistance, reductions in all of which were associated with FEM1B SNP rs10152450 and haplotype GGAAT. With the current data, we cannot distinguish between the following models: FEM1B SNP rs10152450 minor allele or a linked functional variant (i) leads to improved insulin sensitivity, which then leads secondarily to reduced insulin secretion (a sequential model); (ii) primarily reduces insulin secretion, which then results in improved insulin sensitivity (sequential model); (iii) independently modulates insulin secretion and insulin resistance (pleiotropic model). That partial pancreatectomy in dogs (reducing insulin secretion) led to insulin resistance (Matveyenko et al., 2006) argues against the second model; however, as these thoughts involve extrapolation of results between mice, dogs and humans, we cannot rule out any of these models. Similar to FEM1A, the molecular function of FEM1B is unknown, although a role in apoptosis has been proposed (Chan et al., 1999).
The reduced insulin resistance and lower frequency of PCOS observed in rs10152450 minor allele carriers is expected. Reduced insulin secretion may be consistent with protection against PCOS, as insulin exacerbates the PCOS phenotype by potentiating ovarian androgen production and suppressing hepatic SHBG levels. Women with PCOS may exhibit an excessive compensatory insulin secretion response to insulin resistance (Goodarzi et al., 2005); perhaps the FEM1B SNP rs10152450 minor allele or linked variant(s) prevent this from occurring. FEM1B is expressed in pancreatic islet cells (Chan et al., 1999; Lu et al., 2005). We note that in each case of association of a FEM1B SNP or haplotype with HOMA-%B and HOMA-IR, the P-value was more significant for HOMA-%B, consistent with the mouse data that Fem1b primarily affects insulin secretion (Lu et al., 2005).
Of this gene family, the least is known about FEM1C. It is expressed in many tissues (Krakow et al., 2001; Ventura-Holman et al., 2003), but physiologic studies carried out in mammalian systems have not clarified its function. Thus, unlike FEM1A and FEM1B, we were unable to hypothesize an a priori association of FEM1C variants with any of the quantitative features of PCOS. Our results suggest no role for the FEM1C gene in development of PCOS.
The haplotype analyses reported herein were completely consistent with the SNP results, but also led to additional insights and discoveries. Haplotypes uniquely identified by a particular minor allele displayed the same associations as that minor allele. In the case of FEM1A, the haplotype analysis suggested that rs1044386 and rs8111933 act independently, as the minor allele of each was present on different haplotypes that displayed opposite associations with PCOS diagnosis. In FEM1B, the haplotype analysis provided evidence of associations of haplotype TGAGG with increased insulin secretion and insulin resistance, associations not found with individual FEM1B SNPs.
Herein, we report provocative associations of variants in FEM1A and FEM1B with PCOS diagnosis and of FEM1B with insulin-related traits in PCOS. We note that association does not prove causation. This report is valuable in that it identifies novel PCOS candidate genes by studying all members of a particular gene family. Our goal is to encourage other investigators in the field to attempt to replicate these associations; however, to avoid false negative results, such efforts must be done with cohorts of greater numbers of subjects. We also hope that this report stimulates cellular study of the FEM1A and FEM1B genes and proteins, to elucidate on a molecular level their role, if any, in PCOS pathogenesis. If these genes are eventually confirmed as relevant to PCOS pathogenesis, it would represent a very significant finding because the risk increase or reduction was 2-fold for the variants associated with PCOS diagnosis.
Funding
National Institutes of Health grants (R01-HD29364 and K24-HD01346 to R.A.); General Clinical Research Center Grant from the National Center for Research Resources (M01-RR00425); an endowment from the Helping Hand of Los Angeles, Inc.
Supplementary Material
References
- Amato G, Conte M, Mazziotti G, Lalli E, Vitolo G, Tucker AT, Bellastella A, Carella C, Izzo A. Serum and follicular fluid cytokines in polycystic ovary syndrome during stimulated cycles. Obstet Gynecol. 2003;101:1177–1182. doi: 10.1016/s0029-7844(03)00233-3. [DOI] [PubMed] [Google Scholar]
- Araya AV, Aguirre A, Romero C, Miranda C, Molina MC, Ferreira A. Evaluation of tumor necrosis factor alpha production in ex vivo short term cultured whole blood from women with polycystic ovary syndrome. Eur Cytokine Netw. 2002;13:419–424. [PubMed] [Google Scholar]
- Azziz R, Woods KS, Reyna R, Key TJ, Knochenhauer ES, Yildiz BO. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab. 2004;89:2745–2749. doi: 10.1210/jc.2003-032046. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Boots LR, Potter S, Potter HD, Azziz R. Measurement of total serum testosterone levels using commercially available kits: high degree of between-kit variability. Fertil Steril. 1998;69:286–292. doi: 10.1016/s0015-0282(97)00464-0. [DOI] [PubMed] [Google Scholar]
- Chan SL, Tan KO, Zhang L, Yee KS, Ronca F, Chan MY, Yu VC. F1Aalpha, a death receptor-binding protein homologous to the Caenorhabditis elegans sex-determining protein, FEM-1, is a caspase substrate that mediates apoptosis. J Biol Chem. 1999;274:32461–32468. doi: 10.1074/jbc.274.45.32461. [DOI] [PubMed] [Google Scholar]
- Diamanti-Kandarakis E, Alexandraki K, Piperi C, Protogerou A, Katsikis I, Paterakis T, Lekakis J, Panidis D. Inflammatory and endothelial markers in women with polycystic ovary syndrome. Eur J Clin Invest. 2006;36:691–697. doi: 10.1111/j.1365-2362.2006.01712.x. [DOI] [PubMed] [Google Scholar]
- Doniach T, Hodgkin J. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev Biol. 1984;106:223–235. doi: 10.1016/0012-1606(84)90077-0. [DOI] [PubMed] [Google Scholar]
- Ehrmann DA, Schwarz PE, Hara M, Tang X, Horikawa Y, Imperial J, Bell GI, Cox NJ. Relationship of calpain-10 genotype to phenotypic features of polycystic ovary syndrome. J Clin Endocrinol Metab. 2002;87:1669–1673. doi: 10.1210/jcem.87.4.8385. [DOI] [PubMed] [Google Scholar]
- Escobar-Morreale HF, Peral B, Villuendas G, Calvo RM, Sancho J, San Millan JL. Common single nucleotide polymorphisms in intron 3 of the calpain-10 gene influence hirsutism. Fertil Steril. 2002;77:581–587. doi: 10.1016/s0015-0282(01)03206-x. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Thusu K, Abdel-Rahman E, Prabhala A, Tomani M, Dandona P. Elevated serum levels of tumor necrosis factor alpha in normal-weight women with polycystic ovary syndrome. Metabolism. 1999;48:437–441. doi: 10.1016/s0026-0495(99)90100-2. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Abril E, Roca A, Aragon MJ, Figueroa MJ, Velarde P, Royo JL, Real LM, Ruiz A. Comment: CAPN10 alleles are associated with polycystic ovary syndrome. J Clin Endocrinol Metab. 2002;87:3971–3976. doi: 10.1210/jcem.87.8.8793. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Saez ME, Aragon MJ, Galan JJ, Vettori P, Molina L, Rubio C, Real LM, Ruiz A, Ramirez-Lorca R. Specific haplotypes of the CALPAIN-5 gene are associated with polycystic ovary syndrome. Hum Reprod. 2006;21:943–951. doi: 10.1093/humrep/dei443. [DOI] [PubMed] [Google Scholar]
- Goodarzi MO, Azziz R. Diagnosis, epidemiology, and genetics of the polycystic ovary syndrome. Best Pract Res Clin Endocrinol Metab. 2006;20:193–205. doi: 10.1016/j.beem.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Goodarzi MO, Guo X, Taylor KD, Quiñones MJ, Samayoa C, Yang H, Saad MF, Palotie A, Krauss RM, Hsueh WA, et al. Determination and use of haplotypes: ethnic comparison and association of the lipoprotein lipase gene and coronary artery disease in Mexican-Americans. Genet Med. 2003;5:322–327. doi: 10.1097/01.GIM.0000076971.55421.AD. [DOI] [PubMed] [Google Scholar]
- Goodarzi MO, Erickson S, Port SC, Jennrich RI, Korenman SG. Beta-cell function: a key pathological determinant in polycystic ovary syndrome. J Clin Endocrinol Metab. 2005;90:310–315. doi: 10.1210/jc.2004-1006. [DOI] [PubMed] [Google Scholar]
- Gunderson KL, Steemers FJ, Ren H, Ng P, Zhou L, Tsan C, Chang W, Bullis D, Musmacker J, King C, et al. Whole-genome genotyping. Methods Enzymol. 2006;410:359–376. doi: 10.1016/S0076-6879(06)10017-8. [DOI] [PubMed] [Google Scholar]
- Haddad L, Evans JC, Gharani N, Robertson C, Rush K, Wiltshire S, Frayling TM, Wilkin TJ, Demaine A, Millward A, et al. Variation within the type 2 diabetes susceptibility gene calpain-10 and polycystic ovary syndrome. J Clin Endocrinol Metab. 2002;87:2606–2610. doi: 10.1210/jcem.87.6.8608. [DOI] [PubMed] [Google Scholar]
- Hatch R, Rosenfield RL, Kim MH, Tredway D. Hirsutism: implications, etiology, and management. Am J Obstet Gynecol. 1981;140:815–830. doi: 10.1016/0002-9378(81)90746-8. [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PE, et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- Kahsar-Miller MD, Nixon C, Boots LR, Go RC, Azziz R. Prevalence of polycystic ovary syndrome (PCOS) in first-degree relatives of patients with PCOS. Fertil Steril. 2001;75:53–58. doi: 10.1016/s0015-0282(00)01662-9. [DOI] [PubMed] [Google Scholar]
- Knochenhauer ES, Key TJ, Kahsar-Miller M, Waggoner W, Boots LR, Azziz R. Prevalence of the polycystic ovary syndrome in unselected black and white women of the southeastern United States: a prospective study. J Clin Endocrinol Metab. 1998;83:3078–3082. doi: 10.1210/jcem.83.9.5090. [DOI] [PubMed] [Google Scholar]
- Krakow D, Sebald E, King LM, Cohn DH. Identification of human FEM1A, the ortholog of a C. elegans sex-differentiation gene. Gene. 2001;279:213–219. doi: 10.1016/s0378-1119(01)00756-9. [DOI] [PubMed] [Google Scholar]
- Legro RS, Driscoll D, Strauss JF, Fox J, Dunaif A. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci USA. 1998;95:14956–14960. doi: 10.1073/pnas.95.25.14956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JC, Matthews DR, Hermans MP. Correct homeostasis model assessment (HOMA) evaluation uses the computer program. Diabetes Care. 1998;21:2191–2192. doi: 10.2337/diacare.21.12.2191. [DOI] [PubMed] [Google Scholar]
- Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- Lu D, Ventura-Holman T, Li J, McMurray RW, Subauste JS, Maher JF. Abnormal glucose homeostasis and pancreatic islet function in mice with inactivation of the Fem1b gene. Mol Cell Biol. 2005;25:6570–6577. doi: 10.1128/MCB.25.15.6570-6577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque-Ramirez M, San Millan JL, Escobar-Morreale HF. Genomic variants in polycystic ovary syndrome. Clin Chim Acta. 2006;366:14–26. doi: 10.1016/j.cca.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Maher JF, Hines RS, Futterweit W, Crawford S, Lu D, Shen P, Oefner P, Kazi M, Wilson JG, Subauste JS, et al. FEM1A is a candidate gene for polycystic ovary syndrome. Gynecol Endocrinol. 2005;21:330–335. doi: 10.1080/09513590500431458. [DOI] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Matveyenko AV, Veldhuis JD, Butler PC. Mechanisms of impaired fasting glucose and glucose intolerance induced by an approximate 50% pancreatectomy. Diabetes. 2006;55:2347–2356. doi: 10.2337/db06-0345. [DOI] [PubMed] [Google Scholar]
- Mugita N, Kimura Y, Ogawa M, Saya H, Nakao M. Identification of a novel, tissue-specific calpain htra-3; a human homologue of the Caenorhabditis elegans sex determination gene. Biochem Biophys Res Commun. 1997;239:845–850. doi: 10.1006/bbrc.1997.7571. [DOI] [PubMed] [Google Scholar]
- Pearlman WH, Crepy O, Murphy M. Testosterone-binding levels in the serum of women during the normal menstrual cycle, pregnancy, and the post-partum period. J Clin Endocrinol Metab. 1967;27:1012–1018. doi: 10.1210/jcem-27-7-1012. [DOI] [PubMed] [Google Scholar]
- Puder JJ, Varga S, Kraenzlin M, De Geyter C, Keller U, Muller B. Central fat excess in polycystic ovary syndrome: relation to low-grade inflammation and insulin resistance. J Clin Endocrinol Metab. 2005;90:6014–6021. doi: 10.1210/jc.2005-1002. [DOI] [PubMed] [Google Scholar]
- Qin ZS, Niu T, Liu JS. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am J Hum Genet. 2002;71:1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner W, Auchus RJ, Azziz R, Sluss PM, Raff H. Utility, limitations, and pitfalls in measuring testosterone: an Endocrine Society position statement. J Clin Endocrinol Metab. 2007;92:405–413. doi: 10.1210/jc.2006-1864. [DOI] [PubMed] [Google Scholar]
- Sayin NC, Gucer F, Balkanli-Kaplan P, Yuce MA, Ciftci S, Kucuk M, Yardim T. Elevated serum TNF-alpha levels in normal-weight women with polycystic ovaries or the polycystic ovary syndrome. J Reprod Med. 2003;48:165–170. [PubMed] [Google Scholar]
- Steemers FJ, Chang W, Lee G, Barker DL, Shen R, Gunderson KL. Whole-genome genotyping with the single-base extension assay. Nat Methods. 2006;3:31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
- Stewart DR, Dombroski B, Urbanek M, Ankener W, Ewens KG, Wood JR, Legro RS, Strauss JF, Dunaif A, Spielman RS. Fine mapping of genetic susceptibility to polycystic ovary syndrome on chromosome 19p13.2 and tests for regulatory activity. J Clin Endocrinol Metab. 2006;91:4112–4117. doi: 10.1210/jc.2006-0951. [DOI] [PubMed] [Google Scholar]
- Takayama K, Sukhova GK, Chin MT, Libby P. A novel prostaglandin E receptor 4-associated protein participates in antiinflammatory signaling. Circ Res. 2006;98:499–504. doi: 10.1161/01.RES.0000204451.88147.96. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Tucci S, Futterweit W, Concepcion ES, Greenberg DA, Villanueva R, Davies TF, Tomer Y. Evidence for association of polycystic ovary syndrome in caucasian women with a marker at the insulin receptor gene locus. J Clin Endocrinol Metab. 2001;86:446–449. doi: 10.1210/jcem.86.1.7274. [DOI] [PubMed] [Google Scholar]
- Urbanek M, Legro RS, Driscoll DA, Azziz R, Ehrmann DA, Norman RJ, Strauss JF, Spielman RS, Dunaif A. Thirty-seven candidate genes for polycystic ovary syndrome: strongest evidence for linkage is with follistatin. Proc Natl Acad Sci USA. 1999;96:8573–8578. doi: 10.1073/pnas.96.15.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek M, Woodroffe A, Ewens KG, Diamanti-Kandarakis E, Legro RS, Strauss JF, Dunaif A, Spielman RS. Candidate gene region for polycystic ovary syndrome on chromosome 19p13.2. J Clin Endocrinol Metab. 2005;90:6623–6629. doi: 10.1210/jc.2005-0622. [DOI] [PubMed] [Google Scholar]
- Urbanek M, Sam S, Legro RS, Dunaif A. Identification of a polycystic ovary syndrome susceptibility variant in fibrillin-3 and association with a metabolic phenotype. J Clin Endocrinol Metab. 2007;92:4191–4198. doi: 10.1210/jc.2007-0761. [DOI] [PubMed] [Google Scholar]
- Ventura-Holman T, Lu D, Si X, Izevbigie EB, Maher JF. The Fem1c genes: conserved members of the Fem1 gene family in vertebrates. Gene. 2003;314:133–139. doi: 10.1016/s0378-1119(03)00712-1. [DOI] [PubMed] [Google Scholar]
- Vink JM, Sadrzadeh S, Lambalk CB, Boomsma DI. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab. 2006;91:2100–2104. doi: 10.1210/jc.2005-1494. [DOI] [PubMed] [Google Scholar]
- Vollmert C, Hahn S, Lamina C, Huth C, Kolz M, Schopfer-Wendels A, Mann K, Bongardt F, Mueller JC, Kronenberg F, et al. Calpain-10 variants and haplotypes are associated with polycystic ovary syndrome in Caucasians. Am J Physiol Endocrinol Metab. 2007;292:E836–E844. doi: 10.1152/ajpendo.00584.2005. [DOI] [PubMed] [Google Scholar]
- Wallace TM, Levy JC, Matthews DR. Use and abuse of HOMA modeling. Diabetes Care. 2004;27:1487–1495. doi: 10.2337/diacare.27.6.1487. [DOI] [PubMed] [Google Scholar]
- Yang XF, Ren FR, Guo SP. Study on the relationship between serum adiponectin and insulin resistance in women with polycystic ovary syndrome. Zhonghua Fu Chan Ke Za Zhi. 2006;41:261–263. [PubMed] [Google Scholar]
- Zawadzki JK, Dunaif A. Diagnostic criteria for polycystic ovary syndrome: towards a rational approach. In: Dunaif A, Givens JR, Haseltine F, Merriam GR, editors. Polycystic ovary syndrome. Cambridge: Blackwell Scientific Publications; 1992. pp. 377–384. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.