Summary
Streptococcus mutans is particularly well-adapted for high-affinity, high-capacity catabolism of multiple carbohydrate sources. S. mutans EIILev, a fructose/mannose permease encoded by the levDEFG genes, and fruA, which encodes a hydrolase that releases fructose from fructan polymers, are transcriptionally regulated by the LevQRST four-component signal transduction system. Here, we demonstrate that (1) levDEFGX are co-transcribed and the levE/F intergenic region is required for optimal expression of levFGX; (2) D-mannose is a potent inducer of the levD and fruA operons; (3) CcpA regulates levD expression in a carbohydrate-specific manner; (4) deletion of the genes for the fructose/mannose-EII enzymes of S. mutans (manL, fruI, and levD) enhances levD expression; (5) repression of the LevQRST regulon by EII enzymes depends on the presence of their substrates and requires LevR, but not LevQST; and (6) CcpA inhibits expression of the manL and frul genes to indirectly control the LevQRST regulon. Further, the manL, ccpA, frul/fruCD and levD gene products differentially exert control over the cellobiose and lactose operons. Collectively, the results reveal the existence of a global regulatory network in S. mutans that governs the utilization of non-preferred carbohydrates in response to the availability and source of multiple preferred carbohydrates.
Keywords: Sugar:phosphotransferase system, β-D-fructosidase, Catabolite repression, CcpA, Gene regulation
Introduction
Carbohydrate metabolism by Streptococcus mutans, the most common etiologic agent of dental caries, is intimately linked to persistence, growth and pathogenesis by this organism. The phosphoenopyruvate (PEP):sugar phosphotransferase system (PTS) of S. mutans catalyzes high-affinity and high-capacity internalization of a wide variety of carbohydrates (Ajdic and Pham, 2007; Vadeboncoeur and Pelletier, 1997). In addition, the production of extracellular polysaccharides from sucrose enhances colonization, biofilm formation and virulence of the organism. S. mutans produces three glucosyltransferase enzymes that convert dietary sucrose into extracellular homopolymers of glucose (glucans), which function as an adhesive scaffolding for biofilm formation (Banas, 2004; Kuramitsu, 2003). The organism also produces an extracellular fructosyltransferase enzyme that converts sucrose into fructose homopolymers (fructans), which rapidly accumulate in oral biofilms and serve primarily as a storage polymer. Metabolism of fructan polymers by a secreted fructosidase, encoded by the fruA gene, enhances the amount and duration of acid production and, thus, the virulence of the organism (Burne et al., 1987; Burne and Penders, 1992, 1994; Burne et al., 1996; Burne et al., 1999).
Expression of fruA is regulated by induction and is sensitive to carbohydrate catabolite repression (CCR) in the presence of rapidly catabolized sugars, such as glucose. A four-component signal transduction system regulates induction of the fruA gene of S. mutans (Zeng et al., 2006). This complex includes LevQ and LevT, which are predicted to be extracellular sugar-binding proteins, and a two-component system consisting of the sensor kinase LevS and the response regulator LevR. All four gene products are required for the activation of fruA; and low concentrations of fructose, the end product of the enzymatic action of FruA on fructan polymers, serves as the inducing signal. We also noted that LevQRST controlled the activation of the genes for a fructose/mannose PTS Enzyme II complex, encoded by the levD, levE, levF and levG genes located immediately downstream of levQRST. Additionally, expression of fruA was enhanced in a strain lacking an intact LevD protein. Since then, our laboratory has also demonstrated that CCR of fruA can be exerted by an apparent homologue of catabolite control protein A (CcpA) in response to readily metabolizable hexoses via direct binding to a conserved catabolite response element (CRE) in the fruA promoter region (Abranches et al., 2008).
A well-supported model for CCR in low-G+C Gram-positive bacteria has emerged that establishes the PTS as the central mediator of CCR through inducer exclusion and modulation of transcriptional regulation by CcpA (Postma et al., 1993). The form of the general PTS protein HPr that is phosphorylated at serine residue 46 by HPr kinase can alter the activity of non-PTS permeases and function as a co-factor in the binding of CcpA to its target sites (Deutscher and Saier, 1983; Deutscher et al., 1994; Fujita et al., 1995). In addition to transporting sugars, the PTS, and in particular the mannose PTS permease in oral streptococci, influences multiple cellular functions; including regulation of the internalization of carbohydrates (Liberman and Bleiweis, 1984), carbon catabolite repression (Abranches et al., 2003; Abranches et al., 2006; Gauthier et al., 1990; Vadeboncoeur et al., 1983), coordination of carbon and nitrogen metabolism (Chen et al., 1998), and global control of gene expression (Abranches et al., 2006). However, additional complexities in catabolite modification of gene expression have been noted in oral streptococci, such as S. mutans and S. salivarius (Gauthier et al., 1997; Liberman and Bleiweis, 1984; Vadeboncoeur and Pelletier, 1997; Ye et al., 1996); including the discovery that PTS permeases other than EIIABMan can exert negative control over gene expression in response to readily metabolizable carbohydrates (Wen and Burne, 2002; Zeng et al., 2006). Results presented in this report reveal a complex regulatory network consisting of CcpA and four PTS permeases that comprise a hierarchical system for coordinating sugar catabolism by S. mutans in response to carbohydrate source and availability.
Results
Characterization of the genetic organization of the levD operon
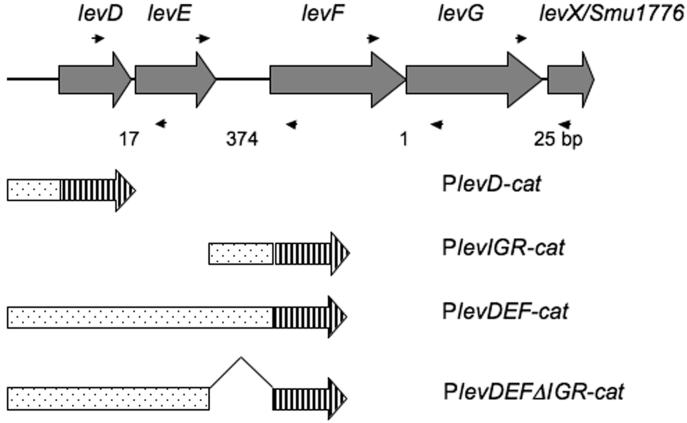
RT-PCR was employed to demonstrate the co-transcription of the levD, -E, -F, and -G genes, and Smu1776 (http://www.oralgen.lanl.gov/), which is designated here as levX because the function for this gene product has not been identified. Four pairs of specific primers (Fig. 1), designed to target DNA sequences encompassing adjacent open reading frames (ORF), were used in PCR reactions after reverse transcription with total RNA from S. mutans UA159. As shown in Supplementary Figure 1, these fragments were readily amplified in the test, but not the control samples, indicative that these 5 genes can be co-transcribed.
Figure 1.
Schematic representation of the levD operon in S. mutans UA159. The cat gene is represented by a barred arrow while the fused fragments are shown as dotted bars aligned with their locations in the levD operon. The size of each intergenic region is indicated by numbers (bp) below the diagram. Small arrows below and above the ORFs denote the primers used in RT-PCR reactions.
Quantitative real-time RT-PCR was utilized to measure the concentrations of the transcripts of each of the five genes. The results (Fig. S2, supplementary material) demonstrated that, for all conditions tested, the levD-G genes were transcribed at a similar level. Also, the amount of transcripts of the genes when fructose or mannose was used to culture the bacteria was approximately 10-fold higher than that found in cells grown in glucose, consistent with observations made using gene fusions (Table 1).
Table 1.
Expression of PlevD-cat and its derivative fusions.
| CAT specific activitya | ||||||
|---|---|---|---|---|---|---|
| Strains | TV-glucose | SD | TV-fructose | SD | TV-mannose | SD |
| SD, standard deviation. | ||||||
| UA159/PlevD-cat | 6.32 | 1.58 | 474.04 | 44.68 | 870.43 | 85.15 |
| levR-/PlevD-cat | 0.18 | 0.04 | 0.03 | 0.05 | 0.24 | 0.07 |
| ccpA-/PlevD-cat | 10.46 | 1.51 | 107.50 | 11.49 | 23.02 | 2.67 |
| UA159/PlevDEF-cat | 2.35 | 0.32 | 545.71 | 28.54 | 1292.88 | 161.59 |
| levR-/PlevDEF-cat | 0.19 | 0.05 | 0.16 | 0.09 | 0.31 | 0.11 |
| UA159/PlevIGR-cat | 0 | 0 | 0 | 0 | 0 | 0 |
| UA159/PlevDEFΔIGR-cat | 1.54 | 0.84 | 235.65 | 9.90 | 849.03 | 74.12 |
values (in the unit of {nmole (mg protein)−1 min−1}) represent averages of at least three separate experiments.
To further dissect the regulation of the levD operon, promoter-cat fusions were constructed using DNA sequences derived from the levD operon (Fig. 1). The intergenic region between the levE and levF genes, which is 374-bp, was used to engineer a fusion designated PlevIGR-cat. This construct produced no detectable CAT activity when tested in the wild-type background under conditions that activate the levD promoter (Table 1). However, when a larger fragment that included the levD promoter, the levD and levE genes, and the levEF intergenic region was fused to cat (PlevDEF-cat), CAT activities in the wild-type background were comparable to those expressed from PlevD-cat (Zeng et al., 2006). Additionally, when tested in the levR mutant background, PlevDEF-cat produced minimal levels of activity. These data demonstrate that transcription of levF, and likely levGX, arises from the same promoter as levD, and that there is no promoter in the levEF intergenic region that is functional under the conditions tested.
To explore the potential role of the 374-bp levEF intergenic region in the expression of this operon, PlevDEFΔIGR-cat was constructed by deleting the intergenic region from the PlevDEF-cat fusion construct (Fig. 1). When assayed in the wild-type background, PlevDEFΔIGR-cat expressed reduced levels of CAT activity compared to the PlevDEF-cat fusion strain grown in TV-glucose or TV-fructose (Table 1). Future studies will explore the mechanism by which the levEF intergenic region affects expression of downstream genes.
Mannose is a second induction signal for the LevQRST regulon
We previously noted that S. mutans cells cultivated in the presence of mannose displayed elevated mannose-PTS activity compared to cells grown in glucose, and that this enhanced activity required the presence of an intact levR gene (Zeng et al., 2006). Further confirmation of this effect was noted in Figure S2 (supplementary material), where growth in mannose resulted in levD operon expression levels comparable to growth on the known inducing carbohydrate, fructose. To investigate the possibility that the LevQRST pathway was responsive to mannose, expression from PlevD-cat and PlevDEF-cat was monitored in the wild-type and LevR-deficient backgrounds (levR-/PlevD-cat and levR-/PlevDEF-cat, respectively) in cells grown to mid-exponential phase in TV supplemented with 0.5% of glucose, fructose or mannose (Table 1). In the wild-type background, PlevD-cat expression was increased by more than 100-fold in TV-mannose compared to cells grown in TV-glucose. CAT activity in the TV-mannose culture was about 2-fold higher than in cells grown in TV-fructose. The CAT activities measured from these gene fusions in the LevR-deficient strain were near the lower limit of detection of the assay. Similar results were obtained with strains lacking the levQ, -S or -T genes, which are other members of the signal transduction complex (Data not shown).
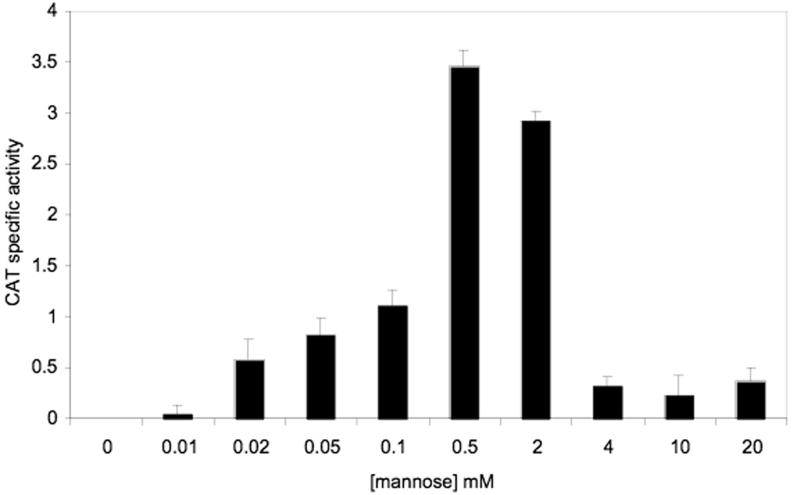
Since fruA expression is also under the control of the LevQRST regulon, CAT activity expressed from a PfruA-cat fusion (Zeng et al., 2006) in the wild-type background was measured in cells grown to early exponential phase in TV-galactose medium after treatment with various concentrations of mannose for 3.5 hours. As shown in Fig. 2, mannose at concentrations as low as 0.01 mM was able to induce the expression of PfruA-cat, with activity plateauing at 0.5 mM and dropping at concentrations of 2 to 4 mM.
Figure 2.
Induction of PfruA-cat by D-mannose. S. mutans strain UA159/PfruA-cat was grown to OD600 = 0.2 in TV supplemented with 0.5% of galactose. Mannose was then added to final concentrations ranging from 0.01 mM to 20 mM and the cultures were incubated for an additional 3.5 hours before harvesting for CAT assays. The results represent the average of three independent cultures and are expressed as nmole (mg protein)−1 (min−1).
Loss of CcpA results in altered expression of the levD operon
We recently reported that CcpA in S. mutans is a repressor of fruA expression in the presence of relatively high levels of glucose, fructose or mannose (Abranches et al., 2008). However, the potential role of CcpA in regulating the levD operon has not been explored in detail. As presented in Table 1, a PlevD-cat fusion in a ccpA mutant background expressed much lower CAT activity in TV-fructose (∼4-fold) or TV-mannose (∼37-fold) than in the wild-type background, suggesting that CcpA is required for optimal expression of levD-X in the presence of inducer. Interestingly, deletion of ccpA also resulted in a modest up-regulation of PlevD-cat expression in glucose-containing medium, indicating that the effects of CcpA on the levD operon depend on the carbohydrate source.
PTS EII enzymes negatively control levD expression
Deletion of the genes encoding the fructose-PTS enzymes II (fruI, fruCD) or mannose-PTS enzymes II (manL) results in derepression of fruA expression (Abranches et al., 2006; Wen and Burne, 2002). Similarly, deletion of levD enhances expression from the levD and fruA promoters (Zeng et al., 2006). Since fruA and levD depend on the LevQRST signal transduction system for activation, negative regulation of these two genes by EII enzymes may involve a common mechanism. As shown in Table 2, mutations in the genes for the fructose PTS permeases (fruI, fruCD), for the IIAB domain of a mannose/fructose PTS (manL), and for the LevD mannose/fructose IIAlev domain, caused enhanced expression from PlevD-cat under inducing conditions.
Table 2.
Expression of PlevD-cat in various EII mutants.
| PlevD-cat CAT specific activitya | ||||||
|---|---|---|---|---|---|---|
| Background | TV-glucose | SD | TV-fructose | SD | TV-mannose | SD |
| In this case, mutations in fruI and fruCD were studied in combination, since our previous work found that loss of either gene resulted in elevated fruA expression, while deletion of both had additive effects on derepression of fruA (Wen and Burne, 2002). | ||||||
| SD, standard deviation. | ||||||
| UA159 | 6.32 | 1.58 | 474.04 | 44.68 | 870.43 | 85.15 |
| levD- | 10.19 | 0.29 | 749.91 | 54.38 | 950.69 | 51.11 |
| fruI- | 48.71 | 7.65 | 814.99 | 36.05 | 679.91 | 42.18 |
| fruCD- | 1.01 | 0.40 | 455.67 | 8.05 | 698.23 | 64.28 |
| fruI-/fruCD- | 79.09 | 0.40 | 787.48 | 21.84 | 681.99 | 11.12 |
| manL- | 21.92 | 1.60 | 458.59 | 5.26 | 2199.88 | 85.68 |
| ptsG- | 6.65 | 0.39 | 394.39 | 9.36 | 698.07 | 53.13 |
values represent averages of at least three separate experiments.
Of particular interest, the effects of the loss of the various PTS permeases on expression of fruA and levD were specific to the carbohydrate source. Compared to the behavior of the gene fusions in the wild-type background, all mutations in the EII-encoding genes of interest resulted in elevated CAT activity expressed from the levD promoter in glucose-grown cultures, with the fruI/fruCD mutant having the greatest level of alleviation. However, when fructose was the growth carbohydrate, alleviation of PlevD-cat expression was seen in the fruI/fruCD and levD mutant backgrounds, but not in the manL mutant background. In contrast, dramatic derepression of PlevD-cat expression was noted in the manL mutant in mannose-grown cultures, whereas loss of fruI/fruCD elicited no derepression in mannose, and loss of levD caused a modest, albeit not statistically-significant, increase in expression in mannose. Importantly, mutation of the ptsG gene, encoding a product involved in maltose transport in S. mutans (Webb et al., 2007), produced no derepression of PlevD-cat, regardless of whether maltose was present in the growth medium (Data not shown). Therefore, the effects seen with ManL, LevD, and FruI/FruCD are not observed with all PTS permeases.
The PTS permeases exert CCR on the fruA operon
To demonstrate that the effects of the EII enzymes of interest on gene regulation and that the dependence on specific substrates was not a peculiarity of the levD operon, we tested the effects of ManL deficiency on fruA expression. When the manL mutant and the wild-type strain were compared for their response of PfruA-cat to fructose using a fructose-pulsing experiment (Table 3), there was much greater derepression of fruA associated with the loss of ManL in TV-glucose medium, compared to when the cells were grown under conditions that poorly induce CCR, i.e. in TV-galactose. When TV medium containing 0.5% fructose was used to culture these two strains, no alleviation of fruA expression was detected in the manL mutant. When TV-mannose was used to grow the cells, fruA expression was greatly derepressed in the manL mutant, compared to the wild-type background. Finally, alleviation of fruA expression in the fruI/fruCD mutant was only evident in TV-fructose cultures.
Table 3.
Expression of PfruA-catfusion.
| Pulsed for 3.5 h with fructose at concentration (mM) | ||||||
|---|---|---|---|---|---|---|
| 0 mM | 0.05 mM | 3 mM | 5 mM | 10 mM | ||
| As CCR of fruA has been shown to be triggered at sugar concentrations at around 2 to 4 mM, we studied the expression of PfruA-cat in both the wild type (UA159) and the manL mutant backgrounds by adding fructose, at concentrations ranging from 50 µM to 10 mM, to early-exponential phase cultures prepared using TV supplemented with 0.5% of glucose or the poorly-repressing substrate galactose, followed by 3.5 h of incubation at 37°C. A similar strategy was utilized to study CCR of fruA by CcpA in S. mutans (Abranches et al., 2008). Also presented are CAT activities of PfruA-cat expressed in the wild type, the manL, and the fruI/fruCD mutant backgrounds, when cells were grown to mid-exponential phase with TV supplemented with 0.5% of fructose or mannose. Values are averages followed by standard deviations in parentheses of at least three independent experiments. | ||||||
| 0.5% glucose | UA159 | 0 | 0 | 0.157 (0.068) | 0.079 (0.040) | 0.180 (0.102) |
| manL- | 0 | 0.189 (0.067) | 3.017 (0.557) | 0.313 (0.256) | 0.195 (0.168) | |
| 0.5% galactose | UA159 | 0 | 2.490 (0.033) | 2.484 (0.403) | 1.147 (0.153) | 1.083 (0.028) |
| manL- | 1.102 (0.086) | 2.615 (0.290) | 2.684 (0.157) | 1.227 (0.025) | 1.074 (0.110) | |
| Grown to mid-exponential phase without fructose pulsing | ||||||
| 0.5% fructose | UA159 | 0.439 (0.056) | ||||
| manL- | 0.458 (0.053) | |||||
| fruI-/fruCD- | 2.336 (0.123) | |||||
| 0.5% mannose | UA159 | 0.253 (0.112) | ||||
| manL- | 26.73 (2.59) | |||||
| fruI-/fruCD- | 0.206 (0.277) | |||||
We further probed the roles of the different EII enzymes in catabolite-dependent regulation of carbohydrate utilization by comparing the growth curves of the manL mutant in TV-medium containing 0.5% of the fructose homopolymer inulin, which is a substrate for FruA, and 0.05% of glucose or fructose, using inocula prepared in glucose or fructose alone, respectively. As shown in Fig. S3 (supplementary material), diauxic growth was evident in the wild-type genetic background when cells were grown on a mixture of glucose and inulin. Consistent with the behavior of the gene fusion strains, deletion of manL diminished the extent of the diauxie. In contrast, when fructose was used instead of glucose as the repressing carbohydrate in these growth assays, the manL mutant presented a diauxic growth curve. Furthermore, when the levD, fruI/fruCD and ccpA mutants were used in the same experiment, their growth behaviors were also consistent with regulation of fruA by EII permeases (see Fig. S3 for details). Collectively, these observations provide further support that multiple EII enzymes, in concert with CcpA, modulate expression of the LevQRST regulon in a carbohydrate source-dependent manner.
Derepression associated with loss of PTS permeases does not occur in a LevR-deficient strain
We examined whether the permeases of interest could exert their influence through the LevQRST signal-transduction pathway. To explore this possibility, additional mutants were engineered in the background of the strain carrying a PlevD-cat fusion, where mutations in fruI/fruCD, levDor manL were combined with deletions of the levR, levS, levQ or levT genes (Table 4). First, in the background of a levR mutant, introduction of mutations in fruI/fruCD, manL or levD failed to significantly alter the expression from the PlevD-cat fusion, although expression levels were very low because of the requirement for LevR for activation of the operons. However, in the levQ mutant, where a similar level of PlevD-cat expression was obtained as in the levR mutant, deletion of manL or fruI/fruCD resulted in apparent derepression of PlevD-cat expression, indicating that these EII enzymes can still exert negative control on the levD promoter in the absence of LevQ. Also, in the levQ mutant background, substantial derepression of levD expression was caused by mutation of manL only when mannose was used to culture the bacteria. Also consistent with data presented above, the fruI/fruCD mutation resulted in higher levels of expression when fructose was used to grow the cells, as compared with mannose. Results similar to that seen in the levQ mutant background were obtained when using the levS mutant that lacks the sensor kinase; i.e. expression of PlevD-cat was increased when manL or fruI/fruCD mutations were introduced into the levS mutant background. The results from the levT mutant were similar to that of the levS and levQ mutants, although an overall higher level of alleviation of expression from Plev-cat expression was caused by the manL or fruI/fruCD mutations in the levT mutant background.
Table 4.
Repression of levD expression by EII enzymes in strains lacking components of the LevQRST signal transduction complex.
| PlevD-cat CAT specific activitya | ||||||
|---|---|---|---|---|---|---|
| Background | TV-glucose | SD | TV-fructose | SD | TV-mannose | SD |
| SD, standard deviation. | ||||||
| levR- | 0.18 | 0.04 | 0.03 | 0.05 | 0.24 | 0.07 |
| levR-/levD- | 0.13 | 0.04 | 0.01 | 0.01 | 0.09 | 0.07 |
| levR-/manL- | 0.17 | 0.06 | 0.03 | 0.03 | 0.00 | 0.00 |
| levR-/fruI-/fruCD- | 0.15 | 0.12 | 0.13 | 0.11 | 0.02 | 0.01 |
| levQ- | 0.12 | 0.11 | 0.00 | 0.00 | 0.29 | 0.29 |
| levQ-/manL- | 0.90 | 0.06 | 0.19 | 0.06 | 12.47 | 2.61 |
| levQ-/fruI-/fruCD- | 1.14 | 0.34 | 2.54 | 1.17 | 0.56 | 0.15 |
| levT- | 3.06 | 0.02 | 0.14 | 0.03 | 1.18 | 0.63 |
| levT-/manL- | 32.24 | 1.91 | 0.45 | 0.08 | 536.93 | 19.80 |
| levT-/fruI-/fruCD- | 5.54 | 1.41 | 44.71 | 9.61 | 1.17 | 0.18 |
| levS- | 3.78 | 1.22 | 0.49 | 0.06 | 1.30 | 0.08 |
| levS-/manL- | 12.28 | 1.75 | 1.41 | 0.27 | 8.12 | 1.66 |
| levS-/fruI-/fruCD- | 9.34 | 0.43 | 8.38 | 0.61 | 1.61 | 0.04 |
values represent averages of at least three separate experiments.
CcpA regulates EII permease gene expression
Although CcpA appears to be required for optimal expression of levD (Table 1), we have been unable to demonstrate that a purified CcpA protein from S. mutans is able to bind the levD promoter region. Specifically, a recombinant His-tagged CcpA protein that binds the CRE in the fruA promoter (Abranches et al., 2008) was used in an electrophoretic mobility shift assay with a ∼0.3 kbp radio-labeled DNA fragment containing the entire levD promoter region, the same DNA fragment that was used to construct the PlevD-cat fusion. No interaction between CcpA and the probe was detected in this assay (Data not shown). Consistent with this observation, there are no CREs within 200-bp 5′ or 3′ of the levD promoter region, suggesting that direct binding of CcpA to the levD promoter may not occur in vivo.
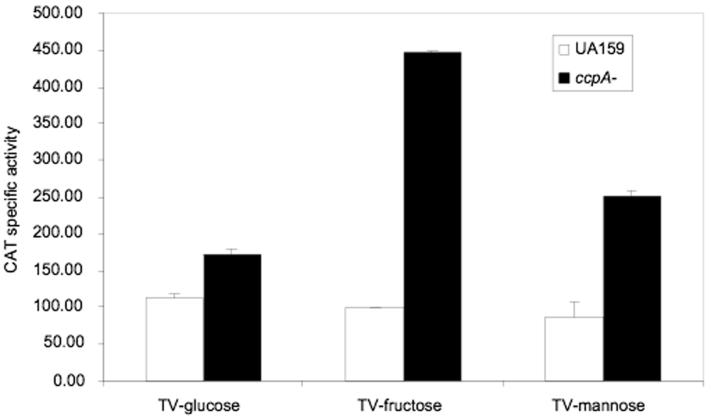
We previously observed that deletion of ccpA resulted in major changes in the transcriptome and metabolic potential of S. mutans (Abranches et al., 2008), so we explored whether CcpA was able to alter the expression of factors that govern levD and fruA expression. In particular, the expression of fruI, fruCD, manL, and levQRST was compared in the wild-type and CcpA-deficient strains. A PmanL-cat reporter fusion was constructed using the manL promoter and used to monitor the expression of manL in the ccpA mutant background and the parental strain. Cells were grown to mid-exponential phase in TV-base medium supplemented with 0.5% glucose, fructose or mannose, and harvested for CAT assays. The results (Fig. 3) indicated that manL expression was derepressed in the ccpA mutant 1.5- to 5-fold, with the highest level of increase seen in the TV-fructose cultures. This finding was confirmed when the levels of manL transcripts were compared in strain UA159 and the ccpA mutant using real-time quantitative PCR (Data not shown). Similarly, when the transcript levels of fruI or fruC were compared in UA159 and the ccpA mutant strain, significant increases in fruC, and especially fruI mRNA, were detected in the ccpA mutant grown in TV-fructose or TV-mannose (Fig. S4 of the supplementary material). For fruI, there was about a 4-fold increase in transcription in TV-fructose cultures due to deletion of ccpA, and about a 50% increase in TV-mannose cultures. However for fruC, the only increase, about 2-fold, came when the cells were grown in TV-mannose medium. Also noteworthy is that while transcription of both fruI and fruC was increased in cells cultured with fructose, compared to mannose-grown cells, the overall level of fruI transcript was about 3-logs greater than that of fruC. The significance of this observation in terms of carbohydrate transport remains to be determined.
Figure 3.
Expression of PmanL-cat in the wild-type (UA159) and ccpA mutant backgrounds. Cells were grown to mid-exponential phase in the specified media before being subjected to CAT assays. See experimental procedures for detail.
The fact that very little derepression in fruC transcription was seen in the ccpA mutant prompted us to study the effects of the fruCD and fruI mutations on levD expression, separately. As shown in Table 2, a mutation in fruI alone resulted in an elevation of expression of PlevD-cat of a magnitude similar to that seen in the fruI/fruCD double mutant, whereas the fruCD mutation alone caused little change in PlevD-cat expression. The same trend was observed when fruA gene expression was analyzed in the individual fruI and fruCD mutants (Wen et al, 2001), despite major differences in the growth media and in the conditions used to induce the operon in this study and that of Wen et al (2001). Therefore, we conclude that FruCD does not play a significant role in the regulation of the levD or fruA operons under the conditions tested, and therefore the primary effectors of catabolite modification are ManL, FruI and LevD.
As FruI and ManL have been implicated in the negative regulation of levD, it is reasonable to interpret that reduced PlevD-cat expression in a ccpA mutant grown in fructose or mannose medium results from elevated expression of the PTS permeases, FruI and ManL. Indeed, the characteristics of expression of the PlevD-cat gene fusion in two additional mutants, manL/ccpA and fruI/fruCD/ccpA, support this idea (Table 5). In particular, a strain carrying a fruI/fruCD double mutation and lacking CcpA displayed dramatic repression of PlevD-cat activity in TV-mannose-grown cells, likely due to the elevated expression of manL. On the other hand, when the ccpA mutation was introduced into a manL mutant strain, a reduction in PlevD-cat expression was seen only in TV-glucose and TV-fructose cultures, presumably related to enhanced expression of fruI. It is also important to note that the expression of levQRST was not altered by the ccpA mutation (Data not shown), so the observed effects of loss of CcpA are not due to down regulation of the primary activator of levD.
Table 5.
Effect of loss of CcpA on expression of PlevD-cat in the background of manL or fruI/fruCD mutants.
| PlevD-cat CAT specific activitya | ||||||
|---|---|---|---|---|---|---|
| Background | TV-glucose | SD | TV-fructose | SD | TV-mannose | SD |
| SD, standard deviation. | ||||||
| UA159 | 6.32 | 1.58 | 474.04 | 44.68 | 870.43 | 85.15 |
| fruI-/fruCD- | 79.09 | 0.40 | 787.48 | 21.84 | 681.99 | 11.12 |
| fruI-/fruCD-/ccpA- | 26.93 | 1.31 | 595.65 | 30.78 | 7.02 | 3.11 |
| manL- | 21.92 | 1.60 | 458.59 | 5.26 | 2199.88 | 85.68 |
| manL-/ccpA- | 1.52 | 0.25 | 34.21 | 9.18 | 1706.42 | 105.49 |
values represent averages of at least three separate experiments.
The role of the EII enzymes in catabolite repression is not restricted to the LevQRST regulon
A major question arising from these findings is whether the ability of ManL, FruI and LevD to exert catabolite control of gene expression is peculiar to the LevQRST regulon, or if this is a general mechanism for repression of genes for the catabolism of non-preferred carbohydrates by glucose, fructose or mannose. In a previously published microarray study (Abranches et al., 2006), a manL mutant strain (JAM1) showed markedly elevated expression of the cellobiose-PTS enzyme II component genes and celA, which encodes a 6-phospho-β-glucosidase that is capable of hydrolyzing β-glucosides, including cellobiose.
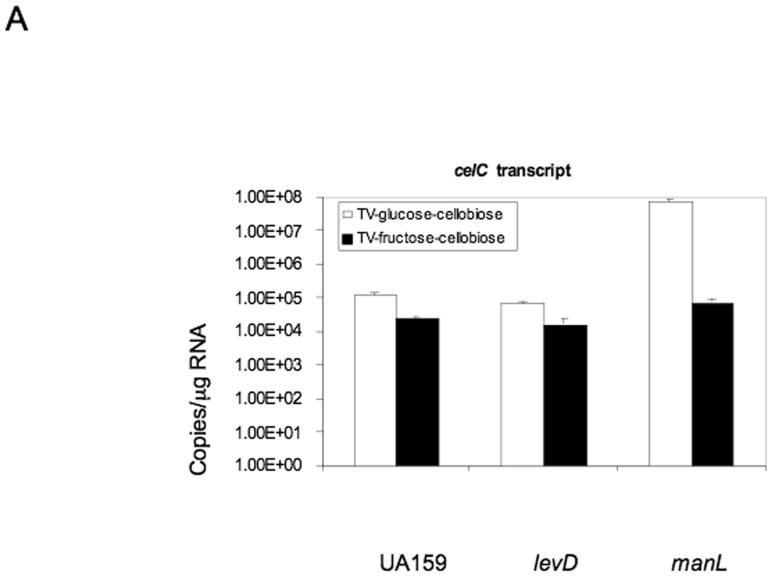
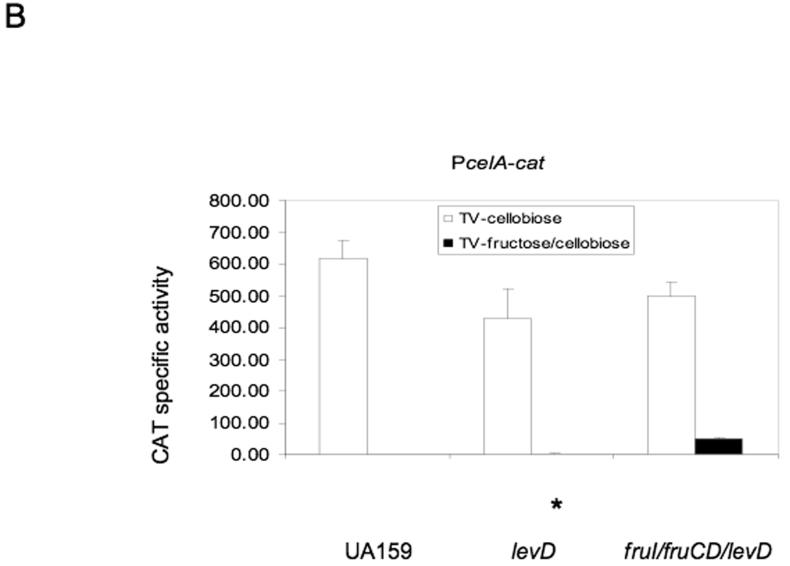
We measured mRNA levels of celC, the gene encoding the domain A of the cellobiose-PTS Enzyme II, in strain UA159 and in the manL and levD mutants. As shown in Fig. 4A, expression of celC was elevated nearly 1000-fold in the manL mutant growing in glucose-cellobiose medium when compared to the wild-type background, whereas only a 3-fold increase was seen in fructose-cellobiose medium. On the other hand, loss of levD showed little change in the level of celC transcript, regardless of whether glucose or fructose was present in the medium. As the utilization of cellobiose by S. mutans requires both cellobiose-PTS activity and a 6-phospho-β-glucosidase (celA), the level of celA expression was probed using a PcelA-cat fusion, constructed in a manner similar to that of the PlevD-cat fusion. As shown in Fig. 4B, inactivation of levD resulted in a slight, but significant, alleviation of expression of celA during growth in the presence of fructose and cellobiose, and only low levels of derepression of celA expression were noted in the strains lacking a single fructose-PTS enzyme, i.e., LevD, FruI or FruCD. However, a levD/fruI/fruCD triple mutant showed significantly increased celA expression compared to the wild-type background when fructose and cellobiose were present. Finally, the loss of ManL caused a dramatic elevation (∼150-fold) in transcription of the lactose utilization operon in S. mutans (data not shown), as measured by quantitative RT-PCR of the mRNA for the lacG gene (coding for phospho-β-galactosidase), adding further support to the idea that this permease participates in global control of CCR in S. mutans.
Figure 4.
The transcript level of celC measured by qRT-PCR (A) and the CAT activity of PcelA-cat (B) in strain UA159 and various mutants. The asterisk indicates P-values of less than 0.05.
We also tested manL and levD mutants in a diauxic growth assay using TV broth supplemented with 0.05% of glucose or fructose and 0.5% of cellobiose. Results of these tests were consistent with the effect of loss of manL or levD on the expression of the cel locus, i.e. the manL mutant showed faster growth on cellobiose than the wild-type strain when cultured in TV-glucose-cellobiose, whereas the levD mutant displayed non-diauxic growth in TV-fructose-cellobiose (see Fig. S5, supplementary material). Similar results were obtained when cellobiose was replaced with lactose as the non-preferred sugar substrate (Fig. S6).
Discussion
Our previous work on fruA and levDEFG (Wen and Burne, 2002; Zeng et al., 2006) has established that a four-component regulatory system, encoded by the levQRST operon immediately upstream of levD, is the primary induction mechanism for the expression of both fruA and levD. In addition to the complexity of induction of these genes, fruA has proven to be a particularly useful model for studying CCR in S. mutans. One longstanding enigma associated with CCR in S. mutans arose from the observations that deletion of ccpA rarely resulted in increased expression of genes that were sensitive to CCR (Griswold et al., 2006; Simpson and Russell, 1998; Wen and Burne, 2002). On the other hand, inactivation of manL did induce a loss of diauxic growth when S. mutans was cultivated on glucose-inulin medium (Abranches et al., 2003). It is now established that CcpA can control CCR of fruA through CRE binding (Abranches et al., 2003).
It was of interest that mannose was at least as effective an inducer of the LevQRST regulon as fructose. The reason why mannose would induce expression of the genes for a mannose transporter (LevDEFG) seems obvious, but the rationale for induction of the fruA operon by mannose is less clear. The fruA operon encodes a levanase/inulinase enzyme (FruA) and a protein with strong homology to known β-fructosidases (FruB), although an enzymatic activity has not been ascribed to FruB. We have not detected an ability of S. mutans UA159 to utilize mannans, and a strain lacking FruB behaves like the parental strain in terms of mannose utilization (Zeng and Burne, Unpublished). Frequently, carbohydrate transporters that internalize fructose also internalize mannose, so it seems that the conservation of the response to mannose by LevQRST could be due to constraints that govern carbohydrate recognition by the complex. Analysis of the carbohydrate recognition capabilities of the four-component signal transduction system are ongoing.
Mannose was also able to repress fruA at concentrations above 4 mM, similar to what was noted for the pattern of fruA induction with fructose (Abranches et al., 2008). Thus, it appears that CCR begins to affect expression of these operons when a readily metabolizable hexose is present at concentrations of about 3∼4 mM. Repression at this concentration could involve CcpA, since mannose enters the Embden-Meyerhoff pathway after isomerization from mannose-6-phosphate to fructose-6-phosphate. Subsequent phosphorylation would yield fructose-1,6-bisphosphate, which activates HPr kinase to produce HPr-SerPO4, which in turn stimulates binding of CcpA to CRE (Deutscher et al., 1995). As is demonstrated in this report, though, there are multiple CcpA-independent pathways for repression of the fruA and levD operons.
A central finding of this study is that inactivation of manL, fruI or levD, but not ptsG, relieved CCR of operons involved in use of non-preferred carbohydrate sources in a carbohydrate source-dependent manner. Based on these results, we propose that the substrate specificity of these permeases is a determining factor in their ability to negatively regulate catabolic operons. Specifically, the current evidence supports that FruI, FruCD, and LevD are mainly required for internalization of fructose, whereas ManL participates primarily in mannose and glucose transport (Abranches et al., 2003; Wen et al., 2001; Zeng et al., 2006). Since derepression of levD expression was seen in glucose cultures of all mutants tested, except ptsG, we postulate that glucose could be a common substrate for these EII enzymes. This idea is supported by enzymatic assays that show changes in the transport of glucose in mutants defective in these particular permeases (Abranches et al., 2003; Wen et al., 2001; Zeng et al., 2006).
A major question arising from this investigation is how the permeases elicit their effects on gene expression. The LevS sensor kinase, LevR response regulator and LevQT sugar binding proteins constitute the complete sensing complex of the LevQRST four-component system (Zeng et al., 2006). One possible mechanism for negative regulation of fruA and levD operon expression could involve direct interaction of the permeases with LevR. In fact, only a LevR-deficient strain showed a lack of response to the elimination of ManL, FruI or LevD, in cells grown in the presence of the cognate substrates for these permeases. Although this finding may be due to the profound lack of activation of the operon in the absence of LevR, strains lacking LevQ, LevS or LevT showed differential expression of fruA and levD depending on whether manL, levD or fruI mutations were present. Thus, these results could indicate that the permeases, directly or indirectly, affect the activity or phosphorylation state of LevR. The lack of an apparent PTS regulatory domain (PRD) (van Tilbeurgh and Declerck, 2001) in LevR suggests that the effects in this particular case may not involve phosphorylation. It should also be noted that there were different degress of derepression in the levS, levT and levQ mutants in response to the loss of ManL, FruI and LevD. Given that LevT and LevQ encode sugar binding proteins with substantial similarity to ABC transporters, the PTS permeases could fine tune the capacity of LevQST to efficiently sense inducers or phosphorylate LevR. Mechanisms for this modulation of activity could be similar to examples of PTS modification of the activity of non-PTS permeases with similarities to LevQ or LevT (Deutscher and Sauerwald, 1986; Romano et al., 1990). Thus, the simplest explanation for our data is that the dephosphorylated forms of the PTS permeases, which would be present if they are engaged in transport of their cognate sugar, act as direct effectors of CCR. In support of this idea, mutants that should have a constitutively phosphorylated ManL protein, by virtue of elimination of the EIICman or by mutagenesis of the histidine residue in the B domain of ManL, also show alleviation of CCR (Zeng and Burne, Unpublished). Although beyond the scope of this communication, we are in the early stages of experiments to assess the potential for direct modulation of LevR activity or phosphorylation state by EII enzymes, and to assess whether the permeases could impact LevS-dependent phosphorylation of LevR through the LevQT sugar binding proteins, by purifying and reconstituting these systems for in vitro analysis.
The model for direct interaction of the permeases as a mechanism for exerting CCR may also apply to the cellobiose or lactose operons. For example, dephosphorylated permeases could interfere with CelR or LacR activity, or the permeases may cause inducer exclusion by negatively affecting the transport of cellobiose or lactose by the PTS permeases that are specific for these sugars, required for induction and encoded within the cel and lac operons, respectively (Sondej et al., 1999). Alternatively, phosphotransfer from the permeases to the CelR or LacR regulators could influence DNA binding activity. Although CelR contains two conserved PRD domains, as well as an EIIA-like domain, LacR has no obvious potential phosphorylation sites. As above, in vitro reconstitution of these systems will be needed to clarify the underlying mechanisms. Given the heterogeneity of the systems involved, CCR by the permeases is likely to involve multiple mechanisms.
As an alternative to a direct influence of the permeases on regulatory proteins or transporters to effect CCR, it should be recognized that the phosphorylation state of the various permeases, which is dependent on the availability of their cognate substrates, could affect the phosphorylation state of Enzyme I or HPr. Similar to the Ser-46 phosphorylated form of HPr, HPr-His15-PO4 has been documented to have multiple regulatory functions, including inducer exclusion and modulation of the phosphorylation state of transcriptional regulators and antiterminators (Deutscher, 2008; Vadeboncoeur and Pelletier, 1997). Thus, the EI-HPr circuit could be the primary pathway that integrates the information from the PTS porters examined in this study. Specifically, direct phosphorylation by EI or HPr, or allosteric interaction of certain forms of HPr with components of the regulatory proteins or transporters, could explain the ability of multiple carbohydrates and permeases to control CCR in S. mutans. The strongest piece of data arguing against a model where the permeases influence expression through EI or HPr is the finding that the inactivation of ptsG, which encodes a maltose transporter, does not affect CCR even when cells are grown on maltose. Still, more information is needed to draw firm conclusions.
In B. subtilis and some other organisms it has been possible to evaluate the role of EI and HPr in CCR in vivo by using mutants with deletions or defects in the ptsH (HPr) or ptsI (EI) genes, but such an approach may not be possible with S. mutans. Although an EI truncation mutant of S. mutans has been isolated that was unable to accumulate sugars via the PTS system (Cvitkovitch et al., 1995), the mutant exhibited extremely slow growth and there were technical considerations that precluded use of such a mutant in these types of experiments. Specifically, the operons here are involved in catabolism or uptake of PTS sugars and, unlike B. subtilis, S. mutans lacks an electron transport chain for energy generation and depends entirely on sugar catabolism for growth. A variety of strategies to replace the entire coding sequence of the ptsI gene with an antibiotic resistance marker have yielded no viable mutants. Similar results were obtained when we tried to replace the HPr coding sequence using an antibiotic resistance gene cassette or to introduce a single copy of mutated versions of the ptsH gene. Therefore, if HPr or EI of S. mutans is directly involved in CCR, it may be necessary to demonstrate their involvement using in vitro systems or a heterologous host.
In conclusion, this study has begun to fill critical gaps in our understanding of the complex regulation of carbohydrate catabolism in S. mutans, a major human pathogen responsible for what is perhaps the most common infectious disease of man. The results presented herein have provided substantial insights into the molecular basis for CcpA-independent CCR and revealed that multiple PTS permeases are utilized by S. mutans in a complex network that appears to be structured to allow the organism to monitor the availability of specific carbohydrates using PTS permeases, while monitoring overall carbohydrate flow through the glycolytic pathway via HPr kinase and CcpA (Warner and Lolkema, 2003). Recently, studies with Gram-positive pathogens have revealed the involvement of some catabolic regulators in control of major virulence factors; for example the link between HPr-Ser-PO4 and virulence in Listeria monocytogenes and Streptococcus pyogenes (Deutscher et al., 2005), and a demonstration of a role for CcpA in virulence regulation in both Streptococcus pneumoniae (Giammarinaro and Paton, 2002) and S. pyogenes (Almengor et al., 2007; Shelburne et al., 2008). Thus, detailed analysis of CCR in S. mutans may have broad relevance to carbon-source dependent virulence regulation in other important human pathogens.
Experimental procedures
Bacterial strains, growth conditions, and reagents
E. coli strains were grown in Luria-Bertani (LB) medium supplemented, when needed, with antibiotics at the following concentrations: ampicillin (Ap, 100 µg ml−1), kanamycin (Km, 40 µg ml−1), erythromycin (Em, 500 µg ml−1), spectinomycin (Sp, 50 µg ml−1), and tetracycline (Tc, 10 µg ml−1). S. mutans UA159 and its derivatives were maintained in brain heart infusion broth (BHI, Difco Laboratories, Detroit, Mich.) at 37°C in 5% CO2 and 95% air with antibiotics added at the following concentrations: Km (700 µg ml−1), Em (5 µg ml−1), Sp (500 µg ml−1), and Tc (10 µg ml −1), when necessary. S. mutans cultures for CAT assay and growth rate tests were done in tryptone-vitamin-base (TV) medium (Burne et al., 1999) with 0.5% of glucose, fructose, or mannose as the carbohydrate source. All chemical reagents and antibiotics were obtained from Sigma Chemical Co. Growth curves of S. mutans UA159 and mutant strains were generated using a Bioscreen C (Oy Growth Curves AB Ltd., Helsinki, Finland), with readings taken every 30 minutes.
DNA manipulation
Standard recombinant DNA techniques were performed to engineer plasmids. All restriction and modifying enzymes were purchased from Invitrogen (Carlsbad, CA) or New England Biolabs (Beverly, Mass.) and used as recommended by the supplier. DNA purification was carried out using Qiaquick DNA purification kits purchased from Qiagen, Inc. (Valencia, CA). All primers (Supplementary Table S1) were synthesized by Integrated DNA Technologies (IDT), Inc. (Coralville, IA). The majority of the gene inactivations were carried out using the PCR-ligation-transformation technique, according to previously published protocols (Lau et al., 2002). Alternatively, constructs for gene inactivation were created by inserting antibiotic-resistance markers into S. mutans DNA cloned on an appropriate vector plasmid, and then used in transformation of S. mutans strains. All mutants were confirmed by PCR followed by DNA sequencing, including determining the sequence of the chromosomal content of the arms used for recombination to ensure no undesired mutations were inserted during PCR or cloning. In some cases, strains carrying multiple mutations were generated by transformation with chromosomal DNA isolated from other mutant strains of S. mutans, followed by selection for the appropriate antibiotic resistance and PCR confirmation.
Promoter-reporter gene fusions were constructed by inserting a promoter-containing sequence, including the cognate ribosome-binding site (RBS), into the integration vector plasmid pJL84 in front of a cat gene from Staphylococcus aureus that lacks a promoter and RBS (Zeng et al., 2006). DNA sequences flanking the promoter-cat fusion were derived from the mtlA-phnA locus, which serves as the integration site after transformation into S. mutans. All gene fusions were confirmed by sequencing before being used to transform S. mutans and the correct conformation of the integration cassette was confirmed by PCR.
CAT assay
Bacterial cells with various cat fusions were grown in TV-base medium, supplemented with various carbohydrates, to exponential phase (OD600 = 0.3 to 0.4 for fructose or glucose cultures, OD600 = 0.1 to 0.2 for mannose cultures), washed once with the same volume of 10 mM Tris buffer (pH 7.8), and then disrupted in a Bead Beater for 30 sec, twice, with a 2-min interval on ice. The cell lysates were centrifuged at 18,000 × g for 10 min and the supernates were recovered and used for measuring CAT activity by the method of Shaw (Shaw, 1979). The concentration of protein in the lysates was determined by the bicinchoninic acid (BCA) assay (Sigma). CAT activity was expressed as nmoles of chloramphenicol acetylated minute−1 × (mg protein)−1.
RNA isolation, RT-PCR and quantitative real-time RT-PCR
Total RNA was extracted from S. mutans cultures by using RNeasy mini kit (Qiagen) according to a protocol adapted for small scale purification (Ahn et al., 2005). Briefly, 10 ml of a mid-exponential phase bacterial culture was harvested, treated with RNAProtect reagent at room temperature for 5 min, and then resuspended in 250 µl of TE (50/10) buffer. A solution containing 5 µl of 20% SDS, 300 µl of acid phenol and 250 µl of glass beads was added to the cells, followed by 30 seconds of bead beating, twice, with a 5 min interval on ice. After 10 min of centrifugation at 14,000 rpm at 4°C, 200 µl of the supernatant fluid was removed from the tube and processed with the RNeasy mini kit, including an on-column DNase I (Qiagen) treatment, according to protocols provided with the kit.
For RT-PCR and quantitative real-time PCR, cDNA templates were generated from 2 µg of total RNA with random hexamers using the Superscript III first-strand synthesis system (Invitrogen) according to instructions from the supplier. At same time, a cDNA control was generated for each sample by omitting the reverse transcriptase in the RT reaction. For RT-PCR assays, specific primers were designed to amplify each DNA fragment overlapping two adjacent open-reading frames, with UA159 chromosomal DNA, cDNA control and cDNA test samples serving as the PCR templates. Real-time PCR reactions were carried out using an iCycler iQ real-time PCR detection system (Bio-Rad, Hercules, CA) and iQSYBR green supermix (Bio-Rad), according to the standard protocol provided by the supplier. Three individual cultures from each strain were used to prepare total RNA and cDNA, and triplicates were included in subsequent reactions for each cDNA sample, along with cDNA control and standard DNA generated with a PCR reaction using the specific primers for each target gene.
Acknowledgments
This work was supported by DE12236 from the National Institute of Dental and Craniofacial Research. Lin Zeng is supported by T32DE007200 training grant from Department of Health and Human Services.
Supplementary Material
References
- Abranches J, Chen YY, Burne RA. Characterization of Streptococcus mutans strains deficient in EIIAB Man of the sugar phosphotransferase system. Appl Environ Microbiol. 2003;69:4760–4769. doi: 10.1128/AEM.69.8.4760-4769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches J, Candella MM, Wen ZT, Baker HV, Burne RA. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J Bacteriol. 2006;188:3748–3756. doi: 10.1128/JB.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches J, Nascimento MM, Zeng L, Browngardt CM, Wen ZT, Rivera MF, Burne RA. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J Bacteriol. 2008;190:2340–2349. doi: 10.1128/JB.01237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SJ, Lemos JA, Burne RA. Role of HtrA in growth and competence of Streptococcus mutans UA159. J Bacteriol. 2005;187:3028–3038. doi: 10.1128/JB.187.9.3028-3038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, Pham VT. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J Bacteriol. 2007;189:5049–5059. doi: 10.1128/JB.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almengor AC, Kinkel TL, Day SJ, McIver KS. The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator Mga in the Group A streptococcus. J Bacteriol. 2007;189:8405–8416. doi: 10.1128/JB.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas JA. Virulence properties of Streptococcus mutans. Front Biosci. 2004;9:1267–1277. doi: 10.2741/1305. [DOI] [PubMed] [Google Scholar]
- Burne RA, Schilling K, Bowen WH, Yasbin RE. Expression, purification, and characterization of an exo-beta-D-fructosidase of Streptococcus mutans. J Bacteriol. 1987;169:4507–4517. doi: 10.1128/jb.169.10.4507-4517.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne RA, Penders JE. Characterization of the Streptococcus mutans GS-5 fruA gene encoding exo-beta-D-fructosidase. Infect Immun. 1992;60:4621–4632. doi: 10.1128/iai.60.11.4621-4632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne RA, Penders JE. Differential localization of the Streptococcus mutans GS-5 fructan hydrolase enzyme, FruA. FEMS Microbiol Lett. 1994;121:243–249. doi: 10.1111/j.1574-6968.1994.tb07105.x. [DOI] [PubMed] [Google Scholar]
- Burne RA, Chen YY, Wexler DL, Kuramitsu H, Bowen WH. Cariogenicity of Streptococcus mutans strains with defects in fructan metabolism assessed in a program-fed specific-pathogen-free rat model. J Dent Res. 1996;75:1572–1577. doi: 10.1177/00220345960750080801. [DOI] [PubMed] [Google Scholar]
- Burne RA, Wen ZT, Chen YY, Penders JE. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J Bacteriol. 1999;181:2863–2871. doi: 10.1128/jb.181.9.2863-2871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Hall TH, Burne RA. Streptococcus salivarius urease expression: involvement of the phosphoenolpyruvate:sugar phosphotransferase system. FEMS Microbiol Lett. 1998;165:117–122. doi: 10.1111/j.1574-6968.1998.tb13135.x. [DOI] [PubMed] [Google Scholar]
- Cvitkovitch DG, Boyd DA, Thevenot T, Hamilton IR. Glucose transport by a mutant of Streptococcus mutans unable to accumulate sugars via the phosphoenolpyruvate phosphotransferase system. J Bacteriol. 1995;177:2251–2258. doi: 10.1128/jb.177.9.2251-2258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Saier MH., Jr. ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc Natl Acad Sci U S A. 1983;80:6790–6794. doi: 10.1073/pnas.80.22.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Sauerwald H. Stimulation of dihydroxyacetone and glycerol kinase activity in Streptococcus faecalis by phosphoenolpyruvate-dependent phosphorylation catalyzed by enzyme I and HPr of the phosphotransferase system. J Bacteriol. 1986;166:829–836. doi: 10.1128/jb.166.3.829-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Reizer J, Fischer C, Galinier A, Saier MH, Jr., Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Kuster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Herro R, Bourand A, Mijakovic I, Poncet S. P-Ser-HPr--a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochim Biophys Acta. 2005;1754:118–125. doi: 10.1016/j.bbapap.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- Gauthier L, Bourassa S, Brochu D, Vadeboncoeur C. Control of sugar utilization in oral streptococci. Properties of phenotypically distinct 2-deoxyglucose-resistant mutants of Streptococcus salivarius. Oral Microbiol Immunol. 1990;5:352–359. doi: 10.1111/j.1399-302x.1990.tb00440.x. [DOI] [PubMed] [Google Scholar]
- Gauthier M, Brochu D, Eltis LD, Thomas S, Vadeboncoeur C. Replacement of isoleucine-47 by threonine in the HPr protein of Streptococcus salivarius abrogates the preferential metabolism of glucose and fructose over lactose and melibiose but does not prevent the phosphorylation of HPr on serine-46. Mol Microbiol. 1997;25:695–705. doi: 10.1046/j.1365-2958.1997.4981870.x. [DOI] [PubMed] [Google Scholar]
- Giammarinaro P, Paton JC. Role of RegM, a homologue of the catabolite repressor protein CcpA, in the virulence of Streptococcus pneumoniae. Infect Immun. 2002;70:5454–5461. doi: 10.1128/IAI.70.10.5454-5461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold AR, Jameson-Lee M, Burne RA. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J Bacteriol. 2006;188:834–841. doi: 10.1128/JB.188.3.834-841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramitsu HK. Molecular genetic analysis of the virulence of oral bacterial pathogens: an historical perspective. Crit Rev Oral Biol Med. 2003;14:331–344. doi: 10.1177/154411130301400504. [DOI] [PubMed] [Google Scholar]
- Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods. 2002;49:193–205. doi: 10.1016/s0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- Liberman ES, Bleiweis AS. Role of the phosphoenolpyruvate-dependent glucose phosphotransferase system of Streptococcus mutans GS5 in the regulation of lactose uptake. Infect Immun. 1984;43:536–542. doi: 10.1128/iai.43.2.536-542.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano AH, Saier MH, Jr., Harriott OT, Reizer J. Physiological studies on regulation of glycerol utilization by the phosphoenolpyruvate:sugar phosphotransferase system in Enterococcus faecalis. J Bacteriol. 1990;172:6741–6748. doi: 10.1128/jb.172.12.6741-6748.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw WV. Chloramphenicol acetyltransferase activity from chloramphenicol-resistant bacteria. Methods Enzymol. 1979;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Shelburne SA, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, Brennan RG, Musser JM. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci U S A. 2008;105:1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CL, Russell RR. Identification of a homolog of CcpA catabolite repressor protein in Streptococcus mutans. Infect Immun. 1998;66:2085–2092. doi: 10.1128/iai.66.5.2085-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondej M, Sun J, Seok YJ, Kaback HR, Peterkofsky A. Deduction of consensus binding sequences on proteins that bind IIAGlc of the phosphoenolpyruvate:sugar phosphotransferase system by cysteine scanning mutagenesis of Escherichia coli lactose permease. Proc Natl Acad Sci U S A. 1999;96:3525–3530. doi: 10.1073/pnas.96.7.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadeboncoeur C, Bourgeau G, Mayrand D, Trahan L. Control of sugar utilization in the oral bacteria Streptococcus salivarius and Streptococcus sanguis by the phosphoenolpyruvate: glucose phosphotransferase system. Arch Oral Biol. 1983;28:123–131. doi: 10.1016/0003-9969(83)90119-x. [DOI] [PubMed] [Google Scholar]
- Vadeboncoeur C, Pelletier M. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol Rev. 1997;19:187–207. doi: 10.1111/j.1574-6976.1997.tb00297.x. [DOI] [PubMed] [Google Scholar]
- van Tilbeurgh H, Declerck N. Structural insights into the regulation of bacterial signalling proteins containing PRDs. Curr Opin Struct Biol. 2001;11:685–693. doi: 10.1016/s0959-440x(01)00267-6. [DOI] [PubMed] [Google Scholar]
- Warner JB, Lolkema JS. CcpA-dependent carbon catabolite repression in bacteria. Microbiol Mol Biol Rev. 2003;67:475–490. doi: 10.1128/MMBR.67.4.475-490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AJ, Homer KA, Hosie AH. A phosphoenolpyruvate-dependent phosphotransferase system is the principal maltose transporter in Streptococcus mutans. J Bacteriol. 2007;189:3322–3327. doi: 10.1128/JB.01633-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen ZT, Browngardt C, Burne RA. Characterization of two operons that encode components of fructose-specific enzyme II of the sugar:phosphotransferase system of Streptococcus mutans. FEMS Microbiol Lett. 2001;205:337–342. doi: 10.1111/j.1574-6968.2001.tb10969.x. [DOI] [PubMed] [Google Scholar]
- Wen ZT, Burne RA. Analysis of cis- and trans-acting factors involved in regulation of the Streptococcus mutans fructanase gene (fruA) J Bacteriol. 2002;184:126–133. doi: 10.1128/JB.184.1.126-133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JJ, Minarcik J, Saier MH., Jr. Inducer expulsion and the occurrence of an HPr(Ser-P)-activated sugar-phosphate phosphatase in Enterococcus faecalis and Streptococcus pyogenes. Microbiology. 1996;142(Pt 3):585–592. doi: 10.1099/13500872-142-3-585. [DOI] [PubMed] [Google Scholar]
- Zeng L, Wen ZT, Burne RA. A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans. Mol Microbiol. 2006;62:187–200. doi: 10.1111/j.1365-2958.2006.05359.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.