Abstract
Staphylococcus aureus produces a virulence factor, protein A (SpA), that contains five homologous Ig-binding domains. The interactions of SpA with the Fab region of membrane-anchored Igs can stimulate a large fraction of B cells, contributing to lymphocyte clonal selection. To understand the molecular basis for this activity, we have solved the crystal structure of the complex between domain D of SpA and the Fab fragment of a human IgM antibody to 2.7-Å resolution. In the complex, helices II and III of domain D interact with the variable region of the Fab heavy chain (VH) through framework residues, without the involvement of the hypervariable regions implicated in antigen recognition. The contact residues are highly conserved in human VH3 antibodies but not in other families. The contact residues from domain D also are conserved among all SpA Ig-binding domains, suggesting that each could bind in a similar manner. Features of this interaction parallel those reported for staphylococcal enterotoxins that are superantigens for many T cells. The structural homology between Ig VH regions and the T-cell receptor Vβ regions facilitates their comparison, and both types of interactions involve lymphocyte receptor surface remote from the antigen binding site. However, T-cell superantigens reportedly interact through hydrogen bonds with T-cell receptor Vβ backbone atoms in a primary sequence-independent manner, whereas SpA relies on a sequence-restricted conformational binding with residue side chains, suggesting that this common bacterial pathogen has adopted distinct molecular recognition strategies for affecting large sets of B and T lymphocytes.
The common bacterial pathogen, Staphylococcus aureus, produces a 42-kDa factor, protein A (SpA), that contains five highly homologous extracellular Ig-binding domains in tandem, designated domains E, D, A, B, and C. Protein A, which exists in both secreted and membrane-associated forms, possesses two distinct Ig-binding activities: each domain can bind Fcγ (the constant region of IgG involved in effector functions) and Fab (the Ig fragment responsible for antigen recognition) (1). The Fcγ binding site has been localized to the elbow region at the CH2 and CH3 interface of most IgG subclasses, and this binding property has been extensively used for the labeling and purification of antibodies (2, 3). The Fab specificity is less well characterized but it has been shown to involve a site on the variable region of the Ig heavy chain (4). Correlation with antibody sequence usage indicates that the Fab binding specificity is restricted to products of the human variable region of the Fab heavy chain VH3 family that represent nearly half of inherited VH genes (5–8) and their homologues in other mammalian species (9, 10). Presumably through interactions with surface membrane-associated VH3-encoded B-cell antigen receptors (11), in vitro stimulation with SpA can contribute to selection of these B cells and promote their production of antibodies that may include rheumatoid factor autoantibodies (12, 13). In vivo exposure to recombinant SpA can result in supraclonal suppression and deletion of B lymphocytes that are susceptible based on their VH usage (14, 15).
Although the mechanism(s) are not defined, experimental models indicate that SpA enhances staphylococcal virulence (16, 17). Many features of the interactions of SpA with host B lymphocytes are akin to those of superantigens for T lymphocytes that cause a variety of inflammatory diseases including toxic shock syndrome, food poisoning, and exfoliative syndromes (18–20), and T-cell superantigens also have been postulated to contribute to the pathogenesis of autoimmune disease (18, 21). These superantigens target T-cell receptors (TcRs) from particular variable β chain (Vβ) families and induce global changes in T lymphocyte repertoires (18).
Here, we report the crystal structure of domain D of SpA complexed with the Fab fragment of a human IgM antibody and describe the key contact residues from both partners in the interaction. The residues in domain D involved in the interaction with Fab are highly conserved in other Ig-binding domains of SpA, and these are distinct from the residues that mediate the binding of an SpA domain and Fcγ (2). In the Fab, the residues in contact with SpA are located in the VH region framework β-strands and the interstrand loops most remote from the antigen combining site. The structure of the complex provides a rationale for the restricted specificity of SpA toward VH3-encoded antibodies, the largest human VH gene family. Hence, elucidation of the structural features of the binding interactions of SpA aids our understanding of the biological properties of a B-cell superantigen. In addition, structural comparisons with the characterized interactions of bacterial T-cell superantigens reveal how proteins produced by the same bacterial pathogen target the two limbs of the adaptive immune system.
Materials and Methods
Crystallization.
The Fab of the VH3–30/1.9III-encoded 2A2 IgM rheumatoid factor was produced by trypsin cleavage of the IgM secreted by a hybridoma created from synovial B cells of a rheumatoid arthritis patient, as described (22). The production of recombinant domain D of SpA used in crystallization was described in Roben et al. (23). In the crystallization screening, various ratios of Fab and domain D were tested, with subsequent optimization of ratios to improve crystal size. Crystals were grown by vapor diffusion at room temperature in sitting drops by mixing a reservoir solution of 21–24% (wt/wt) monomethyl polyethylene glycol 5,000, 100 mM sodium cacodylate, pH 6.5 with an equal volume of the protein solution at 5 mg/ml. Crystals for data collection were enlarged by using streak seeding followed by macroseeding (24). Data were recorded at room temperature from two crystals on a Rigaku rotating anode generator with Supper long mirrors by using a MarResearch image plate detector and processed by using the hkl package (25). These crystals belong to the monoclinic space group P21 with a = 68.5 Å, b = 78.9 Å, c = 163.2 Å, β = 100.7o. The data set used for structural analysis has 46,831 unique reflections in the 20- to 2.7-Å resolution with an overall Rmerge of 6.5% with 87% completeness and a 3.9-fold redundancy. In the 2.8- to 2.7-Å shell, completion is 77% with an <I/σ> of 1.9 and an Rmerge of 37%.
Structure Determination and Refinement.
The structure was solved by molecular replacement by using the program amore (26) and the coordinates of the uncomplexed 2A2 Fab-rheumatoid factor from a previously solved crystal structure (B.J.S., A.L.C., and M.J.T., unpublished work). In the resulting model, the three Fabs in the asymmetric unit had an Rfactor of 38.2% for data in the 15 Å to 4 Å range. After initial refinement of this model, σA-weighted 2Fo−Fc and Fo−Fc electron density maps were examined with the xtalview program (27), which clearly showed electron densities for domain D associated with two of the Fabs. Coordinates from all previously reported SpA domain structures were fitted globally into these densities, with the best fit demonstrated for domain E (28), which then was modified based on the sequence of domain D. For the third Fab, there is no electron density at the corresponding site, and crystal packing precludes the placement of a domain D molecule there.
Noncrystallographic symmetry restraints of 100 kcal mol−1⋅Å-2 were applied on main-chain atoms of the three Fab molecules, using the software package xplor (29). The progress of the refinement was judged by the decrease of Rfree after Powell minimization and temperature factor refinement. Initially, two B factors per residue were used in the refinement, whereas in the last cycles a single B factor per nonhydrogen atom was refined. The final Rwork and Rfree factors are 21.7% and 28.1%, respectively, and the structure displays standard stereochemistry as analyzed by procheck (30) with rms deviations (rmsds) from ideality of 0.008 Å on bond lengths and 1.59° on bond angles. Detectable domain D residues range from Phe-5 to Ala-56 (see Fig. 2A for numbering). A large portion of constant region of one Fab molecule in the asymmetric unit is poorly defined. However, the VH and VL regions, domain D molecules, and in particular the VH-domain D interface, are well defined in the electron density maps.
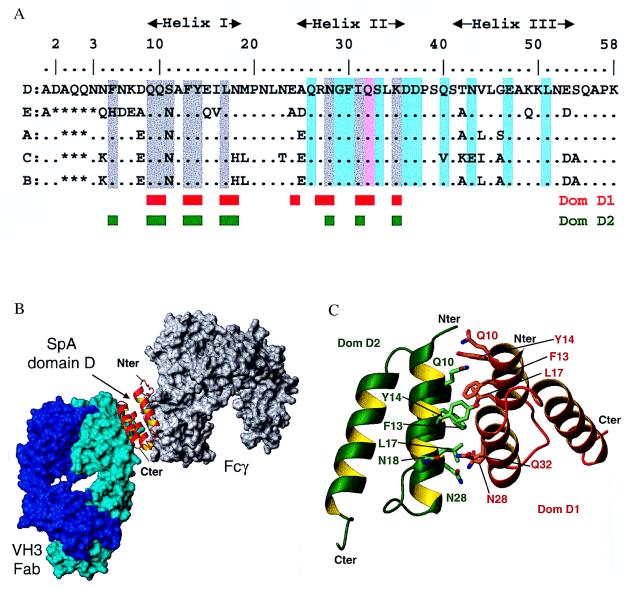
Figure 2.
Interactions of individual SpA domains. (A) Alignment of the amino acid sequences of the five SpA domains. Domain D residues involved in interaction with Fab 2A2 are highlighted in cyan. With the exception of Gln-32 (pink), there is no overlap between the residues involved in Fab interaction and those mediating Fcγ binding (2) (gray highlight). The engineered domain Z differs by the key mutation Gly-29 in Ala and does not bind Fab. The residues involved in the dimer of domain D observed in the asymmetric unit are indicated by red and green boxes. (B) Cross-linking of a VH3 Fab (cyan surface) and a Fcγ (gray surface) by a single domain of SpA (red ribbon). This model is based on the superposition of helix I and II of SpA domains in the Fab-domain D complex reported here and in the previously determined Fcγ-domain B complex (2) (rmsd of 0.73 Å for 140 backbone atoms). (C) Interface between domain D monomers. Schematic view of the interaction between the two domains D observed in the asymmetric unit, dom-D1 (red ribbon) and dom-D2 (green ribbon). Contact residues from both domains are shown in stick representation.
Results and Discussion
Overall Structure of the Fab-Domain D Complex.
The crystal form selected for analysis has two domain D-Fab complexes and one unliganded Fab in the asymmetric unit. The conformations of the variable domain heterodimers in the Fab are unchanged between the two complexed Fab with rmsd of 0.25 Å over 903 backbone atoms. Similarly, both complexed Fab superimpose well on the unliganded Fab with rmsd of 0.14 Å and 0.33 Å over 904 backbone atoms. The domain D assumes the triple α-helical bundle reported for domains B and E (28, 31). All three helices of domain D in the Fab-domain D complexes are clearly defined in electron density maps and superimpose well with the reported domain E structure (28) with rmsd of 0.52 Å over 196 main-chain atoms. For both complexes, the Fab-domain D interaction buries a total solvent accessible surface area of 1,220 Å2 with approximately equal contributions from both molecules, as determined with a 1.4-Å probe. This value is similar to many antigen-antibody complexes and other types of complexes with Ig-binding proteins but lower than the average calculated for reported protein–protein complexes (32).
In the complex, the Fab interacts with helix II and helix III of domain D via a surface composed of four VH region β-strands: B, C", D, and E (Fig. 1 A–C). The major axis of helix II of domain D is approximately 50o to the orientation of the strands, and the interhelical portion of domain D is most proximal to the C" strand. The site of interaction on Fab is remote from the Ig light chain and the heavy chain constant region. The interaction involves the following domain D residues: Gln-26, Gly-29, Phe-30, Gln-32, Ser-33, and Asp-36 of helix II; Asp-37 and Gln-40 in the loop between helix II and helix III; and Asn-43, Glu-47, and Leu-51 of helix III. In the Fab, the interaction is mediated by the heavy chain residues: Gly-H15 and Ser-H17 in the β turn before strand B; Arg-H19 of strand B; Lys-H57 and Tyr-H59 of strand C"; Lys-H64, Gly-H65, and Arg-H66 before strand D; Thr-H68 and Ser-H70 of strand D; Gln-H81 of strand E; Asn-H82a and Ser-H82b after strand E. Six of these residues are in framework region (FR) β strands, whereas the other seven residues are in VH region FR interstrand loops on the side farthest from the antigen binding pocket. Both interacting surfaces are composed predominantly of polar side chains, with three negatively charged residues on domain D and two positively charged residues on the 2A2 Fab buried by the interaction, providing an overall electrostatic attraction between the two molecules.
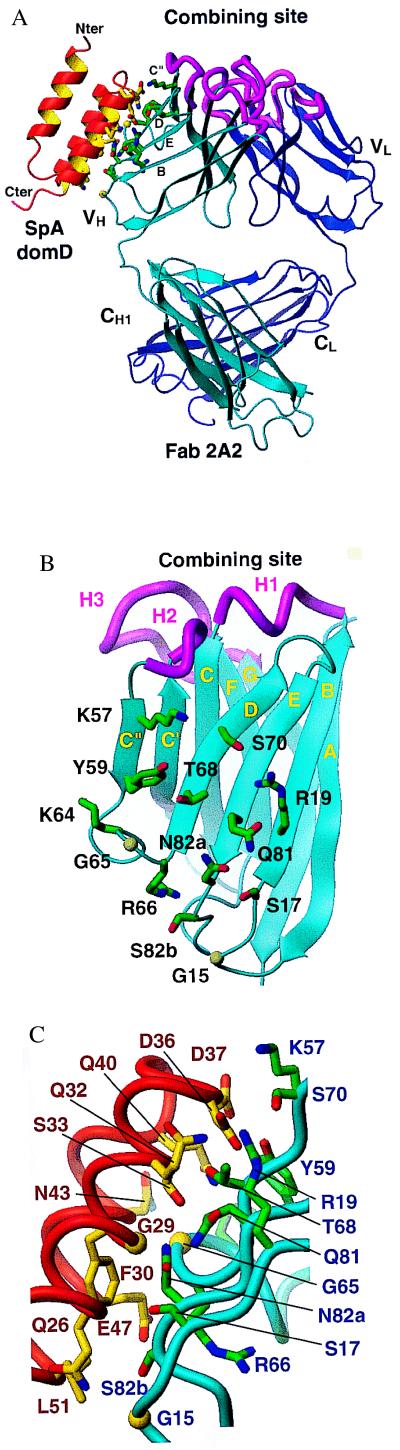
Figure 1.
Schematic representation of the complex between SpA domain D and Fab 2A2 from a human IgM. (A) Side view showing SpA domain D (red) bound to the framework region of the Fab heavy chain (cyan). The VL domain, which is not involved in this interaction, is shown in dark blue. The CDR loops as defined by Chothia and Lesk (33) are highlighted in magenta. (B) Ribbon representation of the VH region of Fab showing the positions of the residues that interact with domain D. (C) Schematic diagram detailing the residues of SpA domain D and Fab 2A2 involved in the interaction. Kabat numbering is used for the VH residues (blue); domain D is numbered (in brown) with the convention used for SpA domains (34). Contact residues are identified if 20 Å2 or more of their surface are buried in the interface and if they make at least one van der Waals contact. All figures were generated by using molmol (35).
Of the five polar interactions identified between Fab and domain D, three are between side chains. A salt bridge is formed between Arg-H19 and Asp-36 and two hydrogen bonds are made between Tyr-H59 and Asp-37 and between Asn-H82a and Ser-33. There are also two hydrogen bonds that are formed between main-chain atoms and side-chain atoms, namely Gly-H15 carbonyl with Gln-26, and Lys-H57 with Asp-36 carbonyl. Except for minor variations, all interactions are common to both complexes in the asymmetric unit. From the analysis of the complex structure it appears that sequence-restricted interactions dominate the contact surfaces between SpA and the VH region.
Domain D superposes well on domain E (28) with an rmsd of 0.52 Å over 196 main-chain atoms. The overall positions of the three helices are very similar with tilt angles of the helix I relative to the two parallel helices II and III of 15o. This interhelical angle is identical in domain Z, which was engineered from domain B, but different from domain B itself where it is 30o (31). However, this variation does not affect the relationship between helices II and III that are responsible for VH binding. Importantly, each of the other SpA domains shares 75–89% sequence identity with domain D, and there is a high conservation of all Fab interacting residues in domains E, A, and B (Fig. 2A). For domain C, Gln-40 is replaced by Val but this substitution appears conservative because at this site the alkyl chain mediates Fab contact, and the same can be said for the Asn-43 to Glu substitution. For the synthetic domain Z, the naturally occurring Gly-29 in domain B was mutated to Ala to improve the stability to hydroxylamine during purification procedures (36). In the solved Fab complex with domain D, the Cα of this glycine is less than 3.5 Å away from Gln-H81 and Asn-H82a, predicting that the Cβ of an alanine at this position would perturb the interaction between the molecules. These observations correlate well with the Fab-binding activities of the various domains, as the synthetic domain Z is devoid of this activity whereas all of the natural domains have similar Fab dissociation constants (Kd) of 0.4–5.0 × 10−6 M (23, 34, 37). Moreover, presumably because of avidity effects, the strength of the interaction between native pentameric SpA and native decavalent VH3 IgM is increased by 2–3 orders of magnitude (23).
Dual Reactivity of an Individual SpA Domain with Fcγ and Fab.
The site responsible for Fab binding is structurally separate from the domain surface that mediates Fcγ binding. As first demonstrated in a crystallographic complex (2) and recently reinvestigated in NMR studies (38), the interaction of Fcγ with domain B primarily involves residues in helix I with lesser involvement of helix II (Fig. 2A). With the exception of the Gln-32, a minor contact in both complexes, none of the residues that mediate the Fcγ interaction are involved in Fab binding. The area buried in the Fcγ-domain B interface is 1,320 Å2, which is comparable to the 1,220 Å2 buried in the current complex with Fab. However, the nature of these buried SpA residues differs significantly, as the Fab binding is dominated by polar contacts whereas the Fcγ interaction is predominantly hydrophobic.
To examine the spatial relationship between these different Ig-binding sites, we superposed the SpA domains in these complexes to construct a model of a complex between an Fab, an SpA domain, and an Fcγ molecule. In this ternary model, we found an rmsd of 0.73 Å for the backbone atoms in helix I and helix II of the SpA domains (Fig. 2B). Here, the Fab and Fcγ form a sandwich about opposite faces of the helix II without evidence of steric hindrance of either interaction. These findings illustrate how, despite its small size (i.e., 56–61 aa), an SpA domain can simultaneously display both activities, explaining experimental evidence that the interactions of Fcγ and Fab with an individual domain are noncompetitive (23, 39).
Interaction Between Domain D Monomers.
In the crystallographic structure, the two molecules of domain D, designated domD-1 and domD-2, interact with one another to form an asymmetric dimer (Fig. 2C). They are related by a rotation of 148o with an interface of 1,300 Å2 composed of equal contributions from each SpA domain. In both domD-1 and domD-2, this interaction involves the same 9 aa (Gln-9, Gln-10, Phe-13, Tyr-14, Leu-17, Asn-18, Asn-28, Ile-31, and Lys-35) that contribute 90% of the buried surface. Completing this interaction are Glu-24, Arg-27, and Gln-32 from domD-1 and Phe-5 from domD-2. These interactions are formed asymmetrically, and further asymmetry is seen in the hydrogen bond between the side chains of Gln-32 of domD-1 and Asn-18 of domD-2, and even in the hydrogen bond between the Asn-28 from both domains.
All hydrophobic and aromatic residues implicated in Fcγ binding are also involved in the van der Waals interactions in the domain D dimer (Fig. 2A). The surface areas buried in these two types of interactions are also comparable. Although only two hydrogen bonds are formed in the dimer, there are four hydrogen bonds in the interaction of domain B and Fcγ (2), suggesting that under certain conditions the latter interaction may be favored. However, we believe that this dimer form is unlikely to represent a random relationship, as the dimensions of the associated interface falls in the upper 3% of what can be considered random crystal contacts (40). Recent NMR studies strengthen our hypothesis, as weak homodimer interactions have been noted for domain B with the involvement of residues from helix I and II (41), which is in good agreement with the crystallographic dimer. Considering that the residues involved in this dimerization are all conserved in each of the five extracellular domains of SpA, should such an interaction occur between the proximal domains of membrane-associated molecules of native SpA, the functional valency for binding of lymphocyte membrane-associated B-cell antigen receptor potentially could be enhanced. The biological relevance of this postulated interaction merits further investigation.
Accessibility of the Antigen Combining Site in the Fab-Domain D Complex.
The crystal structure of the Fab-domain D complex accounts for functional evidence that SpA does not compete with antigen binding (42), as the VH binding surface does not involve the complementarity determining region (CDR) loops that mediate binding of conventional antigens (33, 43). Although two of the VH residues involved in the interaction, Lys-H57 and Tyr-H59, originally were assigned to CDR2 based on primary sequence hypervariability comparisons, these residues are not part of loop H2 and are in fact part of the C" strand (33) with side chains pointing away from the combining site (Fig. 1A). Moreover, in the complex, domain D is more than 10 Å away from VH CDR3, the loop most commonly contributing contacts for antigen recognition and specificity, which is consistent with the lack of correlation between SpA binding activity and CDR3 sequence (7, 8). Based on superposition of the Fab-domain D complex onto reported Fab-antigen complexes, and based on the observation that complexed and uncomplexed Fab molecules in the crystal have identical conformations, it is unlikely that the binding of domain D would alter the combining site or result in steric hindrance so that it would prevent the binding of an antigen, including very large ones (data not shown). Furthermore, because of the spatial relationships between the termini of each of the domains (Fig. 1A) and their associated VH, even binding of intact five-domain SpA would appear unlikely to interfere with antigen binding. We therefore conclude that the manner by which SpA is bound to the VH does not disturb Fab conformation and does not appear to affect accessibility to the antigen binding site.
VH Specificity of the SpA Binding Interaction.
The structure of the VH site responsible for SpA binding explains the high frequency of these binding interactions in the human immune system. About half of a panel of human IgM, and 32–54% of human peripheral blood B cells, have been reported to interact with SpA (5, 8, 44, 45). Among human antibodies studied to date, SpA binding has been restricted to products of the VH3 gene family with no examples from any of the other six families (5–8). Among the 13 VH residues implicated as SpA contacts, VH3 genes include frequent germ-line sequence variations only at position H57 (Table 1 and Fig. 1B). This is not a core residue in the interface, and Lys, Ile, and Thr are permissive at this position (7, 8). The other 12 VH residues in direct contact with domain D are highly conserved in members of this family. In fact, in the 22 potentially functional human VH3 gene segments, there are only two germ-line variations (the conservative change of Lys at H19 in V3–73 and the nonconservative change of Gly at H82a in V3–64) but each of these genes are the source of less than 2% of adult expressed VH3 Ig repertoire (E. Milner, personal communication). In the mouse, homologous genes in the S107, J606, 7183, and VH10/DNA4 families also commonly encode for SpA binding (9, 10). With the exception of the conservative H19 Lys substitution (15), contact residues also are conserved in the murine homologues. Some of these related murine VH genes, including certain 7183 genes, contain a Ser or Thr substitution at H82a that correlates with weaker SpA binding activity. VH10/DNA4 genes also are associated with low binding activity and include an Asp at H65. By comparison, inherited genes of the other nonbinding human and murine VH families each contain two or more residue differences at the 13 VH positions identified as SpA contacts (15). In particular, for the two other large human families, VH1 and VH4, there are nonconservative differences at position H82a (Table 1). VH4 genes also have a nonconservative change at H19. From a structural point of view, a core of critical residues can be defined, consisting of seven residues: Arg/Lys-H19, Gly-H65, Arg-H66, Thr-H68, Ser-H70, Gln-H81, and Asn-H82a (Fig. 1B). Apart from the essential salt bridge formed at position H19, all other residues are virtually inaccessible to solvent, making their replacement by a larger residue highly disruptive. Replacement by a smaller residue, as for Asn-H82a by Ser, is likely to lead to weaker binding. Hence from such considerations, we conclude that this core of seven VH residues constitutes the structural motif for SpA binding, and this conveys the restricted specificity for VH3-encoded Ig and their homologues.
Table 1.
Sequences of human VH gene families at the positions involved in the interface with domain D
| H15 | H17 | H19 | H57 | H59 | H64 | H65 | H66 | H68 | H70 | H81 | H82a | H82b | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VH1 (11) | G | S, T | K | T, A | Y | Q | G, D, E | R | T | T | E | S, R | S, R |
| VH2 (3) | T | T | T | K | Y | K | S, T | R | T | T, S | T | T | N |
| VH3 (22) | G | S | R, K | K, I, T | Y | K | G | R | T | S | Q | N, G | S |
| VH4 (11) | S | T | S | T | Y | K | S | R | T | S | K | S | S |
| VH5 (2) | G | S | K, R | T | Y | Q | G | Q, H | T | S | Q | S | S |
| VH6 (1) | S | T | S | N | Y | K | S | R | T | N | Q | N | S |
| VH7 (1) | G | S | K | P | Y | T | G | R | V | S | Q | C | S |
The numbers in parentheses for each VH group are the numbers of human germ-line V gene sequences reported in the V Base database (Medical Research Council, Centre for Protein Engineering). The VH3 family represents nearly half of all inherited human VH gene segments. Amino acids in italics are represented in less than 10% of functional inherited human genes.
The structure offers the possibility of rationalizing the influence of somatic hypermutations in VH3-encoded IgM upon binding reactivity with SpA. Whereas all tested VH3-germ-line-encoded IgM were found to be reactive with SpA (8), about 20% of adult VH3 IgM and a greater proportion of VH3 IgG lack this reactivity (5–7). In almost every case, the loss of binding activity now can be correlated with identifiable nonconservative mutations at one or more of the VH positions at the interface (data not shown). Although some somatic mutations can be tolerated at position H57, the inversion of charge from Lys to Glu (46) results in loss of activity because of electrostatic repulsion. Although the change from Asn to Ser at position H82a (47) can be considered conservative, this position is part of the core and hence even such a minor change leads to reduced activity.
Structural Comparisons with the Interactions of T-Cell Superantigens.
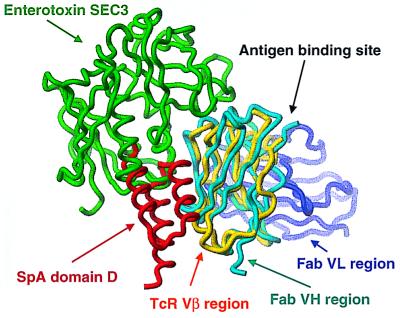
The capacity of SpA to bind a large proportion of the human immune repertoire, expressed either as receptors on B-cells (i.e., B-cell antigen receptor) or in soluble Ig form, has parallels with the abilities of Staphylococcus aureus enterotoxins and other T-cell superantigens to interact with a large repertoire of T lymphocytes through certain TcR Vβ. By virtue of their strong structural similarities, the VH of Fab 2A2 bound to domain D can be superposed onto the Vβ of TcR complexed with the Staphylococcus aureus enterotoxin, SEC3, (48) with an rmsd of 1.6 Å over 393 main-chain atoms (Fig. 3). Although in both cases they are required for antigen recognition, the partner chains in these heterodimeric receptors, the VL of the B-cell antigen receptor and Vα of the TcR, do not directly interact with domain D or SEC3, respectively. Overall, SEC3 is positioned closer to the antigen binding site of the TcR. Only about 25% of the Vβ chain contacts derive from the β-strands, whereas the remainder are from the CDR1, CDR2, and HV4 loops. For characterized staphylococcal T-cell superantigens (21, 48, 49), the interaction with the TcR also differs radically from the domain D-Fab complex as it is mediated primarily through hydrogen bonds with the Vβ region backbone atoms that are contacted in a defined conformationally sensitive distribution. Because this interaction is permissive of amino acid side-chain variations, the promiscuity of these T-cell superantigens enables the targeting of the cellular products of different TcR Vβ families, each of which generally include only 1–2 Vβ genes. In contrast, the SpA-Fab interaction clearly relies on a sequence-restricted binding mode with a structural motif presented by the side chains of highly conserved VH residues. Because these residues are part of a large set of VH genes that are highly expressed in the immune system, SpA targets a large proportion of the B-cell pool.
Figure 3.
Comparison of the interactions of B-cell and T-cell superantigens. The Vβ region (yellow) of the TcR superimposed well on VH (cyan) of Fab 2A2 with an rmsd of 1.6 Å over 393 main-chain atoms. The T-cell superantigen, enterotoxin SEC3, (green) binds to the CDR1, CDR2, and HV4 loops of the Vβ region. SpA (red) binds at framework region 1 and 3, and the carboxyl-terminal portion of the CDR2 (including positions H57 and H59 in the C" strand) of the VH region.
In conclusion, our studies demonstrate that by exploiting a conformational surface on the antigen receptor well represented in the repertoire, a bacterial virulence factor targets host B lymphocytes and their soluble Ig products. Staphylococcus aureus has developed proteins that interact with analogous but not fully equivalent sites in B-cell and T-cell receptors. Therefore, despite prominent differences in the molecular basis of these interactions, they both act to stimulate large proportions of the host's B cells or T cells, resulting in subversion of the immune system.
Acknowledgments
We thank M. Léonetti for helpful discussions and help in the preparation of the manuscript, M. Hamon and G. Konfortova for cell culture, D. Beale for purification of 2A2 IgM and preparation of the Fab fragment, and L. Luo for protein expression and purification. The 2A2 cell line was kindly provided by Prof. R. Maini (Kennedy Institute, London). We thank M.-H. LeDu and M. Knossow for reading of the manuscript, W. Shepard and A. Bentley for assistance at the Laboratoire pour l'Utilisation du Rayonnement Electromagnétique synchrotron facility (Orsay), and A. Ménez for continuous support. B.J.S. and M.J.T. acknowledge support from the Arthritis Research Campaign (U.K.) and are also supported by the Biotechnology and Biological Sciences Research Council (U.K.). G.J.S. was supported by National Institutes of Health Grants R01-AI40305, 5P60-AR40770, and R03-AI46637–01, and a Biomedical Sciences Award from the Arthritis Foundation.
Abbreviations
- SpA
Staphylococcus aureus protein A
- TcR
T-cell receptor
- Fcγ
constant region of IgG
- VH and Vβ
variable region of the Fab heavy chain and TcR β chain, respectively
- CDR
complementarity determining region
- rmsd
rms deviation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1DEE).
References
- 1.Boyle M D P. In: Bacterial Immunoglobulin-Binding Proteins. Boyle M P D, editor. Vol. 1. San Diego: Academic; 1990. pp. 17–28. [Google Scholar]
- 2.Deisenhofer J. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 3.Tashiro M, Montelione G T. Curr Opin Struct Biol. 1995;5:471–481. doi: 10.1016/0959-440x(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 4.Vidal M A, Conde F P. J Immunol. 1985;135:1232–1238. [PubMed] [Google Scholar]
- 5.Sasso E H, Silverman G J, Mannik M. J Immunol. 1989;142:2778–2783. [PubMed] [Google Scholar]
- 6.Sasso E H, Silverman G J, Mannik M. J Immunol. 1991;147:1877–1883. [PubMed] [Google Scholar]
- 7.Sasano M, Burton D R, Silverman G J. J Immunol. 1993;151:5822–5839. [PubMed] [Google Scholar]
- 8.Hillson J L, Karr N S, Oppliger I R, Mannik M, Sasso E H. J Exp Med. 1993;178:331–336. doi: 10.1084/jem.178.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seppala I, Kaartinen M, Ibrahim S, Makela O. J Immunol. 1990;145:2989–2993. [PubMed] [Google Scholar]
- 10.Cary S, Krishnan M R, Marion T, Silverman G J. Mol Immunol. 1999;36:769–776. doi: 10.1016/s0161-5890(99)00085-1. [DOI] [PubMed] [Google Scholar]
- 11.Romagnani S, Giudizi M G, del Prete G, Maggi E, Biagiotti R, Almerigogna F, Ricci M. J Immunol. 1982;129:596–602. [PubMed] [Google Scholar]
- 12.Kristiansen S V, Pascual V, Lipsky P E. J Immunol. 1994;153:2974–2982. [PubMed] [Google Scholar]
- 13.Kozlowski L M, Kunning S R, Zheng Y, Wheatley L M, Levinson A I. J Clin Immunol. 1995;15:145–151. doi: 10.1007/BF01543106. [DOI] [PubMed] [Google Scholar]
- 14.Silverman G J, Nayak J V, Warnatz K, Hajjar F F, Cary S, Tighe H, Curtiss V E. J Immunol. 1998;161:5720–5732. [PubMed] [Google Scholar]
- 15.Cary S, Lee J, Wagenknecht R, Silverman G J. J Immunol. 2000;164:4730–4741. doi: 10.4049/jimmunol.164.9.4730. [DOI] [PubMed] [Google Scholar]
- 16.Foster T J, O'Reilly M, Patel A H, Bramley A J. Antonie Van Leeuwenhoek. 1988;54:475–482. doi: 10.1007/BF00461866. [DOI] [PubMed] [Google Scholar]
- 17.Patel A H, Nowlan P, Weavers E D, Foster T. Infect Immun. 1987;55:3103–3110. doi: 10.1128/iai.55.12.3103-3110.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotzin B L, Leung D Y, Kappler J, Marrack P. Adv Immunol. 1993;54:99–166. doi: 10.1016/s0065-2776(08)60534-9. [DOI] [PubMed] [Google Scholar]
- 19.Bohach G A, Fast D J, Nelson R D, Schlievert P M. Crit Rev Microbiol. 1990;17:251–272. doi: 10.3109/10408419009105728. [DOI] [PubMed] [Google Scholar]
- 20.Papageorgiou A C, Tranter H S, Acharya K R. J Mol Biol. 1998;277:61–79. doi: 10.1006/jmbi.1997.1577. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Llera A, Malchiodi E L, Mariuzza R A. Annu Rev Immunol. 1999;17:435–466. doi: 10.1146/annurev.immunol.17.1.435. [DOI] [PubMed] [Google Scholar]
- 22.Sohi M K, Sutton B J, Corper A L, Wan T, Maini R N, Brown C, Rijnders T, Beale D, Feinstein A, Humphreys A S, Taussig M J. J Mol Biol. 1994;242:706–708. doi: 10.1006/jmbi.1994.1620. [DOI] [PubMed] [Google Scholar]
- 23.Roben P W, Salem A N, Silverman G J. J Immunol. 1995;154:6437–6445. [PubMed] [Google Scholar]
- 24.Stura E A, Wilson I A. J Crystallogr Growth. 1991;110:270–282. [Google Scholar]
- 25.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 27.McRee D E. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 28.Starovasnik M A, Skelton N J, O'Connell M P, Kelley R F, Reilly D, Fairbrother W J. Biochemistry. 1996;35:15558–15569. doi: 10.1021/bi961409x. [DOI] [PubMed] [Google Scholar]
- 29.Brünger A T. x-plorManual. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 30.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:238–291. [Google Scholar]
- 31.Gouda H, Torigoe H, Saito A, Sato M, Arata Y, Shimada I. Biochemistry. 1992;31:9665–9672. doi: 10.1021/bi00155a020. [DOI] [PubMed] [Google Scholar]
- 32.Conte L L, Chothia C, Janin J. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 33.Chothia C, Lesk A M. J Mol Biol. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- 34.Jansson B, Uhlen M, Nygren P A. FEMS Immunol Med Microbiol. 1998;20:69–78. doi: 10.1111/j.1574-695X.1998.tb01112.x. [DOI] [PubMed] [Google Scholar]
- 35.Koradi R, Billeter M, Wuthrich K. J Mol Graphics. 1996;14:29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 36.Nilsson B, Moks T, Jansson B, Abrahmsen L, Elmblad A, Holmgren E, Henrichson C, Jones T A, Uhlen M. Protein Eng. 1987;1:107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- 37.Silverman G J, Pires R, Bouvet J P. J Immunol. 1996;157:4496–4502. [PubMed] [Google Scholar]
- 38.Gouda H, Shiraishi M, Takahashi H, Kato K, Torigoe H, Arata Y, Shimada I. Biochemistry. 1998;37:129–136. doi: 10.1021/bi970923f. [DOI] [PubMed] [Google Scholar]
- 39.Starovasnik M A, O'Connell M P, Fairbrother W J, Kelley R F. Protein Sci. 1999;8:1423–1431. doi: 10.1110/ps.8.7.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janin J. Nat Struct Biol. 1997;12:673–674. doi: 10.1038/nsb1297-973. [DOI] [PubMed] [Google Scholar]
- 41.Karimi A, Matsumura M, Wright P E, Dyson H J. J Pept Res. 1999;54:344–352. doi: 10.1034/j.1399-3011.1999.00115.x. [DOI] [PubMed] [Google Scholar]
- 42.Young W W, Jr, Tamura Y, Wolock D M, Fox J W. J Immunol. 1984;133:3163–3166. [PubMed] [Google Scholar]
- 43.Wilson I A, Stanfield R L. Curr Opin Struct Biol. 1994;4:857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 44.Hakoda M, Kamatani N, Hayashimoto-Kurumada S, Silverman G J, Yamanaka H, Terai C, Kashiwazaki S. J Immunol. 1996;157:2976–2981. [PubMed] [Google Scholar]
- 45.Silverman G J, Sasano M, Wormsley S B. J Immunol. 1993;151:5840–5855. [PubMed] [Google Scholar]
- 46.Randen I, Potter K N, Li Y, Thompson K M, Pascual V, Forre O, Natvig J B, Capra J D. Eur J Immunol. 1993;23:2682–2686. doi: 10.1002/eji.1830231044. [DOI] [PubMed] [Google Scholar]
- 47.Zhang M, Majid A, Bardwell P, Vee C, Davidson A. J Immunol. 1998;161:2284–2289. [PubMed] [Google Scholar]
- 48.Fields B A, Malchiodi E L, Li H, Ysern X, Stauffacher C V, Schlievert P M, Karjalainen K, Mariuzza R A. Nature (London) 1996;384:188–192. doi: 10.1038/384188a0. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Llera A, Tsuchiya D, Leder L, Ysern X, Schlievert P M, Karjalainen K, Mariuzza R A. Immunity. 1998;9:807–816. doi: 10.1016/s1074-7613(00)80646-9. [DOI] [PubMed] [Google Scholar]