Abstract
OBJECTIVE—Humans with functional variants in the melanocortin 4 receptor (MC4R) are obese, hyperphagic, and hyperinsulinemic but have been reported to have no difference in energy expenditure.
RESEARCH DESIGN AND METHODS—We investigated the association of two MC4R variants, Arg165Gln (R165Q) and A insertion at nucleotide 100 (NT100), with adiposity in 3,074 full-heritage Pima Indians, a subset of whom had metabolic measures including 24-h energy expenditure (n = 252) and resting metabolic rate (RMR) (n = 364).
RESULTS—Among the 3,074 subjects, 43 were heterozygous for R165Q and 14 for NT100 (frequency = 0.007 and 0.002). Mean (± SD) BMI was higher among subjects with R165Q (39.3 ± 8.6 kg/m2) or NT100 (41.2 ± 7.8) than subjects without either variant (37.1 ± 8.4) (P = 0.04 and 0.02, adjusted for age, sex, and birth year and accounting for family membership). The 24-h energy expenditure (four with NT100; three with R165Q) or RMR (six with NT100; two with R165Q) was lower in heterozygous subjects but only met statistical significance when heterozygous subjects were combined and compared with subjects without either variant: least-squares means, 2,163 kcal/24 h (95% CI 2,035–2,291) vs. 2,307 kcal/24 h (2,285–2,328), P = 0.03 for 24-h energy expenditure, and 1,617 kcal/24 h (1,499–1,734) vs. 1,754 kcal/24 h (1,736–1,772), P = 0.02 for RMR; adjusted for age, sex, fat-free mass, and fat mass). For RMR, this difference persisted, even after accounting for family membership.
CONCLUSIONS—Pima Indians heterozygous for R165Q or NT100 in MC4R have higher BMIs and lower energy expenditure (by ∼140 kcal/day), indicating that lower energy expenditure was a component of the increased adiposity.
Environment plays an important role in the development of obesity, but considerable evidence exists for a genetic contribution to body weight (1). Several genes including the melanocortin 4 receptor (MC4R) located on chromosome 18q22 have been identified as monogenic causes of obesity in humans (2). Many of the identified variants in MC4R lead to partial or complete loss of receptor activity (3). Mice lacking MC4R are obese, are hyperinsulinemic, have increased food intake (4), and have lower energy expenditure (5). Humans with MC4R mutations are obese, are hyperinsulinemic, and have increased food intake, but no difference in energy expenditure has been reported (3).
The Pima Indians of southern Arizona have a high prevalence of obesity (6). Sequencing of the coding region of MC4R in 300 severely obese and 126 nonobese Pima Indians identified 10 individuals (all obese) heterozygous for a previously characterized G-to-A substitution at nucleotide 165 (R165Q) that leads to partial inactivation of MC4R (3). Three additional individuals (all obese) were found to be heterozygous for a novel single-base insertion (A) at nucleotide 100 (NT100) that predicts a frameshift resulting in a premature STOP (TGA) at codon 37 (7). A premature STOP codon at this position would produce a truncated protein lacking critical ligand binding and transmembrane domains (3). Previously described variants (V103I and I251L) found to be protective against obesity (8,9) were not identified in this population. In the current study, we investigated the prevalence of the MC4R functional missense R165Q and the frameshift mutation in a population-based survey of full-heritage Pima Indians, and their association with BMI and in vivo measures of energy expenditure and insulin action.
RESEARCH DESIGN AND METHODS
Every 2 years since 1965, members of the Gila River Indian Community age ≥5 years are invited to participate in a longitudinal study of diabetes and its complications. This population is dynamic in that individuals may enter and leave over the course of the study. After gaining informed consent, each participant undergoes an examination, including measurements of height and weight and a 75-g oral glucose tolerance test. Diabetes was diagnosed according to 1999 World Health Organization criteria (10). DNA from 3,074 full-heritage Pima Indians from 1,676 sibships was used for genotyping in the present study. An individual's highest recorded BMI (i.e., maximum BMI) during the longitudinal study was used to compare genotypes regardless of diabetes status. Maximum BMI was chosen to attempt to fully capture the propensity to increased adiposity. For all subjects, 59% were female, age (mean ± SD) was 35.4 ± 13.0 years, the mean maximum BMI was 37.2 ± 8.5 kg/m2, and 36% had type 2 diabetes. We also analyzed the subset of individuals for whom we had BMI measurements before diabetes development and identified the maximum BMI from among the nondiabetic exams. This group consisted of 2,603 individuals (mean age 34.6 ± 12.7 years, 58% female, mean maximum BMI 37.3 ± 8.6 kg/m2).
Clinical studies.
Volunteers from the Gila River Indian Community also participate in inpatient studies examining the pathophysiology of type 2 diabetes and obesity. After gaining informed consent, volunteers are admitted to the clinical research unit and are fed a weight-maintaining diet (calories from carbohydrate, fat, and protein 50, 30, and 20%, respectively) and abstain from strenuous exercise. After 3 days, volunteers undergo a 75-g oral glucose tolerance test (after a 12-h overnight fast). Nondiabetic volunteers then undergo tests to assess body composition, insulin action in vivo, and energy expenditure. All studies were approved by the Institutional Review Board of the National Institute of Diabetes and Digestive and Kidney Diseases.
Body composition measurements.
Body composition was measured by underwater weighing with simultaneous determination of residual lung volume by helium dilution (11) or by total-body dual-energy X-ray absorptiometry (DPX-L Lunar Radiation, Madison, WI) (12). Percent body fat, fat-free mass (FFM), and fat mass were calculated; measurements using the two different methods were made comparable using a previously derived equation (13).
Measurement of energy expenditure
24-h respiratory chamber.
The measurement of energy expenditure in the respiratory chamber has previously been described (14). Briefly, volunteers entered the chamber at 0745 h after an overnight fast and remained therein for 23 h. Meals were provided at 0800, 1130, 1700, and 2000 h (evening snack). Because of the confinement within the chamber, only 80% of the calories from the weight-maintaining diet on the metabolic ward were provided in the respiratory chamber. Fresh air was drawn through the chamber, and CO2 production and O2 consumption were measured and calculated every 15 min and extrapolated for a 24-h interval. Energy expenditure over the 24 h (24-h energy expenditure) was calculated as previously described (14). Spontaneous physical activity (SPA) was detected by radar sensors and expressed as percentage of time over the 24-h period in which activity was detected. Sleeping metabolic rate (sleep energy expenditure) was defined as the average energy expenditure of all 15-min periods between 2330 and 0500 h during which spontaneous physical activity was <1.5%. Carbon dioxide production (VCO2) and oxygen consumption (VO2) were calculated for every 15-min interval and extrapolated for the 24-h interval. The 24-h respiratory quotient was calculated as the ratio of the 24-h VCO2 and the 24-h VO2. Carbohydrate and lipid oxidation rates were calculated from the 24-h respiratory quotient accounting for protein oxidation (calculated from 24-h urinary nitrogen excretion) (15).
Resting metabolic rate.
Before starting the euglycemic-hyperinsulinemic clamp, a clear plastic ventilated hood was placed over the volunteer's head. Room air was drawn through the hood, and flow rate was measured. A constant fraction of expired air was withdrawn and analyzed for O2 and CO2 content. The O2 analyzer was a zirconium cell analyzer and the CO2 an infrared analyzer (Applied Electrochemistry, Sunnyvale, CA). The analyzers were connected to a computer that recorded continual integrated calorimetric measurements every 5 min for the hour before the start of the clamp procedure. Energy expenditure was calculated using the equations of Lusk (16) for the 40 min before the start of the clamp.
Measurement of insulin action.
Insulin action was assessed at physiologic insulin concentrations during the hyperinsulinemic-euglycemic clamp technique (17). Briefly, after an overnight fast, primed (30 μCi) continuous (0.3 μCi/min) infusion of [3-3H]glucose infusion was started to determine endogenous glucose output. Two hours after starting the isotope infusion, a primed continuous intravenous insulin infusion was administered for 100 min at a rate of 40 mU/m2 body surface area per min (low dose), followed by a second 100-min infusion at 400 mU/m2 body surface area per min (high dose). These infusions achieved steady-state insulin concentrations of 143 ± 42 and 1,714 ± 1,571 mU/l (mean ± SD), respectively. Plasma glucose concentrations were maintained at ∼100 mg/dl with a variable infusion of 20% dextrose solution. Blood samples for measurement of [3-3H]glucose specific activity were collected at the end of the basal period and every 10 min during the final 40 min of the low-dose insulin infusion. Endogenous glucose output was calculated using Steele's equation (18). As described previously, the rate of total insulin-stimulated glucose disposal (M) was calculated for the last 40 min of the low-dose (M-low) and high-dose (M-high) insulin infusions. M-low was corrected for mean glucose and insulin concentrations and endogenous glucose output during the final 40 min of the insulin infusion (19,20). M-high was corrected for the mean glucose concentration, and endogenous glucose output was assumed to be 0. All measurements derived from the glucose clamp were normalized to estimated metabolic body size (estimated metabolic body size or FFM + 17.7 kg) (21).
Analytical measurements.
Plasma glucose concentration was determined by the glucose oxidase method (Beckman Instruments, Fullerton, CA). Plasma insulin concentrations were measured by the Herbert modification of the method of Yalow and Berson (22,23), by an automated autoanalyzer (ICN Radiochemicals, Costa Mesa, CA), or by an automated immunoassay (Access, Beckman Instruments). Values from the final two assays were regressed to the original assay.
Genotyping.
DNA was genotyped by the method of Taqman Alleleic Discrimination (Applied Biosystems) for the G/A variant that predicts the R165Q and was genotyped by the method of SNPlex (Applied Biosystems) for the single base (A) insertion at nucleotide 100. DNA from all individuals genotyped as heterozygous for either of these mutations was directly sequenced to confirm their genotype using the Big Dye Terminator (Applied Biosystems) on an automated DNA capillary sequencer (model 3730xl; Applied Biosystems). Sequence information for all oligonucleotide primers and probes is available upon request.
Statistical analysis.
In the population study, maximum BMI and weight were adjusted for age, sex, and birth year (to account for population changes in BMI over time) and analyzed for association using generalized estimating equations to account for family membership (by modeling the correlation among family members). Height was analyzed in the same way but adjusted only for sex and birth year. To satisfy the assumptions of linear regression, the logarithm BMI was analyzed in all models; for simplicity of interpretation, unadjusted mean BMI is reported but P values are derived from the models. Cross-sectional data were analyzed for the subset with energy expenditure and insulin action measurements. Linear regression models were used to calculate adjusted least-squares means and 95% CIs for the energy expenditure (24-h energy expenditure, sleep energy expenditure, or RMR) and insulin action (M-low or M-high). Measurements were adjusted for covariates by genotype. M-low and M-high were log transformed to approximate a normal distribution. Energy-expenditure measurements were also adjusted for the same covariates using generalized estimating equations to account for family membership.
RESULTS
Population survey.
In the population study of full-heritage Pima Indians (n = 3,074), 14 individuals had the A insertion at nucleotide 100 (minor allele frequency 0.002) and 43 individuals had the R165Q substitution (minor allele frequency 0.007). Among these full-heritage Pima Indians, none were homozygous for either variant or compound heterozygous for the two variants. BMI (± SD) was higher among subjects heterozygous for the NT100 compared with subjects without this variant (41.2 ± 7.8 vs. 37.1 ± 8.4 kg/m2, P = 0.04). BMI was also higher among subjects heterozygous for R165Q compared with subjects without this variant (39.3 ± 8.6 vs. 37.1 ± 8.4 kg/m2, P = 0.02). Body weight was also significantly higher among the heterozygous subjects. However, heterozygotes for NT100 tended to be shorter (163.3 ± 6.8 vs. 164.6 ± 8.4 cm, P = 0.02), whereas heterozygotes for R165Q were slightly taller (166.8 ± 8.6 vs. 164.6 ± 8.4, P = 0.14). Because of the relatively small number of subjects heterozygous for either variant, BMI was also analyzed after combining heterozygotes for either variant (57 of 3,074 = 1.8% of individuals were heterozygous for either variant). Heterozygotes for either variant had a significantly higher mean BMI than subjects without either variant (39.8 ± 8.6 vs. 37.1 ± 8.4 kg/m2, P = 0.002, adjusted for age, sex, and birth year using generalized estimating equations to account for family membership). If the analysis was restricted to individuals measured when nondiabetic, 46 of 2,649 (1.7%) were heterozygous for either variant, and the difference in BMI versus those without either variant remained (39.5 ± 8.0 vs. 36.2 ± 8.2 kg/m2, P = 0.0003). In the nondiabetic exams for each variant separately, the trend was the same (for NT100 [n = 12], 38.4 ± 6.7 vs. 36.2 ± 8.2 kg/m2, P = 0.12; for the R165Q [n = 34], 39.8 ± 8.6 vs. 36.2 ± 8.2 kg/m2, P = 0.0009).
Energy expenditure and insulin action.
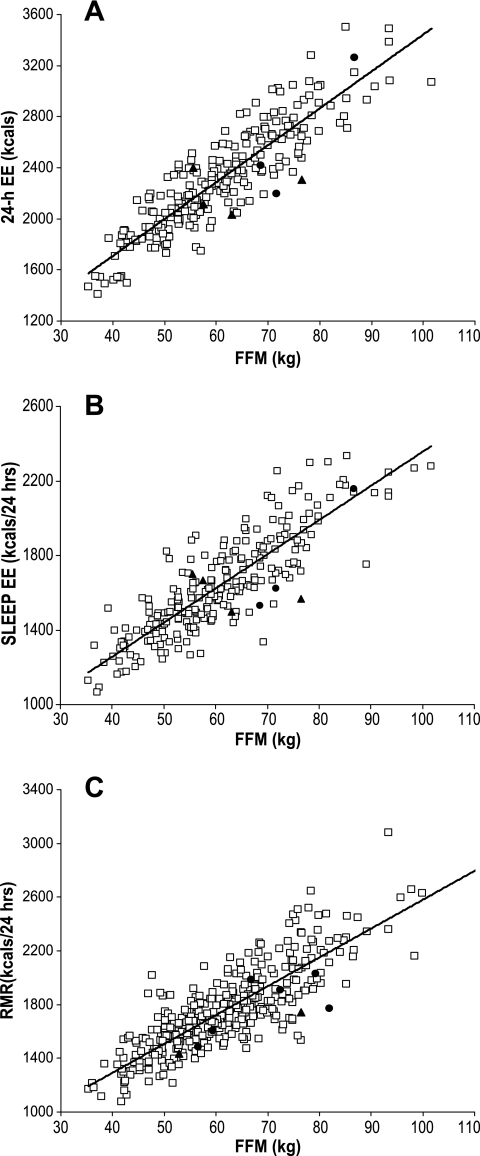
Table 1 shows the baseline variables for the subsets who had measurements of energy expenditure and insulin action displayed for each heterozygote. Because of the smaller numbers in these subsets, differences in adiposity between the variants was not always significant. The same seven heterozygotes are present in the 24-h energy expenditure and sleep energy expenditure subsets. One heterozygote had measurements of 24-h energy expenditure, sleep energy expenditure, and RMR and is included in each analysis. All of the heterozygotes who had measurements of RMR also had measurements of insulin action. Energy expenditure (adjusted for age, sex, FFM, and fat mass) and insulin action (adjusted for age, sex, and percent body fat) measurements are shown in Table 2 for both the individual and combined variants with and without accounting for family membership. For the 24-h energy expenditure measurements, additional adjustment for spontaneous physical activity or energy balance did not alter the results (data not shown). Energy expenditure measures were consistently (although not significantly) lower for each heterozygote. Because of each heterozygote's association with increased BMI in the population study, they were analyzed together (combined group in Table 2). All energy expenditure measurements were significantly lower in the combined group compared with the common variants. After accounting for family membership, the differences were attenuated, although they were still quite significant for RMR. Measures of insulin action did not display a similar consistent pattern, although M-low (but not M-high) was significantly lower in the NT100 heterozygotes. Figure 1 shows plots of measured energy expenditure versus FFM for each variant, demonstrating relatively lower energy expenditure in the majority of the heterozygotes compared with individuals of comparable FFM with the common genotype. There were no differences in 24-h respiratory quotient or lipid oxidation after adjustment for the same covariates (data not shown).
TABLE 1.
Characteristics of subsets of full-heritage Pima Indians with measurements of energy expenditure and insulin action by the MC4R variant
| 24-h energy expenditure |
Sleep energy expenditure |
RMR |
M-low |
M-high |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | NT100 | R165Q | Control | NT100 | R165Q | Control | NT100 | R165Q | Control | NT100 | R165Q | Control | NT100 | R165Q | |
| n (M/F) | 245 (129/116) | 4 (1/3) | 3 (3/0) | 237 (122/115) | 4 (1/3) | 3 (3/0) | 356 (203/153) | 6 (5/1) | 2 (1/1) | 350 (192/158) | 3 (1/2) | 6 (5/1) | 314 (177/137) | 3 (1/2) | 5 (4/1) |
| Age (years) | 26.3 ± 6.6 | 21.2 ± 2.1 | 31.7 ± 6.1 | 26.2 ± 6.5 | 21.2 ± 2.1 | 31.7 ± 6.1 | 26.1 ± 6.0 | 27.9 ± 4.9 | 20.6 ± 2.7 | 26.2 ± 6.0 | 21.6 ± 2.5 | 27.9 ± 4.9 | 26.2 ± 6.1 | 21.6 ± 2.5 | 26.8 ± 4.6 |
| Height (cm) | 166.0 ± 7.9 | 167 ± 5.4 | 175 ± 0.5* | 166.0 ± 7.9 | 167.0 ± 5.4 | 175 ± 0.5* | 166.1 ± 8.1 | 168.3 ± 4.7 | 161 ± 9.5 | 165.8 ± 8.2 | 161.4 ± 6.8 | 168.3 ± 4.7 | 166.0 ± 8.5 | 161.4 ± 6.8 | 168.6 ± 5.2 |
| Weight (kg) | 92.9 ± 24.1 | 99.5 ± 7.4 | 119.4 ± 21.1* | 93.3 ± 24.3 | 99.5 ± 7.4 | 119.4 ± 21.1 | 94.0 ± 22.7 | 103.3 ± 10.9 | 91.4 ± 7.2 | 95.0 ± 23.0 | 90.9 ± 5.2 | 103.4 ± 10.9 | 94.0 ± 22.2 | 90.9 ± 5.2 | 102.1 ± 11.7 |
| BMI (kg/m²) | 33.7 ± 8.6 | 35.8 ± 4.7 | 38.7 ± 6.7 | 33.5 ± 8.6 | 35.8 ± 4.7 | 38.7 ± 6.7 | 34.0 ± 7.8 | 36.5 ± 4.0 | 35.0 ± 1.4 | 34.5 ± 7.7 | 34.9 ± 1.0 | 36.5 ± 4.0 | 34.0 ± 7.4 | 34.9 ± 1.0 | 35.9 ± 4.1 |
| % Fat | 33.3 ± 9.0 | 36.2 ± 10.9 | 36.1 ± 5.7 | 33.5 ± 8.9 | 36.2 ± 10.9 | 36.1 ± 5.7 | 33.1 ± 8.7 | 32.7 ± 8.1 | 29.8 ± 12.8 | 33.8 ± 8.4 | 33.3 ± 10.9 | 32.7 ± 8.0 | 33.4 ± 8.4 | 33.3 ± 10.9 | 34.2 ± 8.0 |
| FFM (kg) | 60.7 ± 12.4 | 63.2 ± 9.4 | 75.7 ± 9.7* | 60.8 ± 12.7 | 63.2 ± 9.4 | 75.7 ± 9.7* | 61.8 ± 12.4 | 69.4 ± 10.3 | 64.7 ± 16.8 | 62.0 ± 12.7 | 61.0 ± 13.5 | 69.4 ± 10.3 | 61.7 ± 12.6 | 61.0 ± 13.5 | 66.9 ± 9.3 |
| Fat mass (kg) | 32.2 ± 14.8 | 36.4 ± 12.4 | 43.7 ± 13.0 | 32.4 ± 14.8 | 36.4 ± 12.4 | 43.7 ± 13.0 | 32.2 ± 13.7 | 33.9 ± 9.6 | 26.7 ± 9.6 | 33.1 ± 13.6 | 29.9 ± 8.7 | 34.0 ± 9.6 | 32.3 ± 13.1 | 29.9 ± 8.7 | 35.2 ± 10.2 |
| Fasting plasma glucose (mmol/l) | 4.88 ± 0.55 | 4.78 ± 0.43 | 4.55 ± 0.61 | 4.94 ± 0.55 | 4.78 ± 0.43 | 4.55 ± 0.61 | 5.00 ± 0.55 | 5.03 ± 0.21 | 4.75 ± 0.82 | 5.00 ± 0.55 | 5.04 ± 0.77 | 5.03 ± 0.21 | 4.98 ± 0.54 | 5.04 ± 0.77 | 4.98 ± 0.21 |
| 2-h plasma glucose (mmol/l) | 6.72 ± 1.67 | 6.69 ± 2.49 | 5.11 ± 1.12 | 6.72 ± 1.67 | 6.69 ± 2.49 | 5.11 ± 1.12 | 6.89 ± 1.72 | 6.90 ± 1.27 | 5.39 ± 2.28 | 6.94 ± 1.67 | 5.98 ± 1.91 | 6.90 ± 1.27 | 6.91 ± 1.71 | 5.98 ± 1.91 | 7.08 ± 1.34 |
Data are means ± SD. Control = neither variant; NT100 = A insertion variant; R165Q = G-to-A substitution.
P < 0.05 for comparison of NT100 or R165Q to common variant using Wilcoxon's rank-sum test.
TABLE 2.
Differences in energy expenditure and insulin action for each MC4R variant and the combined variants
| Control | NT100 | P | R165Q | P | Combined | P | ||
|---|---|---|---|---|---|---|---|---|
| 24-h energy expenditure (kcal) | 2,307 (2,285–2,328) | 2,175 (2,004–2,345) | 0.13 | 2,148 (1,949–2,346) | 0.12 | 2,163 (2,035–2,291) | 0.03 | |
| Family | 2,319 (2,297–2,342) | 2,204 (1,933–2,475) | 0.40 | 2,156 (1,906–2,405) | 0.20 | 2,182 (1,997–2,367) | 0.14 | |
| Sleep energy expenditure (kcal/24 h) | 1,640 (1,623–1,657) | 1,584 (1,452–1,715) | 0.40 | 1,466 (1,313–1,619) | 0.02 | 1,534 (1,435–1,635) | 0.03 | |
| Family | 1,645 (1,628–1,663) | 1,593 (1,409–1,777) | 0.58 | 1,472 (1,321–1,622) | 0.02 | 1,539 (1,416–1,663) | 0.09 | |
| RMR (kcal/24 h) | 1,754 (1,736–1,772) | 1,553 (1,317–1,788) | 0.09 | 1,637 (1,502–1,773) | 0.54 | 1,617 (1,499–1,734) | 0.02 | |
| Family | 1,765 (1,746–1,784) | 1,561 (1,470–1,653) | <0.0001 | 1,646 (1,573–1,719) | 0.002 | 1,625 (1,560–1,689) | <0.0001 | |
| M-low (mg/kgEMBS × min) | 2.49 (2.42–2.56) | 1.87 (1.39–2.50) | 0.06 | 2.65 (2.15–3.26) | 0.56 | 2.36 (1.99–2.39) | 0.56 | |
| Family | 2.47 (2.40–2.54) | 1.85 (1.79–1.90) | <0.0001 | 2.59 (2.15–3.10) | 0.62 | 2.32 (1.93–2.78) | 0.48 | |
| M-high (mg/kgEMBS × min) | 8.32 (8.10–8.56) | 8.24 (6.22–10.93) | 0.95 | 6.84 (5.50–8.51) | 0.08 | 7.34 (6.18–8.82) | 0.16 | |
| Family | 8.27 (8.02–8.51) | 8.21 (7.85–8.57) | 0.79 | 6.74 (5.05–8.97) | 0.18 | 7.24 (5.94–8.83) | 0.19 |
24-h energy expenditure, sleep energy expenditure, and RMR are adjusted for age, sex, fat mass, and fat-free mass in linear regression models and are presented as least-squares means (95% CI). M-low and M-high are adjusted for age, sex, and percent body fat in linear regression models and are presented as geometric means (95% CI) in parentheses. Second row (labeled as family) for each variable represents general estimating equations adjusted for the same variables accounting for family membership. Control = neither variant, NT100 = A insertion variant, R165Q = G-to-A substitution, combined = NT100 or R165Q. P values are for comparison of NT100, R165Q, or combined variants versus subjects with neither variant. EMBS, estimated metabolic body size.
FIG. 1.
A: 24-h energy expenditure (EE) versus FFM. B: Sleep energy expenditure versus FFM. C: RMR versus FFM. □, neither variant; ▴, NT100 variant; •, R165Q variant.
DISCUSSION
Previous studies of the frequency of functional MC4R variants have been performed in groups selected for obesity (24,25), but in a more general population study, these variants were not associated with higher BMI (26). For nonfunctional variants, meta-analysis of a V103I MC4R polymorphism in predominantly Caucasian subjects concluded that the isoleucine allele was modestly associated with protection from obesity (9,27). This locus is monomorphic for the valine allele in the 426 Pima Indians in whom the gene was sequenced. A recent meta-analysis in a large group of Europeans found that a range of functional MC4R variants were more common in obese than lean individuals (28). In a German population study, R165Q was rare (minor allele frequency 1.2 × 10−4) and the individual was not obese (26). In our general population study of full-heritage Pima Indians, the percentage of individuals carrying either the R165Q or the NT100 is 1.8%, and their association with higher BMI was clear. This was even more pronounced in the analysis excluding subjects with type 2 diabetes, presumably because of the influence of diabetes itself or associated medications on weight. Furthermore, in a subset of Pima Indians who also underwent metabolic studies, we found that energy expenditure tended to be lower (although not significantly so) among individuals heterozygous for one of these two variants. The R165Q variant has known functional consequences (3). Although not proven, we propose that the NT100 also has functional consequences because it predicts a truncated receptor lacking critical domains. Therefore, we combined these rare variants to obtain additional statistical power and, in so doing, we have shown for the first time in humans that energy expenditure, adjusted for age, sex, FFM, and fat mass is lower in individuals heterozygous for either variant. The lower energy expenditure (of ∼110–140 kcal/day) was apparent in analyzing 24 h and sleep energy expenditure (both measured in our respiratory chamber) and replicated by measurements of RMR, using a ventilated hood, on a separate day.
In mice, the MC4R is critical in the role of regulating energy homeostasis and is abundant in the hypothalamic paraventricular nucleus. MC4R knockout mice have early-onset obesity, hyperphagia, hyperinsulinemia, and hyperglycemia, with heterozygotes having an intermediate obese phenotype (4). The importance of MC4R variants in humans was documented with discovery of a frameshift mutation associated with autosomal dominantly inherited obesity (29,30). The clinical phenotype of these individuals (including both homozygous and heterozygous subjects) included early-onset obesity and excessive hunger during childhood with food-seeking behavior (2). Children with MC4R variants ate nearly three times as many calories as their unaffected siblings in studies evaluating ad libitum food intake (3), although this hyperphagia decreased toward adulthood. In these same studies, RMR, measured by indirect calorimetry, was not different based on comparisons with age- and sex-specific equations corrected for FFM (2,3).
Our data indicate that reduced energy expenditure (by ∼110–140 kcal/day) is part of the clinical phenotype of adults heterozygous for rare obesity-associated MC4R variants. Mice with MC4R knockout mutations have reduced energy expenditure (5), but this is not due to reduced core body temperature or an inability to thermoregulate. MC4R knockout mice also have lower energy expenditure compared with leptin knockout (OB/OB) mice (27). Although data on the association between MC4R variants and measured energy expenditure is limited in humans, the Val103Ile MC4R polymorphism, which has been negatively associated with obesity (27,31), was also associated with higher RMRs in a separate study (32). Although RMR was higher in this prior report, BMI was not different between individuals with the Val versus the 103lle allele, and individuals with the 103Ile allele tended to gain more weight over time (32). Furthermore, unlike our studies, RMR was not performed after an inpatient period of weight stabilization. Therefore, increased food intake could account for the increased RMR. Despite recent evidence in mice for a role of MC4R in lipid metabolism (33), we did not find differences in respiratory quotient or lipid oxidation in MC4r heterozygotes.
Evidence for a direct effect of MC4R on energy expenditure comes from rodent studies. In particular, MC4R is important in activating the sympathetic nervous system (34,35). MC4R mediates the central response to leptin-induced expression of uncoupling protein 1 (UCP1) in brown adipose tissue of rats (35), and sympathetic nerve activity in rats increased when an MC4R agonist was given (34). Activation of the sympathetic nervous system increases basal energy metabolism (36); therefore, loss of function in MC4R with resulting lower sympathetic tone would explain the lower energy expenditure in these individuals. It is not clear, however, if the lower energy expenditure in these adults precedes (and is therefore causative of their increased weight or whether this lower energy expenditure represents a failure to adapt to increased body size (thus making it difficult for these individuals to lose weight). Leptin concentrations and sympathetic nervous system activity increase with body size, thereby increasing energy expenditure and braking further weight gain (37).
Although this analysis concentrates on the differences noted in energy expenditure, increased food intake (hyperphagia) may still account for a portion (perhaps even most) of the weight gain and adiposity among individuals with these variants. Our study did not include measurement of ad libitum food intake, and more careful under- or overfeeding studies would be needed to elucidate the relative contribution of each factor. In addition, while the energy-expenditure differences were relatively large, the EE measurements were performed on individuals on weight-maintaining diets in an inpatient setting, and were confirmed using measurements done at separate times on different individuals (only one individual with a variant was in the group with both 24-h energy expenditure and RMR), given the small numbers, these differences should be interpreted with caution. The adjusted mean difference in energy expenditure between heterozygotes and those with the common genotype was 110–140 kcal/day. If this deficit was maintained over a year, it would amount to ∼40,150–51,000 kcal difference per year.
Our data indicate that full-heritage Pima Indians heterozygous for either a frameshift or a functional missense MC4R variant have higher BMI values and lower 24-h, sleeping, and resting energy expenditure. This is one of the clearest examples in humans of a genotype resulting in reduced energy expenditure and provides a link between the melanocortin system and energy expenditure. Further understanding of how this system affects energy expenditure may provide important clues for advances in obesity treatment and prevention.
Acknowledgments
This study was supported by the intramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. L.M. was supported by a grant from the American Diabetes Association.
We thank the members of the Gila River Indian Community for their support and the staff of the Phoenix Epidemiology and Clinical Research Branch.
Published ahead of print at http://diabetes.diabetesjournals.org on 3 October 2008.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Barsh GS, Farooqi IS, O’Rahilly S: Genetics of body-weight regulation. Nature 404 :644 –651,2000 [DOI] [PubMed] [Google Scholar]
- 2.Farooqi IS, Yeo GS, Keogh JM, Aminian S, Jebb SA, Butler G, Cheetham T, O’Rahilly S: Dominant and recessive inheritance of morbid obesity associated with melanocortin 4 receptor deficiency. J Clin Invest 106 :271 –279,2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farooqi IS, Keogh JM, Yeo GS, Lank EJ, Cheetham T, O’Rahilly S: Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med 348 :1085 –1095,2003 [DOI] [PubMed] [Google Scholar]
- 4.Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, Gu W, Kesterson RA, Boston BA, Cone RD, Smith FJ, Campfield LA, Burn P, Lee F: Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 88 :131 –141,1997 [DOI] [PubMed] [Google Scholar]
- 5.Ste ML, Miura GI, Marsh DJ, Yagaloff K, Palmiter RD: A metabolic defect promotes obesity in mice lacking melanocortin-4 receptors. Proc Natl Acad Sci U S A 97 :12339 –12344,2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pavkov ME, Hanson RL, Knowler WC, Bennett PH, Krakoff J, Nelson RG: Changing patterns of type 2 diabetes incidence among Pima Indians. Diabetes Care 30 :1758 –1763,2007 [DOI] [PubMed] [Google Scholar]
- 7.Ma L, Tataranni PA, Bogardus C, Baier LJ: Melanocortin 4 receptor gene variation is associated with severe obesity in Pima Indians. Diabetes 53 :2696 –2699,2004 [DOI] [PubMed] [Google Scholar]
- 8.Stutzmann F, Vatin V, Cauchi S, Morandi A, Jouret B, Landt O, Tounian P, Levy-Marchal C, Buzzetti R, Pinelli L, Balkau B, Horber F, Bougneres P, Froguel P, Meyre D: Non-synonymous polymorphisms in melanocortin-4 receptor protect against obesity: the two facets of a Janus obesity gene. Hum Mol Genet 16 :1837 –1844,2007 [DOI] [PubMed] [Google Scholar]
- 9.Young EH, Wareham NJ, Farooqi S, Hinney A, Hebebrand J, Scherag A, O’Rahilly S, Barroso I, Sandhu MS: The V103I polymorphism of the MC4R gene and obesity: population based studies and meta-analysis of 29 563 individuals. Int J Obes (Lond) 31 :1437 –1441,2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization: Definition, Diagnosis and Classification of Diabetes Mellitus and its Complications. Geneva, World Health Org., Department of Noncommunicable Disease Surveillance,1999
- 11.Goldman RFBER: A method for underwater weighing and the determination of body density. In Techniques for Measuring Body Composition. Brozek J, Herschel A, Ed. Washington, DC, National Academy of Sciences, National Research Council,1961. , p.78 –106
- 12.Mazess RB, Barden HS, Bisek JP, Hanson J: Dual-energy x-ray absorptiometry for total-body and regional bone-mineral and soft-tissue composition. Am J Clin Nutr 51 :1106 –1112,1990 [DOI] [PubMed] [Google Scholar]
- 13.Tataranni PA, Ravussin E: Use of dual-energy X-ray absorptiometry in obese individuals. Am J Clin Nutr 62 :730 –734,1995 [DOI] [PubMed] [Google Scholar]
- 14.Ravussin E, Lillioja S, Anderson TE, Christin L, Bogardus C: Determinants of 24-hour energy expenditure in man: methods and results using a respiratory chamber. J Clin Invest 78 :1568 –1578,1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jequier E, Acheson K, Schutz Y: Assessment of energy expenditure and fuel utilization in man. Annu Rev Nutr 7 :187 –208,1987 [DOI] [PubMed] [Google Scholar]
- 16.Lusk G: Animal calorimetry: analysis of oxidation of mixtures of carbohydrate and fat. J Biol Chem 59 :41 –42,1924 [Google Scholar]
- 17.Bogardus C, Lillioja S, Mott D, Reaven GR, Kashiwagi A, Foley JE: Relationship between obesity and maximal insulin-stimulated glucose uptake in vivo and in vitro in Pima Indians. J Clin Invest 73 :800 –805,1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altszuler N, de Bodo RC, Steele R, Wall JS: Measurement of size and turnover rate of body glucose pool by the isotope dilution method. Am J Physiol 187 :15 –24,1956 [DOI] [PubMed] [Google Scholar]
- 19.Best JD, Taborsky GJ Jr, Halter JB, Porte D Jr: Glucose disposal is not proportional to plasma glucose level in man. Diabetes 30 :847 –850,1981 [DOI] [PubMed] [Google Scholar]
- 20.Lillioja S, Mott DM, Howard BV, Bennett PH, Yki-Jarvinen H, Freymond D, Nyomba BL, Zurlo F, Swinburn B, Bogardus C: Impaired glucose tolerance as a disorder of insulin action: longitudinal and cross-sectional studies in Pima Indians. N Engl J Med 318 :1217 –1225,1988 [DOI] [PubMed] [Google Scholar]
- 21.Lillioja S, Bogardus C: Obesity and insulin resistance: lessons learned from the Pima Indians. Diabete Metab Rev 4 :517 –540,1988 [DOI] [PubMed] [Google Scholar]
- 22.Herbert V, Lau KS, Gottlieb CW, Bleicher SJ: Coated charcoal immunoassay of insulin. J Clin Endocrinol Metab 25 :1375 –1384,1965 [DOI] [PubMed] [Google Scholar]
- 23.Yalow RS, Berson SA: Immunoassay of endogenous plasma insulin in man. J Clin Invest 39 :1157 –1175,1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buono P, Pasanisi F, Nardelli C, Ieno L, Capone S, Liguori R, Finelli C, Oriani G, Contaldo F, Sacchetti L: Six novel mutations in the proopiomelanocortin and melanocortin receptor 4 genes in severely obese adults living in southern Italy. Clin Chem 51 :1358 –1364,2005 [DOI] [PubMed] [Google Scholar]
- 25.Rong R, Tao YX, Cheung BM, Xu A, Cheung GC, Lam KS: Identification and functional characterization of three novel human melanocortin-4 receptor gene variants in an obese Chinese population. Clin Endocrinol (Oxf)65 :198 –205,2006 [DOI] [PubMed] [Google Scholar]
- 26.Hinney A, Bettecken T, Tarnow P, Brumm H, Reichwald K, Lichtner P, Scherag A, Nguyen TT, Schlumberger P, Rief W, Vollmert C, Illig T, Wichmann HE, Schafer H, Platzer M, Biebermann H, Meitinger T, Hebebrand J: Prevalence, spectrum, and functional characterization of melanocortin-4 receptor gene mutations in a representative population-based sample and obese adults from Germany. J Clin Endocrinol Metab 91 :1761 –1769,2006 [DOI] [PubMed] [Google Scholar]
- 27.Heid IM, Vollmert C, Hinney A, Doring A, Geller F, Lowel H, Wichmann HE, Illig T, Hebebrand J, Kronenberg F: Association of the 103I MC4R allele with decreased body mass in 7937 participants of two population based surveys. J Med Genet 42 :e21 ,2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stutzmann F, Tan K, Vatin V, Dina C, Jouret B, Tichet J, Balkau B, Potoczna N, Horber F, O’Rahilly S, Farooqi IS, Froguel P, Meyre D: Prevalence of melanocortin-4 deficiency in European population and their age-dependant penetrance in multi-generational pedigrees. Diabetes 57 :2511 –2518,2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaisse C, Clement K, Guy-Grand B, Froguel P: A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet 20 :113 –114,1998 [DOI] [PubMed] [Google Scholar]
- 30.Yeo GS, Farooqi IS, Aminian S, Halsall DJ, Stanhope RG, O’Rahilly S: A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat Genet 20 :111 –112,1998 [DOI] [PubMed] [Google Scholar]
- 31.Geller F, Reichwald K, Dempfle A, Illig T, Vollmert C, Herpertz S, Siffert W, Platzer M, Hess C, Gudermann T, Biebermann H, Wichmann HE, Schafer H, Hinney A, Hebebrand J: Melanocortin-4 receptor gene variant I103 is negatively associated with obesity. Am J Hum Genet 74 :572 –581,2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutanen J, Pihlajamaki J, Karhapaa P, Vauhkonen I, Kuusisto J, Moilanen ML, Laakso M: The Val103Ile polymorphism of melanocortin-4 receptor regulates energy expenditure and weight gain. Obes Res 12 :1060 –1066,2004 [DOI] [PubMed] [Google Scholar]
- 33.Nogueiras R, Wiedmer P, Perez-Tilve D, Veyrat-Durebex C, Keogh JM, Sutton GM, Pfluger PT, Castaneda TR, Neschen S, Hofmann SM, Howles PN, Morgan DA, Benoit SC, Szanto I, Schrott B, Schurmann A, Joost HG, Hammond C, Hui DY, Woods SC, Rahmouni K, Butler AA, Farooqi IS, O’Rahilly S, Rohner-Jeanrenaud F, Tschop MH: The central melanocortin system directly controls peripheral lipid metabolism. J Clin Invest 117 :3475 –3488,2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haynes WG, Morgan DA, Djalali A, Sivitz WI, Mark AL: Interactions between the melanocortin system and leptin in control of sympathetic nerve traffic. Hypertension 33 :542 –547,1999 [DOI] [PubMed] [Google Scholar]
- 35.Satoh N, Ogawa Y, Katsuura G, Numata Y, Masuzaki H, Yoshimasa Y, Nakao K: Satiety effect and sympathetic activation of leptin are mediated by hypothalamic melanocortin system. Neurosci Lett 249 :107 –110,1998 [DOI] [PubMed] [Google Scholar]
- 36.Astrup A: The sympathetic nervous system as a target for intervention in obesity. Int J Obes Relat Metab Disord 19 (Suppl. 7):S24 –S28,1995 [PubMed] [Google Scholar]
- 37.Jequier E: Leptin signaling, adiposity, and energy balance. Ann N Y Acad Sci 967 :379 –388,2002 [DOI] [PubMed] [Google Scholar]