Abstract
In this study 231 Y chromosomes from 12 populations were typed for four diagnostic SNPs to determine haplogroup membership and 43 Y chromosomes from three of these populations were typed for eight Simple Tandem Repeats (STRs) to determine haplotypes. These data were combined with previously published data, amounting to 724 Y chromosomes from 26 populations in North America, and analyzed to investigate the geographic distribution of Y chromosomes among Native North Americans and to test the Southern Athapaskan migration hypothesis. The results suggest that European admixture has significantly altered the distribution of Y chromosomes in North America and because of this caution should be taken when inferring prehistoric population events in North America using Y chromosome data alone. However, consistent with studies of other genetic systems, we are still able to identify close relationships among Y chromosomes in Athapaskan from the Subarctic and the Southwest, suggesting that a small number of proto-Apachean migrants from the Subarctic founded the Southwest Athapaskan populations.
Keywords: Y chromosome, Mitochondrial DNA, Founder effect, European contact, Native American, Migration, Diffusion, Native American
The geographic distribution of the Athapaskan language family in North America is hypothesized to result from long distance population movements (Basso, 1983). Previous studies of albumin variants (Smith et al, 2000) and mitochondrial DNA (mtDNA) (Malhi et al., 2003) have tested the migration hypothesis of Southern Athapaskan (Apachean) speakers. These studies show genetic patterns consistent with an Athapaskan migration to the Southwest from the Subarctic, followed by extensive admixture with groups in the Southwest. However, mtDNA is maternally inherited and therefore only reflects female population history. If male population history differs from female population history, it will not be reflected in patterns of mtDNA variation. For example, differences in male and female post-marital residence patterns may be investigated by comparing mtDNA and Y chromosome variation, as recently conducted by Bolnick et al. (2006) in Eastern North America. Therefore, we undertake a study of Y chromosome variation among Athapaskan-speakers to gain insight into the population history of males in this group, which will broaden our understanding of the processes of population movements in prehistoric North America.
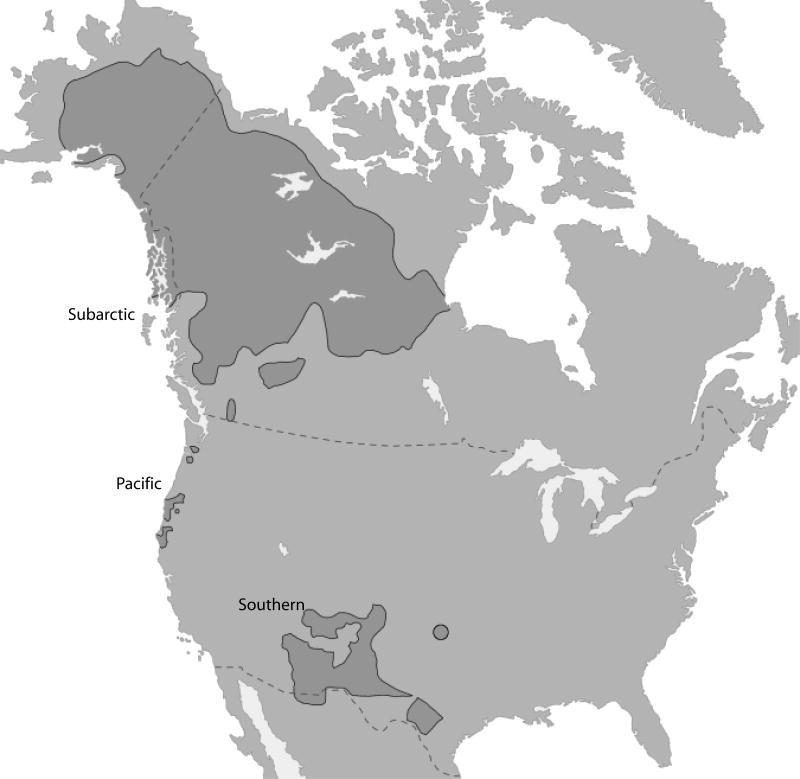
The majority of the speakers of Athapaskan languages are located in the Subarctic (Alaska and Canada), with the rest dispersed along the Pacific coastline, from Oregon to California, and into the Southwest (Figure 1). The Apacheans (the Navajo and Apache) are widely dispersed throughout the central region of the Southwest and speak languages that are closely related to Chipewyan, an Athapaskan language found in the Subarctic (Hoijer, 1956). Hoijer (1956) hypothesized that the homeland of proto-Athapaskan was in the Subarctic and that the distribution of Athapaskan languages along the Pacific coast and in the Southwest was the result of one or more migrations. Sapir (1936) demonstrated that the cultural significance of four Navajo words is indicative of a northern homeland for Southern Athapaskans and hypothesized that the presence of Athapaskans in the Southwest is the result of one or more migrations from the north. Most archaeologists believe Apacheans were present in the Southwest no earlier than around 500 years ago (Cordell, 1997). In accordance with this recent arrival, glottochronological estimates place the divergence of Proto-Apachean from Northern Athapaskan languages around 1,000 to 1,300 years ago and the earliest divergence of Apachean languages around 400 to 600 years ago (Hoijer, 1956; Hymes, 1957). Hence, the Southern Athapaskan migration is thought to have been a relatively recent long distance and rapid population movement from the Subarctic to the Southwest of North America.
Figure 1.
Distribution of Athapskan Language Family in North America. Modified from http://en.wikipedia.org/wiki/Image:Na-Dene_langs.png with reference to Campbell (1997) and Goddard (1996).
In agreement with the archaeological and linguistic conclusions, studies of protein polymorphisms have indicated that, relative to other Native American groups, Apacheans and Subarctic Athapaskans share recent common ancestry (Suarez et al., 1985). However, studies of mtDNA variation among Native North Americans have revealed that Apacheans are polymorphic with regard to mtDNA haplogroups, unlike Subarctic Athapaskans, who are monomorphic for haplogroup A. Approximately half of the maternal lineages sampled in the Apache belong to haplogroup A; while the others are primarily from haplogroups B and C. Furthermore, many of the mtDNA haplotypes in the Apache are shared with non-Athapaskan speaking populations in the Southwest. This suggests that many of the maternal lineages in the Navajo and Apache were acquired through admixture with non-Athapaskan groups in the Southwest (Lorenz and Smith, 1996; Smith et al., 2000; Malhi et al., 2003). In contrast, mtDNA haplotypes that are common in Athapaskans from the Subarctic are not common in non-Athapaskan groups in the Southwest; thus far, haplotypes common in Subarctic Athapaskans have only been observed in the Yavapai (Monroe, 2006) and, at a low frequency, in the Zuni (Malhi et al., 2003). Further supporting this unequal pattern of gene flow, a study of albumin variants (Smith et al., 2000) demonstrated that Southern Athapaskans exhibit a relatively high frequency of Albumin*Mexico, a variant found only in populations of the Southwest and Mesoamerica. However, Southern Athapaskans are the only Southwest populations to exhibit Albumin*Naskapi, an albumin variant that is also found in Subarctic populations (Smith et al., 2000).
A study of the mtDNA haplotypes observed in the Apachean and Subarctic Athapaskans identified a pattern consistent with a founder effect for Southern Athapaskans. The Apachean exhibit a high frequency of a derived subclade of haplogroup A that likely originated in the Subarctic (defined as the A2a clade by Tamm et al. 2007), however, few individuals belong to this sublcade in Subarctic populations. Based on these results, Malhi et al. (2003) suggested that the proto-Apacheans migrated to the Southwest in a single migration that included a relatively small number of females. After arriving in the Southwest, the Southern Athapaskans acquired a large number of non-Athapaskan maternal lineages through admixture with females from Southwestern tribes, but most Southwest groups did not correspondingly acquire as many maternal lineages from the Southern Athapaskans. The result of this process was a spread of Athapaskan culture in the Southwest, which has made the Navajo and Apache among the largest tribes in Native North America.
Previous studies have suggested that the distribution of Y chromosome variation among Native North Americas has been strongly influenced by (Zegura et al. 2004, Hammer et al. 2006, Bolnick et al. 2006) European admixture, in addition to other effects of European contact, such as high mortality and the relocation of indigenous individuals (Spicer, 1962). These effects may limit our ability to make inferences about prehistoric population events in North America from Y chromosome analysis alone. Hence, prior to analyzing Y chromosome variation in Athapaskans, we assess the influence of European admixture on the distribution of Y chromosome variation among Native North Americans. We then study the male population history of Athapaskans within the context of patterns of Y chromosome variation across North America and results from corresponding mtDNA studies. Specifically, we test the hypothesis that Athapaskans in the Subarctic and in the Southwest share Y chromosomes, to the exclusion of non-Athapaskans in the Southwest, as a result of recent common ancestry. This result would be consistent with a migration of Athapaskans from the Subarctic to the Southwest (Hoijer, 1956; Basso, 1983; Lorenz and Smith, 1996; Smith et al., 2000; Malhi et al., 2003; Hunley and Long, 2005).
Materials and Methods
Populations studied and datasets
Two separate datasets, a Haplogroup set and a Haplotype set, were analyzed in this study. The membership in Y chromosome haplogroup Q, C, or R was determined for 231 males from 12 populations in this study, including 15 Dogrib, 23 San Carlos Apache (SC Apache), 13 Jemez, 38 Akimal O'ohdam (Pima), 13 Tohono O'ohdam (Papago), 15 Seri, 20 Tarahumara, 37 Cora, 18 Huichol, 7 Nahua of San Pedro Atocpan (Nahua), 10 Nahua of Cuetzalan (Nahua Cuetzalan), and 22 Mixtec (Mixa). All samples are covered by IRB protocols at the University of California, Davis, and have been described in Smith et al. (2000) except the Seri (Infante et al., 1999), Tarahumara, Cora, Nahua Atocpan and Huichol (Kemp 2006), and Mixe and Mixtec (Hollenbach et al., 2001). These data were combined for analysis with existing Y chromosome haplogroup data from Lell et al. (2002), Bortolini et al. (2003), Zegura et al. (2004), and Bolnick et al. (2006), resulting in a Haplogroup data set totaling 724 males from 26 Native North American populations. A description of the samples used in the Haplogroup data set is given in Table 1. Only samples that were determined to belong to haplogroups Q (Q-M3 and Q-M242), C, or R were included in the Haplogroup data set.
Table 1.
Description of the Haplogroup data set. Language family based on Campbell et al. (1997).
| Tribe | Language Family | Latitude | Longitude | N | Q | C | R | Reference |
|---|---|---|---|---|---|---|---|---|
| SC Apache | Athapaskan | 33.35 | -110.45 | 23 | 20 | 2 | 1 | This study |
| Pima | Uto-Aztecan | 31.40 | -113.40 | 62 | 58 | 0 | 4 | Zegura et al. 2004, this study (38) |
| Papago | Uto-Aztecan | 30.45 | -111.00 | 13 | 8 | 0 | 5 | This study |
| Jemez | Tanoan | 35.57 | -105.67 | 13 | 13 | 0 | 0 | This study |
| Tarahumara | Uto-Aztecan | 28.50 | -106.25 | 20 | 19 | 0 | 1 | This study |
| Seri | Isolate | 29.98 | -111.25 | 15 | 15 | 0 | 0 | This study |
| Nahua Atocpan | Uto-Aztecan | 19.03 | -99.08 | 7 | 6 | 0 | 0 | This study |
| Nahua Cuetzalan | Uto-Aztecan | 19.02 | -99.07 | 10 | 10 | 0 | 0 | This study |
| Cora | Uto-Aztecan | 21.73 | -105.00 | 37 | 37 | 0 | 0 | This study |
| Huichol | Uto-Aztecan | 22.00 | -104.33 | 18 | 18 | 0 | 0 | This study |
| Mixtec | Otomanguen | 17.15 | -97.45 | 22 | 20 | 0 | 2 | This study |
| Mixe | Mixe-Zoquean | 17.01 | -95.54 | 12 | 12 | 0 | 0 | Zegura et al. 2004 |
| Zapotec | Otomanguean | 17.30 | -96.20 | 16 | 12 | 0 | 1 | Zegura et al. 2004 |
| Chipewyan | Athapaskan | 59.55 | -107.30 | 48 | 15 | 3 | 30 | Bortolini et al. 2003 |
| Dogrib | Athapaskan | 63.08 | -117.06 | 15 | 4 | 5 | 6 | This study |
| Tanana | Athapaskan | 65.17 | -152.08 | 11 | 5 | 5 | 1 | Zegura et al. 2004 |
| Apache | Athapaskan | 33.35 | -110.45 | 94 | 75 | 14 | 5 | Zegura et al. 2004 |
| Navajo | Athapaskan | 33.83 | -109.96 | 75 | 72 | 1 | 2 | Zegura et al. 2004 |
| Seminole | Muskogean | 28.33 | -81.23 | 20 | 9 | 0 | 10 | Lell et al. 2002 |
| TM Chippewa | Algonquian | 48.84 | -99.74 | 34 | 3 | 0 | 30 | Bolnick et al. 2006 |
| W Chippewa | Algonquian | 45.07 | -91.27 | 29 | 8 | 0 | 20 | Bolnick et al. 2006 |
| Chey/Arap | Algonquian | 41.14 | -104.82 | 50 | 33 | 8 | 8 | Bolnick et al. 2006 |
| SWS Sioux | Macro-Siouan | 45.66 | -97.05 | 26 | 14 | 8 | 4 | Bolnick et al. 2006 |
| Stillwell Cherokee | Macro-Siouan | 34.26 | -85.16 | 30 | 15 | 0 | 14 | Bolnick et al. 2006 |
| Choctaw | Muskogean | 31.88 | -88.32 | 12 | 11 | 0 | 1 | Bolnick et al. 2006 |
| Creek | Muskogean | 32.46 | -84.99 | 12 | 9 | 1 | 2 | Bolnick et al. 2006 |
The Haplotype data set is comprised of both previously published and newly presented Y chromosome haplotype data from a total of 355 males representing 16 Native North American populations (Table 2). Haplotypes were determined with the genotype data for the following eight STRs: DYS19, DYS390, DYS391, DYS392, DYS393, DYS389I, DYS389II, and DYS439. Forty-three males from three populations (22 San Carlos Apache, 9 Dogrib, and 12 Seri) were genotyped for these eight STRs in the present study. These data were combined with published haplotype data from 169 males representing three populations from Zegura et al. (2004) and 143 males from 10 populations in Kemp et al. (submitted). All individuals that did not belong to haplogroup Q and C were excluded from the Haplotype data set because these haplotypes are likely the result of non-native admixture (Tarazona-Santos and Santos, 2002; Zegura et al., 2004; Bolnick et al, 2006). Data generated in the present study and that reported by Zegura et al. (2004) are given in Appendix A.
Table 2.
Sample diversity and sample size of Haplotype data set.
| Population | N | Reference |
|---|---|---|
| Pima | 29 | Kemp et al. submitted |
| Cora | 30 | Kemp et al. submitted |
| Huichol | 11 | Kemp et al. submitted |
| Mixe | 5 | Kemp et al. submitted |
| Mixtec | 20 | Kemp et al. submitted |
| Nahua Atocpan | 5 | Kemp et al. submitted |
| Tarahumara | 17 | Kemp et al. submitted |
| Zapotec | 6 | Kemp et al. submitted |
| Nahua Cuetzalan | 8 | Kemp et al. submitted |
| SC Apache | 22 | This study |
| Dogrib | 9 | This study |
| Seri | 12 | This study |
| Jemez | 12 | Kemp et al. submitted |
| Navajo | 71 | Zegura et al. 2004 |
| Apache | 88 | Zegura et al. 2004 |
| Tanana | 10 | Zegura et al. 2004 |
DNA extraction and genotyping
DNA was extracted from 200 μl of serum or from buccal swabs using the Qiagen Blood Amp Kit. The DNA extracts were ‘whole genome amplified’ using degenerate oligonucleotide primers (DOP) to increase the amount of DNA template (Cheung and Nelson, 1996) and analyzed for the following binary single nucleotide polymorphisms (SNP): Q-M3, Q-M242 (Seielstad et al., 2003), C-RPS4Y711 (Bergen et al., 1999) and R-M173 (Underhill et al., 2000) to assign each sample to haplogroup Q, C, or R, respectively.
PCR amplifications for the SNP genotyping were carried out in 20 μl reactions containing 1.5 μl of DNA template, 200 μM of each dNTP, 1.2 mM MgCl2, 0.1 μM of each primer, 0.5 units Platinum Taq, and 2.0 μl of the 10X buffer supplied by the manufacturer (Invitrogen). A program of 35 cycles of 1 minute (min) of melting at 94°C, 1 min annealing at 61.0°C for Q-M3 (annealing at 56°C for Q-M242 and at 59° C for M-3), and 1 min extension at 72°C was used for PCR amplification in a Perkin-Elmer 9700 thermocycler. A program of 60 cycles of 15 seconds (sec) of melting at 94°C, 20 sec annealing at 58°C for C-RPS4Y711 and at 50°C for R-M173, and 15 sec extension at 72°C was used for the remaining PCR amplifications. Six microliters of PCR product were electrophoresed in 6% polyacrylamide gels, and the size of each PCR product was determined by comparison to a known standard after staining with ethidium bromide.
Six μl of PCR product tested for the presence of mutations diagnostic of lineages Q-M3, Q-M242, and C-RPS4Y711 were incubated overnight at 37°C with five units of the appropriate restriction endonuclease (as described in Bolnick et al. 2006) and analyzed by electrophoresis as above. Fourteen μl of R-M173 PCR product were purified with Exo I following the supplier's recommendations. Eight μl of purified product were submitted to the CA&ES sequencing facility, on the UC Davis campus, for forward and reverse sequencing on an automatic ABI 3730 genetic analyzer to identify the point mutation that defines R-M173.
Eight Y-STR loci were typed using the Reliagene Y-Plex™ Kit, which includes DYS19, DYS389I and II, DYS390, DYS391, DYS392, DYS393, DYS438 and DYS439. PCR reactions were carried out in 12 μl containing 0.6 μM of each primer, 200 μM of each dNTP, 1.7 mM MgCl2, 0.3 units Platinum Taq, 1.25 ul of the 10X buffer supplied by the manufacturer (Invitrogen), and 2 μl of DNA template. A program of 35 cycles of 1 min of melting at 94°C, 1 min annealing at 55°C and 1 min extension at 72°C was used for PCR amplification of the STRs. Two or three microliters of Reliagene Y-Plex™ 12 kit PCR product were loaded on an ABI Prism 310 Genetic Analyzer (Perkin Elmer). The samples were injected for 22 sec at 15,000 V for 25 min with a run temperature of 60°C using a 310 Genetic Analyzer POP™4. The allele sizes were determined with GeneScan 3.1 software.
Statistical analysis
Two previous studies (Zegura et al., 2004; Bolnick et al., 2006) have demonstrated that frequencies of Native North American Y chromosome haplogroups exhibit a significant correlation with geography. To visualize patterns of Y chromosome haplogroup structure across North America, we interpolated haplogroup frequency using kriging, a commonly used geostatistical interpolation technique based on the weighted linear combination of point data. This same methodology was used to visualize patterns of mtDNA haplogroup structure in North America (Malhi et al., 2002). Kriging uses fitted variogram models to characterize spatial structure in the data (Jensen, 2007). The most parsimonious variogram model was chosen based on least squares fitting. Due to areas of North America that lack adequate data, only results that were supported by additional evidence are discussed here.
Genetic distance (FST and RST) were estimated using ARLEQUIN 2.0 (Schneider et al., 2000). The congruence between the two genetic distance matrices was measured using Kendall's W with the program CADM (Legendre, 2004). Principal coordinates were calculated from estimates of FST using the program SPSS. Due to the small sample sizes of some of the groups used in this analysis, FST was used instead of RST to be more conservative in our analysis (Caglia et al., 2003).
To investigate the relationship between haplotypes sampled from Southwest Athapaskans and those sampled from Subarctic Athapaskans, a median joining network of haplogroup C haplotypes, defined by eight STRs (those used in the Haplotype data set) was constructed using the program NETWORK (Bandelt, 1999). The contributions of STRs to the network were inversely weighted with allele frequency variances as in Bolnick et al. (2006).
Results
Haplogroup frequency distribution in North America
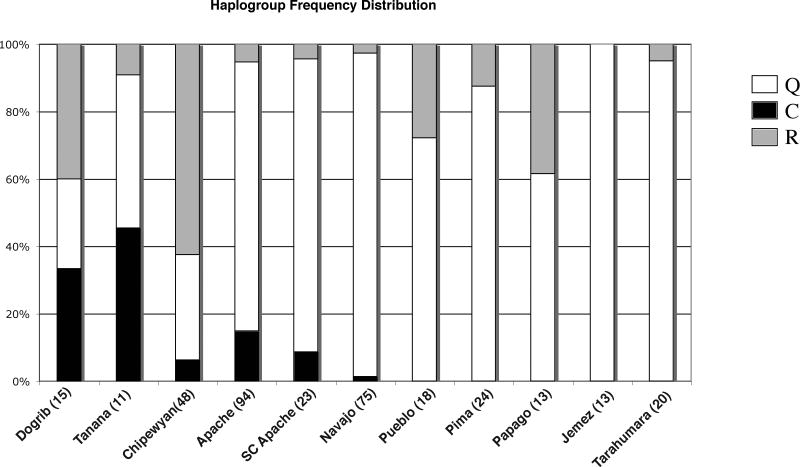
Gene map interpolations (Figures 2A, 2B, and 2C) indicate that the frequency of haplogroup Q is highest in Southwestern North America/Mesoamerica. The frequency of haplogroup C is highest in Northwestern North America and the frequency of haplogroup R, the presence of which is attributed to European admixture, reaches its maximum in Northeastern North America. Seventy-three percent of the populations analyzed exhibited haplogroup R, which in which ranges in frequency from 4% to 88% (Table 1).
Figure 2.
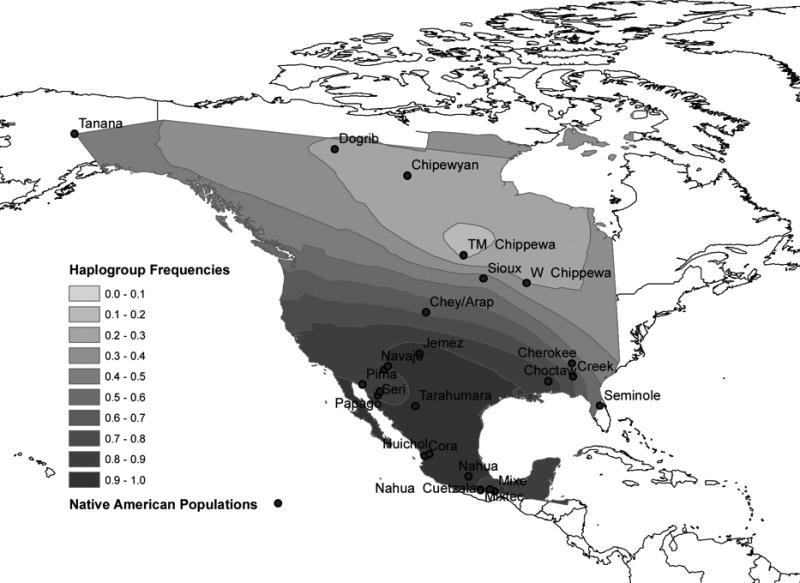
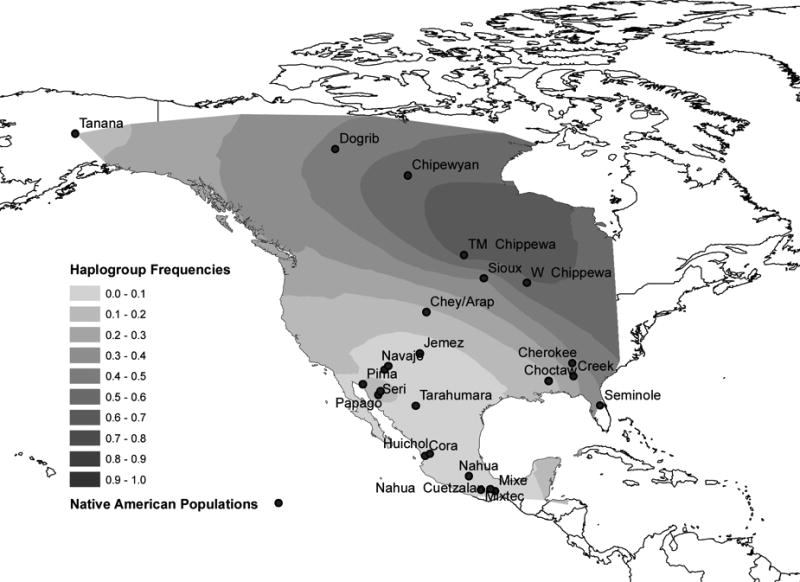
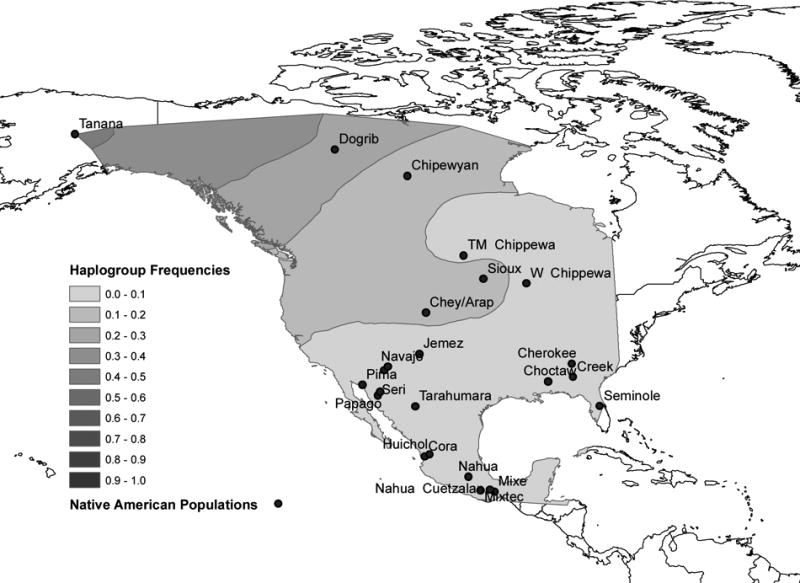
Figure 2a. Frequency of Haplogroup Q in North America.
Figure 2b. Frequency of Haplogroup R in North America.
Figure 2c. Frequency of Haplogroup C in North America.
Haplotype analysis among Athapaskans
Matrices of FST and RST show a high congruence (Kendall's W = 0.829). In the Southwest, only the Athapaskans exhibit haplotypes that belong to haplogroup C. (Table 1, Figure 3). The Apachean groups cluster more closely with Southwestern and Mesoamerican groups than with other Athapaskan populations in the principal coordinates analysis (Figure 4).
Figure 3.
Y chromosome haplogroup frequency distribution for Athapaskan and Southwest populations.
Figure 4.
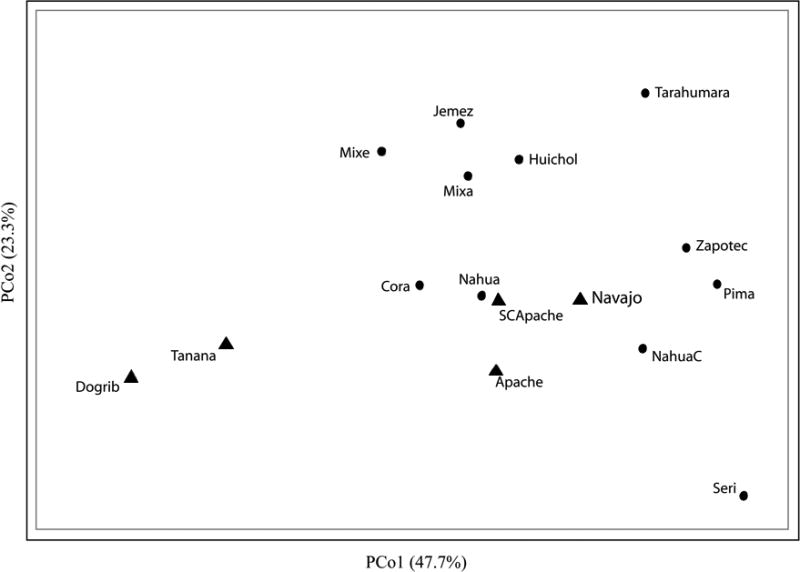
Principal coordinates analysis of the Haplotype dataset based on 8 STRs. Athapaskans are represented by ▲.
The median joining network for haplogroup C haplotypes (Figure 5) reveals that a majority of the haplotypes found in the Apachean populations belong to a derived subclade, as also reported by Zegura et al. (2004). Subarctic Athapaskans are not found in the derived clade in this study or in Zegura et al. (2004).
Figure 5.
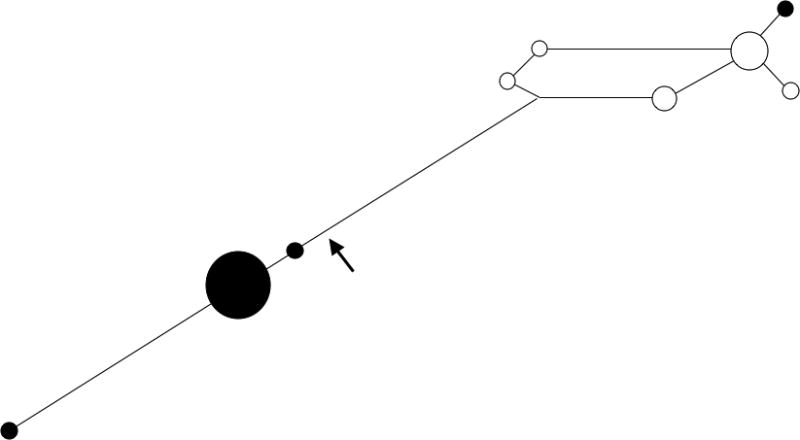
Haplogroup C Network of Athapaskan-speakers based on 8 STRs. Arrow indicates the branch leading to the Apachean subclade. The size of each circle corresponds to the number of individuals belonging to that haplotype. Black circles represent haplotypes identified in Apachean individuals. White circles represent haplotypes identified in Subarctic Athapaskan individuals. The lengths of the lines correspond to the number of mutational differences between haplotypes.
Discussion
European admixture
The influence of European admixture is evidenced by the strong gradient of haplogroup R from northeastern to southwestern North America. This gradient corroborates findings from previous studies of Y chromosome variation in Native Northeastern America (Hammer et al., 2006; Bolnick et al., 2006). The gradient might result from the earlier occurrence of European contact in Northeastern North America, which has provided a longer period of time for admixture to occur. Unlike mtDNA analysis (Smith et al., 1999), Y chromosome analysis shows that Native Americans harbor a high frequency of alleles of European ancestry, likely a result of European male introgression following the time of European contact. In addition, it is hypothesized that European contact altered the population structure of indigenous males in other ways as well, as historical records document a high mortality and forced relocations of males beginning shortly after initial European contact (Spicer, 1962). This study provides evidence of the former, and places future studies of Y chromosomes from prehistoric Native American populations in a key position testing these hypotheses.
Southern Athapaskan male population history
Previous mtDNA studies of Athapaskan groups have demonstrated that a high frequency of Apacheans belonged to a specific subclade of haplogroup A (defined as the A2a clade by Tamm et al. 2007), which exhibits transitions at nucleotide positions (nps) 16192, 16233, and 16331 (Malhi et al., 2003). This pattern is consistent with a founder effect and suggests that Apachean maternal lineages may be traced to a small number of females. Similarly, the Y chromosome data analyzed in this study reflect a pattern consistent with a founder effect, suggesting that Apachean paternal lineages also may be traced to a small number of males. Y chromosome haplogroup C is observed at a moderate frequency in the Subarctic Athapaskan groups and at a low frequency in the Navajo and Apache, but is otherwise absent from the Southwest. Nearly all Navajo and Apache Y chromosomes within haplogroup C belong to a specific, well-defined subclade (Zegura et al., 2004). Hence, it is likely that ancestral Subarctic Athapaskan-speakers provided the source for Y chromosome haplogroup C as well as the mtDNA A2a subclade in Apachean groups.
However, Apachean groups cluster with other Southwest and Mesoamerican groups in the principal coordinates analysis, rather than with Athapaskans from the Subarctic. This suggests that the majority of non-C Y chromosomes in the Navajo and Apache were contributed by non-Athapaskan populations in the Southwest, which mirrors the presence in the Apachean of mtDNA lineages belonging to haplogroups B and C. It is also possible that the effects of European contact may have significantly skewed the frequency of haplotypes in the Southwest.
The founder effect exhibited by the Apachean Y chromosome data is compatible with several different scenarios. It is possible that a large group of emigrants began the journey from the Subarctic but only a small proportion of this group was successful in reaching the Southwest. Alternatively, a large group of closely related migrants could have migrated to the Southwest. However, there is no archaeological evidence of a large migration of Athapaskans to the Southwest (Basso, 1983; Cordell, 1997), despite the relatively recent occurrence of the migration and extensive archaeological investigations in the Southwest over the past century. Nor is the scenario of a large number of migrants reaching the Southwest compelling given the mtDNA evidence that Apacheans spread their language and culture predominantly by assimilating native Southwestern individuals. Rather, if a large group of migrants reached the Southwest with a successful culture, one might expect to find evidence of population growth in the data. In light of the mtDNA, archaeological, and linguistic evidence, the scenario of a small number of male and female migrants traveling from the Subarctic to the Southwest and subsequently spreading their language and culture by assimilation of Southwest individuals is the most likely explanation for the patterns observed in the Y chromosome data in this study.
The idea that a very small population of Apachean speakers was responsible for the geographically extensive spread of Apachean languages is in some ways counter-intuitive because it implies a level of language/cultural assimilation often thought to correspond to a situation in which the donor group is viewed as being culturally dominant or superior to the indigenous groups (Spicer, 1962). It is scarcely conceivable that the occupants of the large Puebloan agricultural centers regarded the comparatively meager material and social culture of Apachean hunter-gatherers sufficiently superior to abandon their own lifeways. Nor can the capture of individuals from these well-defended centers have been a major source of Apachean recruitment. The more likely source was very small social units living on the Puebloan fringe, partly supporting themselves by trade with major population centers (Çordell, 1997). These socially and economically marginalized peoples may have assimilated with less difficulty into Apachean society. Disruption within the Pueblo world caused by environmental deterioration and social conflict during the 13th and 14th centuries would have added to this fringe Puebloan population, the likely source of the Southwestern genetic markers that came to dominate Apachean populations. Archaeological investigation of fringe groups and material culture surrounding major Puebloan centers could provide a test of this proposed fringe Puebloan absorption scenario.
Conclusion
The results of this study suggest that the Y chromosome population structure of Native North America was significantly altered by European admixture. European admixture has resulted in a decreasing gradient of haplogroup R from Northeastern to Southwestern North America. The large effect of European admixture suggests that caution should be used when inferring prehistoric population events in Native North Americans from Y chromosome data alone.
In addition, we identified closely related Y chromosomes among Athapaskans in the Subarctic and the Southwest to the exclusion of non-Athapskans in the Southwest. Consistent with mtDNA studies of Athapaskan-speakers, the Y chromosome analysis identifies a pattern consistent with a founder effect in Apachean groups. Together with archaeological and linguistic evidence, the genetic (Y chromosome and mtDNA) data suggests a scenario of a small number of male and female migrants traveling from the Subarctic to the Southwest and subsequently spreading their language and culture by assimilating members of native Southwestern tribes.
This study provides a clear example of how linguistic and genetic variation were decoupled and evolved independently in a population as predicted by Boas (1911). It is through the interdisciplinary comparison of linguistic, archaeological and genetic information that we were able to infer the processes (e.g. long-distance migration, gene flow, cultural diffusion) that likely resulted in the current distribution of Southern Athapaskan languages. As with this study, additional interdisciplinary efforts will likely provide insight into the biological and cultural evolution of human populations.
Acknowledgments
We are indebted to the numerous personnel of Indian Health Services Facilities, where most of the samples studied were obtained, as well as to individuals who provided and authorized use of their samples in this study. This study was supported by National Science Foundation grant 0422144 to R.S.M., B.M.K., D.G.S., and A.R. and by NIH grant RR05090 to D.G.S. We thank DGAPA, Universidad Nacional Autonoma de Mexico who provided support to A.G.O. and to the late John McDonough, Robert L. Bettinger and Graciela Cabana for helpful comments.
Appendix A
| Sample ID | Population | Haplogroup | DYS19 | DYS390 | DYS391 | DYS392 | DYS393 | DYS389-I | DYS389-II |
|---|---|---|---|---|---|---|---|---|---|
| IM022 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM025 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM046 | Apache | C | 15 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM049 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM053 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM057 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM079 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM092 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM093 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM095 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM103 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM112 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM116 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| IM123 | Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| 32 | SC Apache | C | 15 | 22 | 9 | 11 | 12 | 13 | 30 |
| 396 | SC Apache | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| AR01 | Navajo | C | 16 | 23 | 10 | 11 | 13 | 13 | 28 |
| AK02 | Tanana | C | 15 | 23 | 9 | 11 | 12 | 13 | 30 |
| AK14 | Tanana | C | 15 | 23 | 9 | 11 | 12 | 13 | na |
| AK20 | Tanana | C | 15 | 23 | 9 | 11 | 12 | 13 | 29 |
| AK24 | Tanana | C | 15 | 23 | 9 | 11 | 12 | 13 | 31 |
| AK28 | Tanana | C | 15 | 23 | 9 | 11 | 12 | 13 | 29 |
| 11 | Dogrib | C | 15 | 23 | 9 | 11 | 12 | 13 | 30 |
| 31 | Dogrib | C | 15 | 23 | 9 | 11 | 12 | 13 | 30 |
| 52 | Dogrib | C | 15 | 23 | 9 | 11 | 12 | 13 | 29 |
| 59 | Dogrib | C | 15 | 23 | 9 | 11 | 12 | 13 | 30 |
| 60 | Dogrib | C | 15 | 23 | 9 | 11 | 12 | 13 | 30 |
| IM026 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM027 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 14 | 32 |
| IM028 | Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| IM029 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM031 | Apache | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| IM032 | Apache | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| IM033 | Apache | Q | 13 | 24 | 10 | 14 | 14 | 13 | 30 |
| IM035 | Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| IM037 | Apache | Q | 13 | 22 | 11 | 14 | 13 | 13 | 30 |
| IM038 | Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 30 |
| IM039 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM040 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM042 | Apache | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| IM044 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM045 | Apache | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| IM047 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM050 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM051 | Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| IM054 | Apache | Q | 13 | 22 | 10 | 14 | 13 | 13 | 30 |
| IM056 | Apache | Q | 13 | 24 | 10 | 14 | 13 | 13 | 30 |
| IM058 | Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| IM060 | Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 30 |
| IM064 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM066 | Apache | Q | 13 | 24 | 10 | 14 | 12 | 13 | 30 |
| IM067 | Apache | Q | 13 | 24 | 10 | 16 | 13 | 12 | 28 |
| IM068 | Apache | Q | 14 | 24 | 10 | 16 | 13 | 12 | 28 |
| IM069 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM073 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM077 | Apache | Q | 13 | 24 | 10 | 14 | 13 | 13 | 30 |
| IM080 | Apache | Q | 14 | 24 | 9 | 14 | 14 | 13 | 30 |
| IM081 | Apache | Q | 13 | 23 | 10 | 14 | 14 | 13 | 31 |
| IM089 | Apache | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| IM091 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM098 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM101 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 14 | 32 |
| IM102 | Apache | Q | 13 | 24 | 10 | 15 | 14 | 13 | 29 |
| IM104 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM106 | Apache | Q | 13 | 23 | 10 | 12 | 13 | 12 | 31 |
| IM107 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM109 | Apache | Q | 13 | 24 | 10 | 14 | 12 | 13 | 30 |
| IM110 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM118 | Apache | Q | 13 | 24 | 10 | 15 | 14 | 13 | 30 |
| IM120 | Apache | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| IM122 | Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 30 |
| IM124 | Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| NC001 | Apache | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| IM041 | Apache | Q | 14 | 25 | 10 | 14 | 13 | 13 | 30 |
| IM043 | Apache | Q | 13 | 24 | 10 | 14 | 13 | 13 | 30 |
| IM048 | Apache | Q | 13 | 24 | 10 | 14 | 14 | 13 | 29 |
| IM055 | Apache | Q | 13 | 24 | 10 | 14 | 14 | 13 | 29 |
| IM083 | Apache | Q | 13 | 24 | 10 | 14 | 14 | 13 | 29 |
| IM097 | Apache | Q | 14 | 24 | 10 | 16 | 13 | 13 | 30 |
| IM105 | Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 29 |
| IM113 | Apache | Q | 13 | 24 | 10 | 14 | 14 | 13 | 29 |
| IM021 | Apache | Q | 14 | 23 | 10 | 14 | 13 | 13 | 32 |
| IM024 | Apache | Q | 15 | 23 | 10 | 14 | 13 | 13 | 29 |
| IM036 | Apache | Q | 13 | 24 | 10 | 16 | 13 | 14 | 30 |
| IM059 | Apache | Q | 13 | 24 | 10 | 14 | 14 | 13 | 29 |
| IM061 | Apache | Q | 13 | 24 | 10 | 14 | 14 | 13 | 29 |
| IM065 | Apache | Q | 13 | 23 | 11 | 16 | 13 | 13 | 29 |
| IM070 | Apache | Q | 13 | 25 | 10 | 14 | 14 | 13 | 29 |
| IM082 | Apache | Q | 14 | 23 | 10 | 14 | 13 | 13 | 32 |
| IM084 | Apache | Q | 13 | 24 | 11 | 15 | 14 | 13 | 30 |
| IM085 | Apache | Q | 13 | 24 | 10 | 14 | 13 | 12 | 29 |
| IM086 | Apache | Q | 14 | 25 | 10 | 16 | 13 | 13 | 30 |
| IM087 | Apache | Q | 14 | 25 | 10 | 16 | 13 | 13 | 30 |
| IM088 | Apache | Q | 14 | 25 | 10 | 16 | 13 | 13 | 30 |
| IM090 | Apache | Q | 14 | 25 | 10 | 16 | 13 | 13 | 30 |
| IM094 | Apache | Q | 14 | 25 | 10 | 16 | 13 | 13 | 30 |
| IM096 | Apache | Q | 13 | 24 | 10 | 14 | 14 | 13 | 29 |
| IM099 | Apache | Q | 14 | 23 | 10 | 14 | 13 | 13 | 32 |
| IM108 | Apache | Q | 14 | 24 | 10 | 16 | 13 | 13 | 30 |
| IM114 | Apache | Q | 14 | 23 | 10 | 14 | 13 | 13 | 32 |
| IM115 | Apache | Q | 14 | 24 | 10 | 15 | 13 | 14 | 30 |
| 23 | SC Apache | Q | 13 | 24 | 11 | 14 | 14 | 13 | 31 |
| 26 | SC Apache | Q | 13 | 24 | 10 | 15 | 14 | 13 | 29 |
| 77 | SC Apache | Q | 13 | 23 | 10 | 14 | 14 | 13 | 30 |
| 124 | SC Apache | Q | 13 | 24 | 11 | 16 | 13 | 13 | 30 |
| 166 | SC Apache | Q | 14 | 23 | 10 | 14 | 13 | 13 | 30 |
| 220 | SC Apache | Q | 13 | 24 | 11 | 16 | 13 | 13 | 30 |
| 228 | SC Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| 239 | SC Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 30 |
| 243 | SC Apache | Q | 13 | 24 | 11 | 14 | 14 | 13 | 31 |
| 248 | SC Apache | Q | 13 | 24 | 11 | 14 | 14 | 13 | 30 |
| 258 | SC Apache | Q | 13 | 24 | 10 | 14 | 13 | 13 | 30 |
| 263 | SC Apache | Q | 13 | 23 | 11 | 16 | 13 | 13 | 29 |
| 318 | SC Apache | Q | 13 | 24 | 10 | 14 | 14 | 13 | 29 |
| 400 | SC Apache | Q | 13 | 24 | 10 | 14 | 14 | 13 | 29 |
| 406 | SC Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 30 |
| 416 | SC Apache | Q | 14 | 25 | 10 | 14 | 14 | 14 | 31 |
| 459 | SC Apache | Q | 13 | 24 | 11 | 13 | 14 | 14 | 30 |
| 495 | SC Apache | Q | 14 | 23 | 10 | 14 | 13 | 13 | 30 |
| 510 | SC Apache | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| 524 | SC Apache | Q | na | 23 | 10 | na | 13 | 13 | 31 |
| NC2 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| NC5 | Navajo | Q | 13 | 23 | 10 | 14 | 13 | 15 | 31 |
| NC6 | Navajo | Q | 13 | 25 | 10 | 14 | 12 | 14 | 31 |
| NC13 | Navajo | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| NC24 | Navajo | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| NC29 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 14 | 32 |
| NC30 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 14 | 32 |
| NC43 | Navajo | Q | 13 | 23 | 10 | 14 | 13 | 15 | 31 |
| NC045 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 14 | 32 |
| NC100 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 14 | 32 |
| NC101 | Navajo | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| NC104 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 12 | 30 |
| NC107 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| NC109 | Navajo | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| NC113 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| NC118 | Navajo | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| NC119 | Navajo | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| Nj371 | Navajo | Q | 13 | 24 | 10 | 16 | 13 | 12 | 28 |
| Nj373 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 12 | 30 |
| Nj391 | Navajo | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| Nj395 | Navajo | Q | 13 | 26 | 10 | 14 | 13 | 13 | 30 |
| Nj402 | Navajo | Q | 13 | 23 | 10 | 14 | 14 | 14 | 30 |
| Nj414 | Navajo | Q | 15 | 23 | 10 | 14 | 14 | 14 | 31 |
| Nj416 | Navajo | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| Nj422 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| Nj445 | Navajo | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| Nj473 | Navajo | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| NM09 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| NM11 | Navajo | Q | 13 | 23 | 10 | 14 | 13 | 14 | 30 |
| NM13 | Navajo | Q | 13 | 24 | 10 | 16 | 13 | 12 | 29 |
| NM14 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 14 | 32 |
| NM15 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 12 | 29 |
| NM16 | Navajo | Q | 13 | 24 | 10 | 16 | 13 | 12 | 28 |
| NM18 | Navajo | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| NM19 | Navajo | Q | 13 | 24 | 10 | 16 | 13 | 12 | 28 |
| NM20 | Navajo | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| NM22 | Navajo | Q | 13 | 23 | 10 | 15 | 13 | 14 | 31 |
| NM24 | Navajo | Q | 13 | 24 | 10 | 16 | 13 | 12 | 29 |
| NM25 | Navajo | Q | 13 | 23 | 10 | 14 | 13 | 14 | 30 |
| NC105 | Navajo | Q | 15 | 23 | 10 | 14 | 13 | 14 | 32 |
| NC124 | Navajo | Q | 14 | 25 | 10 | 14 | 13 | 13 | 29 |
| NM04 | Navajo | Q | 14 | 25 | 10 | 16 | 13 | 13 | 30 |
| NM05 | Navajo | Q | 14 | 24 | 10 | 15 | 13 | 13 | 29 |
| NM06 | Navajo | Q | 13 | 24 | 10 | 14 | 13 | 13 | 28 |
| NAV01 | Navajo | Q | 13 | 24 | 10 | 14 | 13 | 13 | 30 |
| NAV10 | Navajo | Q | 14 | 25 | 10 | 14 | 13 | 13 | 29 |
| NC011 | Navajo | Q | 13 | 23 | 12 | 16 | 13 | 13 | 29 |
| NC041 | Navajo | Q | 14 | 23 | 10 | 14 | 13 | 13 | 31 |
| NC042 | Navajo | Q | 14 | 25 | 10 | 14 | 13 | 13 | 29 |
| NC044 | Navajo | Q | 13 | 24 | 10 | 14 | 13 | 13 | 30 |
| NC102 | Navajo | Q | 14 | 25 | 10 | 14 | 13 | 13 | 29 |
| NC103 | Navajo | Q | 13 | 24 | 10 | 14 | 13 | 13 | 30 |
| NC106 | Navajo | Q | 14 | 23 | 10 | 14 | 13 | 14 | 32 |
| NC110 | Navajo | Q | 13 | 24 | 10 | 14 | 13 | 13 | 28 |
| NC111 | Navajo | Q | 13 | 24 | 10 | 14 | 13 | 13 | 30 |
| NC112 | Navajo | Q | 14 | 25 | 10 | 14 | 13 | 13 | 29 |
| NC114 | Navajo | Q | 14 | 23 | 10 | 14 | 13 | 13 | 31 |
| NC115 | Navajo | Q | 14 | 25 | 10 | 14 | 13 | 13 | 30 |
| NC120B | Navajo | Q | 13 | 24 | 10 | 14 | 13 | 13 | 28 |
| NC123 | Navajo | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| Nj439 | Navajo | Q | 13 | 24 | 10 | 14 | 13 | 13 | 30 |
| Nj446 | Navajo | Q | 13 | 24 | 11 | 16 | 13 | 12 | 29 |
| Nj481 | Navajo | Q | 14 | 25 | 10 | 14 | 13 | 13 | 30 |
| Nj495 | Navajo | Q | 13 | 24 | 10 | 14 | 13 | 13 | 30 |
| NM01 | Navajo | Q | 15 | 23 | 10 | 14 | 13 | 13 | 30 |
| NM03 | Navajo | Q | 13 | 24 | 10 | 14 | 13 | 13 | 30 |
| NM08 | Navajo | Q | 14 | 23 | 10 | 14 | 13 | 14 | 32 |
| NM10 | Navajo | Q | 13 | 22 | 11 | 14 | 13 | 14 | 30 |
| NM12 | Navajo | Q | 14 | 23 | 10 | 14 | 13 | 14 | 33 |
| NM23 | Navajo | Q | 13 | 23 | 11 | 16 | 13 | 13 | 29 |
| AK18 | Tanana | Q | 13 | 24 | 10 | 14 | 12 | 13 | 30 |
| AK31 | Tanana | Q | 13 | 24 | 10 | 14 | 14 | 14 | 31 |
| AK32 | Tanana | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| AK33 | Tanana | Q | 13 | 23 | 10 | 14 | 13 | 13 | 32 |
| AK36 | Tanana | Q | 13 | 22 | 10 | 14 | 13 | 13 | 30 |
| 1 | Dogrib | Q | 13 | 23 | 10 | 14 | 13 | 13 | 30 |
| 8 | Dogrib | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| 54 | Dogrib | Q | 13 | 23 | 10 | 14 | 13 | 13 | 31 |
| 57 | Dogrib | Q | 13 | 23 | 10 | 14 | 13 | 14 | 32 |
| Seri 59 | Seri | Q | 13 | 24 | 9 | 15 | 13 | 12 | 31 |
| Seri 58 | Seri | Q | 13 | 24 | 9 | 15 | 13 | 13 | 31 |
| Seri 9 | Seri | Q | 13 | 24 | 9 | 15 | 13 | 13 | 31 |
| Seri 102 | Seri | Q | 13 | 24 | 9 | 15 | 13 | 13 | 31 |
| Seri 57 | Seri | Q | 13 | 24 | 10 | 15 | 13 | 12 | 28 |
| Seri 23 | Seri | Q | 13 | 24 | 10 | 16 | 13 | 12 | 28 |
| Seri 29 | Seri | Q | 13 | 24 | 10 | 16 | 13 | 12 | 28 |
| Seri 40 | Seri | Q | 13 | 24 | 10 | 16 | 13 | 12 | 28 |
| Seri 79 | Seri | Q | 13 | 24 | 10 | 16 | 13 | 12 | 28 |
| Seri 86 | Seri | Q | 13 | 24 | 10 | 16 | 13 | 12 | 28 |
| Seri 36 | Seri | Q | 13 | 24 | 10 | 16 | 13 | 13 | 30 |
| Seri41 | Seri | Q | 13 | 24 | 10 | 14 | 13 | 13 | 30 |
References
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Basso KH. Western Apache. In: Ortiz A, editor. Handbook of North American Indians: Volume 10 Southwest. Smithsonian Institution; Washington D.C.: 1983. pp. 139–152. [Google Scholar]
- Bergen AW, Wang CY, Tsai J, Jefferson K, Dey C, Smith KD, Park SC, Tsai SJ, Goldman D. An Asian-Native American paternal lineage identified by RPS4Y resequencing and by microsatellite haplotyping. Ann Hum Genet. 1999;63:63–80. doi: 10.1046/j.1469-1809.1999.6310063.x. [DOI] [PubMed] [Google Scholar]
- Boas F. Handbook of American Indian languages. Bureau of American Ethnology, Bulletin 40; Washington, D.C.: 1911. Introduction; pp. 5–83. [Google Scholar]
- Bolnick DA, Bolnick DL, Smith DG. Asymmetric male and female genetic histories among Native Americans from Eastern North America. Mol Biol Evol. 2006;11:2161–74. doi: 10.1093/molbev/msl088. [DOI] [PubMed] [Google Scholar]
- Bortolini MC, Salzano MF, Thomas MG, Stuart S, Nasanen SP, Bau CH, Hutz MH, Layrisse Z, Petzl-Erler ML, Tsuneto LT, Hill K, Hurtado AM, Castro-de-Guerra D, Torres MM, Groot H, Michalski R, Nymadawa P, Bedoya G, Bradman N, Labuda D, Ruiz-Linares A. Y-chromosome evidence for differing ancient demographic histories in the Americas. Am J Hum Genet. 2003;73:524–39. doi: 10.1086/377588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglia A, Tofanelli S, Coia V, Boschi I, Pescarmona M, Spedini G, Pascali V, Paoli G, Destro-Bisol G. A study of Y-chromosome microsatellite variation in sub-Saharan Africa: a comparison between Fst and Rst genetic distances. Hum Biol. 2003;75:313–330. doi: 10.1353/hub.2003.0041. [DOI] [PubMed] [Google Scholar]
- Campbell L. American Indian Languages: The Historical Linguistics of Native America. Oxford University Press; Oxford; 1997. [Google Scholar]
- Cheung VG, Nelson SF. Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc Natl Acad Sci USA. 1996;93:14676–9. doi: 10.1073/pnas.93.25.14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell LS. Archaeology of the Southwest. San Diego, CA: Academic Press; 1997. [Google Scholar]
- Goddard I. The classification of the native languages of North America. In: Godard I, editor. Handbook of North American Indians: languages 17. Washington, DC: Smithsonian Institution; 1996. pp. 290–324. [Google Scholar]
- Hammer MF, Chamberlain VF, Kearney VF, Stover D, Zhang G, Karafet T, Walsh B, Redd AJ. Population structure of Y chromosome SNP haplogroups in the United States and forensic implications for constructing Y chromosome STR databases. Forensic Sci Int. 2006;164:45–55. doi: 10.1016/j.forsciint.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Hoijer H. The chronology of the Athapaskan languages. Int J Anthropol Linguist. 1956;22:219–232. [Google Scholar]
- Hollenbach JA, Thomson G, Cao K, Fernandez-Vina M, Erlich HA, Bugawan TL, Winkler C, Winter M, Klitz W. HLA diversity, differentiation, and haplotype evolution in Mesoamerican Natives. Hum Immunol. 2001;62:378–390. doi: 10.1016/s0198-8859(01)00212-9. [DOI] [PubMed] [Google Scholar]
- Hunley K, Long JC. Gene flow across linguistic boundaries in Native North American populations. Proc Natl Acad Sci USA. 2005;102:1312–7. doi: 10.1073/pnas.0409301102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymes DH. A note on Athapaskan glottochronology. Int J Am Linguist. 1957;23:291–297. [Google Scholar]
- Infante E, Olivo A, Alaez C, Williams F, Middleton D, de la Rosa G, Pujol MJ, Duran C, Navarro JL, Gorodezky C. Molecular analysis of HLA class I alleles in the Mexican Seri Indians: implications for their origin. Tissue Antigens. 1999;54:35–42. doi: 10.1034/j.1399-0039.1999.540104.x. [DOI] [PubMed] [Google Scholar]
- Jensen JR. Remote Sensing of the Environment: An Earth Resource Perspective. Prentice Hall; New Jersey: 2007. [Google Scholar]
- Kemp BM. Mesoamerica and Southwest prehistory, and the entrance of humans into the Americas: mitochondrial DNA evidence Department of Anthropology. University of California; Davis: 2006. [Google Scholar]
- Legendre P. Congruence among distance matrices: Program CADM. Department of Biological Sciences. University of Montreal; 2004. [Google Scholar]
- Lell JT, Sukernik RI, Starikovskaya YB, Su B, Jin L, Schurr TG, Underhill PA, Wallace DC. The dual origin and Siberian affinities of Native American Y chromosomes. Am J Hum Genet. 2002;70:192–206. doi: 10.1086/338457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz JG, Smith DG. Distribution of four founding mtDNA haplogroups among Native North Americans. Am J Phys Anthropol. 1996;101:307–329. doi: 10.1002/(SICI)1096-8644(199611)101:3<307::AID-AJPA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Malhi RS, Eshleman JA, Greenberg JA, Weiss DA, Schultz BA, Kemp BM, Kaestle FA, Lorenz JG, Johnson JR, Smith DG. The structure and diversity within New World mtDNA haplogroups: Implications for the prehistory of North America. Am J Hum Genet. 2002;70:905–919. doi: 10.1086/339690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi RS, Mortensen HM, Eshleman JA, Lorenz JG, Kaestle FA, Johnson JR, Gorodesky C, Smith DG. Native American mtDNA prehistory in the American Southwest. Am J Phys Anthropol. 2003;120:108–124. doi: 10.1002/ajpa.10138. [DOI] [PubMed] [Google Scholar]
- Monroe C, Kemp BM, Smith DG. Mitochondrial DNA variation of Yuman speaking populations. American Association of Physical Anthropologists; Alaska: 2006. [Google Scholar]
- Sapir E. Internal linguistic evidence suggestive of the northern origin of Navaho. Am Anthropol. 1936;38:224–235. [Google Scholar]
- Schneider S, Roessli D, Exoffier DL. Arlequin version 2.0: a software for population genetic data analysis. Geneva, Switzerland: Genetics and Biometry Laboratory, University of Geneva; 2000. [Google Scholar]
- Seielstad M, Yuldasheva N, Singh N, Underhill P, Oefner P, Shen P, Wells RS. A novel Y-chromosome variant puts an upper limit on the timing of first entry into the Americas. Am J Hum Genet. 2003;73:700–5. doi: 10.1086/377589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG, Malhi RS, Eshleman J, Lorenz JG, Kaestle FA. Distribution of mtDNA haplogroup X among Native North Americans. Am J Phys Anthropol. 1999;110:271–284. doi: 10.1002/(SICI)1096-8644(199911)110:3<271::AID-AJPA2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Smith DG, Lorenz J, Rolfs BK, Bettinger RL, Green B, Eshleman J, Schultz B, Malhi R. Implications of the distribution of albuman Naskapi and albumin Mexico for New World prehistory. Amer J Phys Anthropol. 2000;111:557–572. doi: 10.1002/(SICI)1096-8644(200004)111:4<557::AID-AJPA10>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Spicer EH. Cycles of Conquest: The Impact of Spain, Mexico, and The United States on the Indians of the Southwest, 1533-1960. University of Arizona Press; tuscon, Arizona: 1962. [Google Scholar]
- Suarez B, Crouse J, O'Rourke D. Genetic variation in North Amerindian populations: the geography of gene frequencies. Am J Phys Anthropol. 1985;67:217–232. doi: 10.1002/ajpa.1330670307. [DOI] [PubMed] [Google Scholar]
- Tamm E, Kivisild T, Reidla M, Metspalu M, Smith DG, Mulligan CJ, Bravi CM, Rickards O, Martinez-Labarga C, Khusnutdinova EK, Fedorova SA, Golubenko MV, Stepanov VA, Gubina MA, Zhadanov SI, Ossipova LP, Damba L, Voevoda MI, Dipierri JE, Villems R, Malhi RS. Beringian Standstill and Spread of Native American Founders. PLoS ONE. 2007;2(9):e829. doi: 10.1371/journal.pone.0000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona-Santos E, Santos FR. The peopling of the Americas: a second major migration? Am J Hum Genet. 2002;70:1377–1380. doi: 10.1086/340388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Zemans R, Oefner PJ, Cavalli-Sforza LL. A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc Natl Acad Sci USA. 1996;93:196–200. doi: 10.1073/pnas.93.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Shen AA, Lin J, et al. Y chromosome sequence variation and the history of human populations. Nat Genet. 2000;26:358–361. doi: 10.1038/81685. 21 co-authors. [DOI] [PubMed] [Google Scholar]
- Zegura SL, Karafet TM, Zhivotovsky LA, Hammer MF. High-resolution SNPs and microsatellite haplotypes point to a single, recent entry of Native American Y chromosomes into the Americas. Mol Biol Evol. 2004;21:164–75. doi: 10.1093/molbev/msh009. [DOI] [PubMed] [Google Scholar]