Abstract
An integrated metabolic profile reflects the combined influence of genetic, epigenetic, and environmental factors that affect the candidate pathway of interest. Recent evidence suggests that some autistic children may have reduced detoxification capacity and may be under chronic oxidative stress. Based on reports of abnormal methionine and glutathione metabolism in autistic children, it was of interest to examine the same metabolic profile in the parents. The results indicated that parents share similar metabolic deficits in methylation capacity and glutathione-dependent antioxidant/detoxification capacity observed in many autistic children. Studies are underway to determine whether the abnormal profile in parents reflects linked genetic polymorphisms in these pathways or whether it simply reflects the chronic stress of coping with an autistic child.
Keywords: Autism, homocysteine, glutathione, DNA methylation, parents
INTRODUCTION
Autism is a behaviorally-defined neurodevelopmental disorder that is usually diagnosed in early childhood and is characterized by impairment in reciprocal communication and speech, social withdrawal, and repetitive hyper-focused behaviors. The 10-fold increase in autism diagnosis in the last decade now affecting ∼1 in 150 children in the US has justifiably raised great public health concern (CDC 2005). Although both genetic and environmental factors are thought to be involved in the genesis of autism, none as yet have been reproducibly identified. Metabolic dysfunction has not been extensively studied in autistic children despite the fact that chronic biochemical imbalance is often a primary factor in the development of complex neurologic disease (Pennington et al. 2007; Kilbourne et al. 2007; Giordano et al. 2007).
In a recent study, we measured baseline levels of methionine transmethylation and transsulfuration metabolites in plasma from 80 autistic children (James et al. 2006). Briefly, we showed that the mean ratio of plasma S-adenosylmethionine (SAM) to S-adenosylhomocysteine (SAM/SAH ratio), an index of cellular methylation capacity, was 50% that in 75 age-matched control children. We also demonstrated a decrease in glutathione (GSH), the major intracellular antioxidant and an increase in oxidized glutathione (GSSG), resulting in a ∼3-fold reduction in the ratio of reduced (active) GSH to oxidized (inactive) glutathione (GSH/GSSG). Glutathione precursors were also lower suggesting inadequate GSH synthesis. These new findings are of concern because they indicate a significant decrease in methylation capacity (↓SAM/SAH) and redox potential (↓GSH/GSSG) and an increase in oxidative stress (↑GSSG). Evidence from several other laboratories similarly indicates that biomarkers of oxidative stress may be increased in some autistic children. (Ming et al. 2005; Chauhan et al. 2004; Sogut et al. 2003; Zoroglu et al. 2004; Yao et al. 2006b)
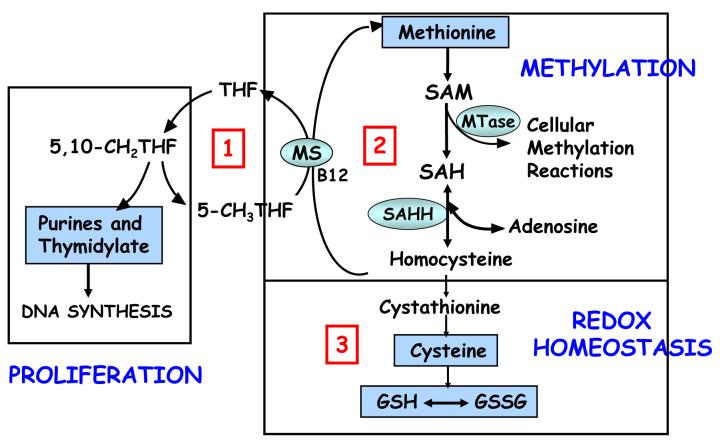
Figure 1 is an overview of the three interdependent pathways involved in folate-dependent methionine transmethylation and transsulfuration. Pathway 1 is the folate cycle, pathway 2 is the methionine cycle, and Pathway 3 is the transsulfuration pathway leading to glutathione synthesis. The functional importance of these pathways is underscored by their essentiality for error-free DNA synthesis (Pathway 1); for cellular methylation reactions including DNA, RNA, protein, and phospholipid methylation (Pathway 2); and for the maintenance of cellular redox and detoxification capacity (Pathway 3). The GSH/GSSG ratio reflects the redox potential of the intracellular environment which is critical for maintenance of normal redox signaling, antioxidant and detoxification capacity (Wu et al. 2004; Filomeni et al. 2002; Pastore et al. 2003). Glutathione is present in millimolar concentrations inside the cell and is the major determinant of intracellular redox homeostasis and cell detoxification/antioxidant capacity. Cell functions affected by glutathione deficit include cell proliferation (e.g., immune function, DNA synthesis and repair), essential methylation (e.g., DNA, RNA, protein, phospholipid, neurotranmittors, creatine) and redox homeostasis (e.g., cell signaling, detoxification, stress response, cell cycle progression, and apoptosis).
Figure 1.
A diagram of tetrahydrofolate (THF)-dependent methionine transmethylation and glutathione synthesis. The methionine cycle (transmethylation) involves the regeneration of methionine from homocysteine via the B12-dependent transfer of a methyl group from 5-methyl-tetrahydrofolate (5-CH3THF) via the methionine synthase (MS) reaction. Methionine is then activated to S-adenosylmethionine (SAM), the methyl donor for multiple cellular methyltransferase (MTase) reactions and the methylation of essential molecules such as DNA, RNA, proteins, phospholipids, creatine, and neurotransmittors. The transfer of the methyl group from SAM results in the demethylated product S-adenosylhomocysteine (SAH). The reversible hydrolysis of SAH to homocysteine and adenosine by the SAH hydrolase (SAHH) reaction completes the methionine cycle. Homocysteine can then be either remethylated to methionine or irreversibly removed from the methionine cycle by cystathionine beta synthase (CBS). This is a one-way reaction that permanently removes homocysteine from the methionine cycle and initiates the transsulfuration pathway for the synthesis of cysteine and glutathione. Glutathione is shown in its active reduced form (GSH) and inactive oxidized disulfide form (GSSG).
DNA methylation is a post-translational modification of DNA that orchestrates a wide variety of functions including differentiation, tissue-specific gene expression, chromatin structure, imprinted gene expression, silencing of parasitic DNA, and X-chromosome inactivation (Razin 1998; Reik and Dean 2001; Robertson and Jones 2000; Hermann et al. 2004). Folate-dependent transmethylation metabolism is integrally involved in establishing and maintaining patterns of DNA methylation (Friso and Choi 2002; Niculescu and Zeisel 2002). DNA hypomethylation is an “epigenetic” modification that is associated with nutritional and genetic deficiencies in methionine metabolism (Jacob et al. 1998; Yi et al. 2000; Fuso et al. 2005; Friso et al. 2002c) aging (Friso and Choi 2002), cancer (Dreosti 1998), and toxic environmental exposures (Feil 2006a; Abdolmaleky et al. 2004). Epigenetic change refers to heritable alteration in gene expression without a change in DNA sequence. The ability of gene expression to adapt and respond to developmental cues and environmental exposures is primarily due to post-translational modifications of DNA methylation and histone methylation/acetylation that alter transcription factor accessibility (Abdolmaleky et al. 2004; Feil 2006b).
The possibility that autism may involve an epigenetic component is supported by reports indicating that only 60-90% of monozygotic twins are concordant for autism (Kates et al. 2004). The fact that the concordance is not 100% strongly suggests that epigenetic DNA alterations and/or gene-environment interactions contribute to the development of autism (Beaudet 2002; Badcock and Crespi 2006). Moreover, the frequency of autism is increased in children with genetic diseases that alter DNA methylation density or function including Fragile X disease, Angelman Syndrome, and Rett Syndrome (Schanen 2006; Lopez-Rangel and Lewis 2006; Samaco et al. 2005). Impaired methylation and glutathione depletion have been documented in several neuropsychiatric disorders including schizophrenia (Regland et al. 1994; Yao et al. 2006a), bipolar disorder (Kuratomi et al. 2007; Frey et al. 2007), and autism (James et al. 2004; James et al. 2006). Based on these considerations and the abnormal transmethylation and transsulfuration metabolites observed in children with autism, it was of considerable interest to examine these pathways in the parents of autistic children as well as their global DNA methylation status.
METHODS
Study Participants
Case parents were mothers and fathers of children who had been diagnosed with Autistic Disorder based on Diagnostic and Statistical Manual of Mental Disorders-Fourth Edition (DSM-IV) and the Childhood Autistic Rating Scales (CARS). A total of 86 parents participated in the study consisting of 46 mothers and 40 fathers. Of the 86 parents, 72 were mother-father pairs (36 pairs), and ranged in age from 21 to 45 years (mean age ± SD: 30 ± 5). The ethnicity distribution among case parents was 87% White, 9% Asian, and 4% African American. Control parents for the metabolic study consisted of 200 mothers with a mean age of 28 (range 17-43 years) who were control participants in an ongoing case-control study of maternal risk factors for congenital heart defects (Hobbs et al. 2005a). The ethnicity among the control parents was 75% White, 7% Hispanic, 16% African American, and 2% Asian. All case and control parents were Arkansas residents. Although over-the-counter multivitamin supplement intake was significantly higher among control compared to case mothers (51% vs. 20%), metabolite levels were not significantly altered by vitamin intake within either group (p>0.05 for all metabolites). The protocol was approved by the Institutional Review Board at the University of Arkansas for Medical Sciences and all participants signed informed consent.
Sample treatment and HPLC methodology
Fasting blood samples were collected before 9:00 am into EDTA-Vacutainer tubes and immediately chilled on ice before centrifuging at 4000 × g for 10 minutes at 4°C. Aliquots of plasma were transferred into cryostat tubes and stored at -80°C for approximately 2-4 weeks until extraction and HPLC quantification. For the determination of total homocysteine (tHcy), methionine, cysteine, and total glutathione, 50 μl freshly prepared 1.43 M sodium borohydride solution containing 1.5 μM.
EDTA, 66 mM NaOH and 10 μl isoamyl alcohol were added to 200 μl plasma to reduce all sulfhydryl bonds and the samples were incubated at 40°C in a shaker for 30 minutes. To precipitate proteins, 250 μl ice cold 10% meta-phosphoric acid was added, mixed well, and the sample was incubated for an additional 10 minutes on ice. After centrifugation at 18,000 × g for 15 minutes at 4°C, the supernatant was filtered through a 0.2 μm nylon membrane filter (PGC Scientific, Frederic, MD) and a 20 μl aliquot was injected into the HPLC system.
For determination of SAM, SAH and free oxidized (GSSG) and free reduced (GSH) glutathione, 100 μl of 10% meta-phosphoric acid was added to 200 μl plasma to precipitate protein; the solution was mixed well, and incubated on ice for 30 minutes. After centrifugation for 15 minutes at 18,000 g at 4°C, supernatants were passed through a 0.2 μm nylon membrane filter and 20 μl was injected into the HPLC system.
The methodological details for HPLC elution and electrochemical detection have been described previously (Melnyk et al. 2000; Melnyk et al. 1999). The analyses were performed using HPLC with a Shimadzu solvent delivery system (ESA model 580) and a reverse phase C18 column (3 μm; 4.6 x 150 mm, MCM, Inc., Tokyo, Japan) obtained from ESA, Inc. (Chemsford, MA). The plasma extract was directly injected onto the column using Beckman Autosampler (model 507E). All plasma metabolites were quantified using a model 5200A Coulochem II and CoulArray electrochemical detection systems (ESA, Inc., Chelmsford, MA) equipped with a dual analytical cell (model 5010), a 4 channel analytical cell (model 6210) and a guard cell (model 5020). The concentrations of plasma metabolites were calculated from peak areas and standard calibration curves using HPLC software. Although it was not technically feasible to run all case and control samples simultaneously, the -80°C storage interval was consistently between 2 and 4 weeks to minimize potential metabolite interconversion.
HPLC determination of 5-methylcystosine and total cystosine in DNA
Genomic DNA was extracted from primary lymphocytes using the Puregene Blood Kit from Gentra Systems, Inc. (Minneapolis, MN). RNase A (Sigma, St. Louis, MO) was added to a final concentration of 0.02 mg/mL and DNA incubated at 37°C for 15 minutes. The purified DNA was digested into component nucleotides using Nuclease P1, snake venom phosphodieasterase, and alkaline phosphatase as previously described (Friso et al. 2002a). Briefly, DNA was denatured by heating for 3 minutes at 100° C and rapidly chilled in an ice water bath. One-tenth volume of 0.1 M ammonium acetate, pH 5.3, was added to 2 units of nuclease P1 (Sigma, St. Louis, MO) for every 0.5 A260 unit of DNA and the mixture incubated at 45°C for 2 h. Subsequently, 1/10 volume of 1 M ammonium bicarbonate and 0.002 units of venom phophodiesterase I (Sigma, St. Louis, MO) were added and the mixture incubated for 2 h at 37° C. To the mixture, 0.5 units of alkaline phosphatase (Sigma, St. Louis, MO) was then added and the incubation continued for an additional hour. The digested nucleotides were stored at - 20°C until HPLC analysis.
Statistical Analysis
Case-control differences were calculated using the Student t test with statistical significance set at p≤ 0.05. Linear regression analysis was used to calculate the correlation coefficients between selected metabolites and level of 5-methylcytosine using software within Microsoft Excel. Odds ratios (OR) for case compared to control mothers were calculated as an estimate of association between metabolite levels and the relative risk of being a mother of an autistic child. Throughout, a two-tailed p value of 0.05 was interpreted as a statistically significant difference.
RESULTS
Plasma transmethylation metabolite levels among cases and controls
The data presented in Table 1 indicate that plasma levels of methionine and SAM were not statistically different among case parents and controls. However, the mean levels of homocysteine and SAH were significantly elevated in case mothers compared to control mothers. Statistical evaluation of case fathers was not possible because blood samples from control fathers were not available from this cohort. Elevations in mean homocysteine and/or SAH levels were not statistically different between case mothers and fathers. [Table 1]
TABLE 1. Comparison of plasma metabolite concentrations in the transmethylation pathway between case and control parents.
| Plasma Transmethylation Metabolites (Mean ± SD) | Control Mothers N=200 | Case Mothers N=46 | Case Fathers N=40 |
|---|---|---|---|
| Methionine (μMol/L) | 26 ± 4.7 | 23 ± 4.1 | 26 ± 5.5 |
| SAM (nMol/L) | 83 ± 14 | 78 ± 20 | 85 ± 16 |
| SAH (nMol/L) | 23 ± 8.1 | 33 ± 14a | 37 ± 15 |
| % of subjects with SAH > 30 μMol/L | 14.9% | 54.3%a | 59% |
| SAM/SAH Ratio | 4.0 ± 1.4 | 2.8 ± 1.6a | 2.7 ± 1.4 |
| Homocysteine (μMol/L) | 7.42 ± 1.7 | 9.8 ± 3.6a | 10.6 ± 2.5 |
P<0.001 Case vs. Control mothers
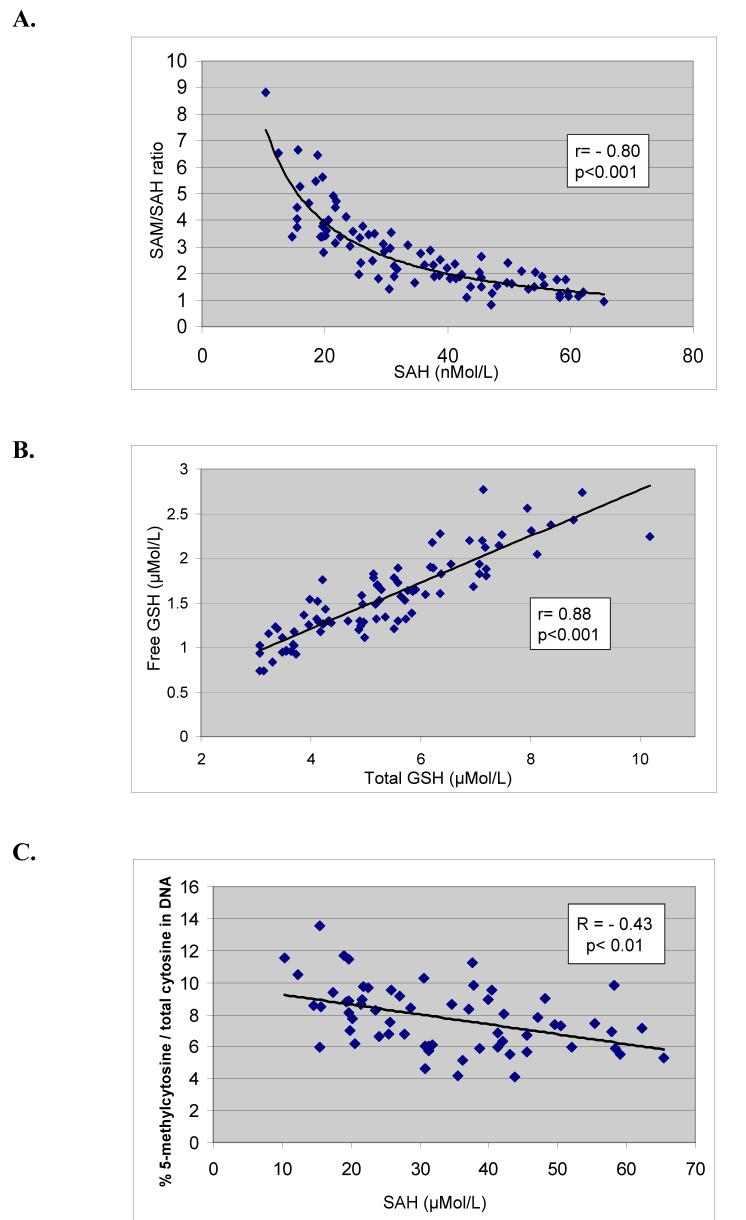
Because SAH is a potent product inhibitor of the DNA methyltransferase, the results were further stratified by SAH levels. Previous studies have shown that SAH levels >30 μMol/L are associated with significant DNA hypomethylation (Yi et al. 2000). As shown in Table 1, among the 200 controls, 14.9% had SAH levels > 30 μMol/L whereas 56.4% of the 86 case parents had SAH levels >30 μMol/L. Both mother and father contributed a blood sample in 72 of the 86 participants (36 pairs). In 39% of the parent pairs (14/36), both parents had SAH levels >; 30 μMol/L. Whereas mean SAM levels were not different between cases and controls, the ratio of SAM/SAH among the case parents was significantly decreased primarily reflecting the increase in SAH levels. Figure 2 demonstrates the strong and significant negative correlation between SAH levels and the SAM/SAH ratio among case parents (r = -0.80; p<0.001).
Figure 2.
A. A scatterplot showing the correlation between plasma levels of SAH and the ratio of SAM/SAH among the case parents indicating that the SAM/SAH ratio is primarily driven by the concentration of SAH. B. A scatterplot showing the strong correlation between free and total plasma glutathione levels among case parents. C. A scatterplot showing the correlation between plasma SAH levels and DNA hypomethylation (% 5-methylcytosine/total cytosine in DNA).
Plasma transsulfuration metabolite levels among cases and controls
Total GSH reflects combined protein-bound and free GSH after reduction of disulfide bonds whereas free GSH reflects GSH remaining after protein precipitation. Relative to control levels, parents of autistic children were found to have significant decreases in both total and free GSH levels and in the GSH/GSSG ratios whereas the levels of the oxidized disulfide GSSG were significantly increased (Table 2). Cysteine levels among the case parents were not significantly different from controls. Figure 3 shows a strong and significant correlation between plasma total GSH and free GSH among case parents (r=0.88; p<0.001). [Table 2]
TABLE 2. Comparison of the plasma metabolite concentrations between case and control parents.
| Plasma Transsulfuration Metabolites (Mean ± SD) | Control Mothers N=200 | Case Mothers N=46 | Case Fathers N=40 |
|---|---|---|---|
| Cysteine (μMol/L) | 229 ± 22 | 227 ± 34 | 243 ± 356 |
| Total GSH (μMol/L) | 7.3 ± 1.6 | 5.1 ± 1.4a | 5.7 ± 1.6 |
| Free GSH (μMol/L) | 2.6 ± 0.7 | 1.5 ± 0.4a | 1.6 ± 0.5 |
| GSSG (μMol/L) | 0.24 ± 0.07 | 0.32 ± 0.08a | 0.30 ± 0.09 |
| Total GSH/GSSG Ratio | 31 ± 10 | 17 ± 8.3a | 20.2 ± 7.2 |
| Free GSH/GSSG Ratio | 10.7 ± 4.2 | 5.1 ± 2.2a | 5.6 ± 2.2a |
Case vs. Control mothers; p< 0.001
DNA methylation and metabolite correlations
The percent 5-methylcytosine/total cytosine in digested DNA from case and control parents is presented in Table 3. Data are presented for combined parents because there is no known gender effect on DNA methylation. DNA from parents with SAH levels >30 μMol/L was more hypomethylated than parents with normal SAH levels (5.9% vs. 7.0% 5-methylcytosine/cytosine; p < 0.001). There was no statistical difference in DNA methylation between parents or controls who had SAH levels <30 μMol/L. The scatterplot in Figure 4 shows the negative correlation between SAH and % 5-methylcytosine in DNA (r = -0.43; p<0.01) consistent with the inhibitory effect of SAH on the DNA methyltransferase. There was no correlation between 5-methylcytosine and SAM or homocysteine (data not shown). [Table 3]
TABLE 3. Comparison of the percent methylcytosine/total cytosine in DNA stratified by SAH levels.
| Group | N | %-Methylcytosine/Cytosine (Mean ± SD) |
|---|---|---|
| Combined Parents | 86 | 6.4±1.3b |
| Controls with SAH<30 μMol/L | 15 | 6.6±0.5b |
| Parents with SAH<30 μMol/L | 38 | 7.0±1.5b |
| Parents with SAH>30 μMol/L | 48 | 5.9±1.2a |
P<0.001: SAH>30 vs. SAH<30
Not statistically different
SAM/SAH and GSH/GSSG and maternal odds ratios
The data in Table 4 present the calculated odds ratios (OR) and 95% confidence intervals (CI) associated with maternal SAH levels and ratios of SAM/SAH and GSH/GSSG independently and combined. An elevation in SAH >30 μmol/L or a SAM/SAH ratio <2.5 increased the likelihood of being a mother with an autistic child by 7.3-fold and 10.7-fold, respectively. In the transsulfuration pathway, mothers whose GSH/GSSG ratio was < 20 had a 15.2-fold increase in the likelihood of being a mother of a child with autism. Most remarkably, among mothers who exhibited both SAM/SAH ratio <2.5 and a GSH/GSSG ratio <20 (41% of mothers), the odds ratio was 46. Odds ratios were not calculated for case fathers because metabolic data from control fathers was not available. [Table 4]
TABLE 4. Odds ratios and 95% confidence intervals for the association between maternal plasma biomarkers and the likelihood of being a mother of an autistic child.
| Stratified Group | Control Mothersa N (%) | Case Mothersb N (%) | Odds Ratio (95% CI) |
|---|---|---|---|
| SAH >30μMol/L) | 28 (14) | 25 (54.3) | 7.3 (3.6, 14.8) |
| SAM/SAH <2.5 | 20 (10) | 25 (54.3) | 10.7 (5.1, 22.5) |
| tGSH/GSSG <20 | 22 (11) | 30 (65.2) | 15.2 (7.2, 32.1) |
| SAM/SAH <2.5 and tGSH/GSSG<20 | 3 (1.5) | 19 (41.3) | 46.2 (12.8, 166.5) |
N=200
N=46
DISCUSSION
The results of this study demonstrate for the first time that some parents of children with autism exhibit significant metabolic deficits in methylation capacity (↓SAM/SAH) and in glutathione-mediated antioxidant and detoxification capacity (↓GSH/GSSG). A remarkably similar metabolic imbalance was recently observed in many autistic children (James et al. 2004; James et al. 2006). Preliminary evidence in the autistic children suggests that genetic polymorphisms affecting enzymes in transmethylation and transsulfuration metabolic pathways may contribute, in part, to the metabolic imbalance (James et al. 2006). The observation that both parents and children with autism share a common metabolic phenotype is consistent with an underlying genetic component. A heritable metabolic phenotype would provide a convenient target in the choice of candidate genes that predispose to complex disease and these studies in autistic families are currently underway.
Abnormal levels of metabolites in the both transmethylation and transsulfuration pathways were surprisingly prevalent among case parents (Tables 1 and 2). Both parents had significant increases in mean plasma homocysteine and SAH levels and significant decreases in the SAM/SAH ratio, total and free GSH, and the GSH/GSSG ratio. Whether the parental metabolic abnormalities are genetically-based or the result of adult dietary deficiencies and/or chronic pro-oxidant environmental exposures cannot be determined from the present data. Further, because the blood samples were taken 3-7 years after the index pregnancy, it is not possible to know whether the maternal metabolic imbalance was present during the index pregnancy and fetal development. This is a common dilemma in studies that attempt to define maternal risk factors for adverse birth outcomes. However, it is generally accepted that adult homocysteine levels and dietary patterns during the reproductive years tend to be relatively stable (Cuco et al. 2006). For example, maternal homocysteine levels measured after pregnancy were found not to differ from pre-pregnancy values (Walker et al. 1999). Glutathione levels in healthy adults plateau between 20 and 40 years of age suggesting that adult values are also relatively stable during childbearing years (Ono et al. 2001). Nonetheless, a prospective study would be required to determine whether the observed abnormal metabolic profile is present during pregnancy and whether it could provide a maternal risk factor for having a child with autism.
Accumulating evidence suggests that epigenetic alterations may be involved in the etiology of complex neuropsychiatric diseases that do not follow classic Mendelian inheritance and are influenced by gene-environment interactions (Abdolmaleky et al. 2004; Mattson 2003; Weaver et al. 2007). For example, aberrant DNA methylation in the promoter regions of CNS reelinRLN) and COMT genes has been implicated in schizophrenia and bipolar disorder (Abdolmaleky et al. 2004; Abdolmaleky et al. 2005; Abdolmaleky et al. 2006). Based on indirect evidence, several reports have speculated that epigenetic mechanisms may contribute to the development or predisposition to autism (Schanen 2006; Nakayama et al. 2006; Lopez-Rangel and Lewis 2006). Associated with the elevated homocysteine and SAH among the case parents was a significant decrease in genome-wide DNA methylation expressed as the percent 5-methylcytosine/total cytosine in DNA (Table 3). It is well established that an increase in homocysteine levels will induce the reversal of the SAH hydrolase (SAHH) reaction causing a concomitant elevation in SAH levels (James et al. 2002; Perna et al. 2003; Castro et al. 2003). In previous studies, we and others demonstrated that plasma homocysteine and SAH levels are positively associated with genome-wide DNA hypomethylation that is most likely due to product inhibition of the DNA methyltransferase by SAH (James et al. 2002; Caudill et al. 2001; Yi et al. 2000; McKeever et al. 1995). Elevated plasma homocysteine, SAH and DNA hypomethylation are common features of autoimmune disease (Richardson 2003) and as such may be contributing factors to the increased frequency of autoimmune disease in parents of autistic children. Further, dysregulation of imprinted gene expression and maternal valproic acid exposure have been associated with elevated SAH and DNA hypomethylation and both have been implicated as risk factors for autism (Ingrosso et al. 2000; Alonso-Aperte et al. 1999; Hogart et al. 2007; Ingram et al. 2000).
The finding of genome-wide DNA hypomethylation in a significant subset of autism parents with elevated SAH provides experimental data to support earlier speculation of epigenetic mechanisms that may be associated with autism. Elevated maternal homocysteine can cross the placental membrane (Guerra-Shinohara et al. 2002) and are correlated with fetal homocysteine levels(Murphy et al. 2004). Elevation in fetal homocysteine and SAH could theoretically alter fetal methylation patterns and induce inappropriate gene expression during development that could affect predisposition to autism. In a recent study, higher maternal plasma homocysteine at preconception, prior to fetal neurogenesis, was inversely associated with cognitive achievement in the offspring (Murphy et al. 2007). Also of related interest, elevated maternal homocysteine during the third trimester was recently found to be a risk factor for schizophrenia (Bleich et al. 2007). These studies raise concern that maternal homocysteine levels should be carefully monitored and controlled especially with high risk pregnancies.
The significant difference in metabolic profiles between control and case parents was an unexpected finding. Because of the differences in mean levels of SAH, SAM/SAH and GSH/GSSG ratio between case mothers and controls, odds ratios were calculated to give an indication of the association between these metabolite levels and the likelihood of being a mother of an autistic child. Among case mothers, 54% (25/46) had SAH levels >30 μmol/L or a SAM/SAH ratio <2.5 which was associated with 7.3-fold and 10.7-fold increased risk respectively of being a mother with an autistic child. Mothers whose GSH/GSSG ratio was < 20 had a 15.2-fold increase in the likelihood of being a mother of a child with autism. Among case mothers, 41% (19/46) exhibited a simultaneous decrease in both SAM/SAH and GSH/GSSG ratios resulting in a remarkable 46-fold increase in the likelihood of being a mother of an autistic child (Table 3).
Regardless of the origin, the finding that many parents of autistic children have an abnormal metabolic profile has important health implications in that an elevation in homocysteine and SAH levels and a decrease in SAM/SAH and GSH/GSSG ratios are well-established risk factors for heart disease, autoimmune disease, structural birth defects, and neurodegenerative disease (Morrison et al. 1999; De Bree et al. 2002; Lazzerini et al. 2007; Fidelus and Tsan 1987; Hobbs et al. 2005a; Hobbs et al. 2005b; Mattson and Shea 2003; Bains and Shaw 1997). Multiple studies have demonstrated that nutritional supplementation with B vitamins can lower homocysteine and SAH levels and that antioxidant supplementation can increase glutathione levels (Brattström et al. 1998; Zaidi et al. 2005; Ullegaddi et al. 2006). If future studies determine that a similar metabolic imbalance is present before and/or during an autism pregnancy, similar intervention strategies to normalize the maternal metabolic profile could prevent placental transfer and fetal exposure to abnormal metabolite levels.
A decrease in the GSH/GSSG redox ratio has been associated with several genetic polymorphisms, chronic dietary deficiencies, and pro-oxidant environmental exposures (Hayes and Strange 2000; Lyons et al. 2000; Greene 1995; Li et al. 2007). Glutathione can also be decreased by chronic psychological stress and severe anxiety (Eskiocak et al. 2005; Chakraborti et al. 2007). The two pathways of transmethylation and transsulfuration are metabolically interdependent such that chronic deficit in glutathione will feed back to inhibit SAM synthesis (Corrales at al. 1991) and create a chronic self-perpetuating cycle that progressively decreases GSH levels. The fact that both pathways were adversely affected in many parents suggests that the initiating factors were chronic in nature.
In summary, we have uncovered a significant metabolic imbalance in transmethylation and transsulfuration pathways in many parents that is similar to the imbalance previously observed in many autistic children. Linkage studies are underway to determine whether the shared metabolic profiles have a genetic component and psychological testing is being done to determine whether the parent profile could reflect, in part, the chronic psychological stress of coping with an autistic child. Although potentially significant, the results of this study should be considered preliminary until confirmed in subsequent studies.
Acknowledgements
The authors would like to express their gratitude to the participating families affected by autism in Arkansas without whom this study would not have been possible. This research was supported, in part, with funding from the National Institute of Child Health and Development (RO1 HD051873) to SJJ, and by grants from the University of Arkansas for Medical Sciences Children’s University Medical Group and the Arkansas Biosciences Institute (SJJ).
References
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, Smith CL, Shafa R, Aeali B, Carnevale J, Pan H, Papageorgis P, Ponte JF, Sivaraman V, Tsuang MT, Thiagalingam S. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum.Mol.Genet. 2006;15:3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, Shafa R, Glatt SJ, Nguyen G, Ponte JF, Thiagalingam S, Tsuang MT. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am.J.Med.Genet.B Neuropsychiatr.Genet. 2005;134:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- Abdolmaleky HM, Smith CL, Faraone SV, Shafa R, Stone W, Glatt SJ, Tsuang MT. Methylomics in psychiatry: Modulation of gene-environment interactions may be through DNA methylation. Am.J Med.Genet. 2004;127B:51–59. doi: 10.1002/ajmg.b.20142. [DOI] [PubMed] [Google Scholar]
- Alonso-Aperte E, Ubeda N, Achon M, Perez-Miguelsanz J, Varela-Moreiras G. Impaired methionine synthesis and hypomethylation in rats exposed to valproate during gestation. Neurology. 1999;52:750–756. doi: 10.1212/wnl.52.4.750. [DOI] [PubMed] [Google Scholar]
- Badcock C, Crespi B. Imbalanced genomic imprinting in brain development: an evolutionary basis for the aetiology of autism. J.Evol.Biol. 2006;19:1007–1032. doi: 10.1111/j.1420-9101.2006.01091.x. [DOI] [PubMed] [Google Scholar]
- Bains JS, Shaw CA. Neurodegenerative disorders in humans: the role of glutathione in oxidative stress-mediated neuronal death. Brain Res.Brain Res.Rev. 1997;25:335–358. doi: 10.1016/s0165-0173(97)00045-3. [DOI] [PubMed] [Google Scholar]
- Beaudet AL. Is medical genetics neglecting epigenetics? Genet.Med. 2002;4:399–402. doi: 10.1097/00125817-200209000-00013. [DOI] [PubMed] [Google Scholar]
- Bleich S, Frieling H, Hillemacher T. Elevated prenatal homocysteine levels and the risk of schizophrenia. Arch.Gen.Psychiatry. 2007;64:980–981. doi: 10.1001/archpsyc.64.8.980. [DOI] [PubMed] [Google Scholar]
- Brattström L, Landgren F, Israelsson B, Lindgren A, Hultberg B, Andersson A, Cuskelly G, McNulty H, Strain SS, McPartlin J, Weir DG, Scott JM, Den Heijer M, Brouwer IA, Blom HJ, Bos GM, Spaans A, Rosendaal FR, Thomas CM, Haak HL, Wijermans PW, Gerrits WB, Naurath HJ, Joosten E. Lowering blood homocysteine with folic acid based supplements: meta-analysis of randomised trials. Br.Med.J. 1998;316:894–898. [Google Scholar]
- Castro R, Rivera I, Struys EA, Jansen EEW, Ravasco P, Camilo ME, Blom HJ, Jakobs C, De Almeida IT. Increased, homocysteine and S- adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clinical Chemistry. 2003;49:1292–1296. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- Caudill MA, Wang JC, Melnyk S, Pogribny IP, Jernigan S, Collins MD, Swendseid ME, Cogger EA, James SJ. Intracellular S- adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl deficient cystathionine β-synthase heterozygous mice. Journal of Nutrition. 2001;131:2811–2818. doi: 10.1093/jn/131.11.2811. [DOI] [PubMed] [Google Scholar]
- CDC 2005 http://www.medicalhomeinfo.org/health/Autism%20downloads/AutismAlarm.pdf.
- Chakraborti A, Gulati K, Banerjee BD, Ray A. Possible involvement of free radicals in the differential neurobehavioral responses to stress in male and female rats. Behav.Brain Res. 2007;179:321–325. doi: 10.1016/j.bbr.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Chauhan V, Brown WT, Cohen I. Oxidative stress in autism: increased lipid peroxidation and reduced serum levels of ceruloplasmin and transferrin--the antioxidant proteins. Life Sci. 2004;75:2539–2549. doi: 10.1016/j.lfs.2004.04.038. [DOI] [PubMed] [Google Scholar]
- Corrales F, Ochoa P, Rivas C, Martin-Lomas M, Mato JM, Pajares MA. Inhibition of glutathione synthesis in the liver leads to S-adenosyl-L-methionine synthetase reduction. Hepatology. 1991;14:528–533. [PubMed] [Google Scholar]
- Cuco G, Fernandez-Ballart J, Sala J, Viladrich C, Iranzo R, Vila J, Arija V. Dietary patterns and associated lifestyles in preconception, pregnancy and postpartum. Eur.J.Clin.Nutr. 2006;60:364–371. doi: 10.1038/sj.ejcn.1602324. [DOI] [PubMed] [Google Scholar]
- De Bree A, Verschuren WM, Kromhout D, Kluijtmans LA, Blom HJ. Homocysteine determinants and the evidence to what extent homocysteine determines the risk of coronary heart disease. Pharmacol.Rev. 2002;54:599–618. doi: 10.1124/pr.54.4.599. [DOI] [PubMed] [Google Scholar]
- Dreosti IE. Nutrition, cancer, and aging. Ann.New York Acad.Sci. 1998;854:371–377. [PubMed] [Google Scholar]
- Eskiocak S, Gozen AS, Yapar SB, Tavas F, Kilic AS, Eskiocak M. Glutathione and free sulphydryl content of seminal plasma in healthy medical students during and after exam stress. Hum.Reprod. 2005;20:2595–2600. doi: 10.1093/humrep/dei062. [DOI] [PubMed] [Google Scholar]
- Feil R. Environmental and nutritional effects on the epigenetic regulation of genes. Mutat.Res. 2006a;600:46–57. doi: 10.1016/j.mrfmmm.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Feil R. Environmental and nutritional effects on the epigenetic regulation of genes. Mutat.Res. 2006b;600:46–57. doi: 10.1016/j.mrfmmm.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Fidelus RK, Tsan MF. Glutathione and lymphocyte activation: a function of ageing and auto-immune disease. Immunology. 1987;61:503–508. [PMC free article] [PubMed] [Google Scholar]
- Filomeni G, Rotilio G, Ciriolo MR. Cell signalling and the glutathione redox system. Biochem.Pharmacol. 2002;64:1057–1064. doi: 10.1016/s0006-2952(02)01176-0. [DOI] [PubMed] [Google Scholar]
- Fowden AL, Sibley C, Reik W, Constancia M. Imprinted genes, placental development and fetal growth. Horm.Res. 2006;65(Suppl 3):50–58. doi: 10.1159/000091506. [DOI] [PubMed] [Google Scholar]
- Frey BN, Andreazza AC, Kunz M, Gomes FA, Quevedo J, Salvador M, Goncalves CA, Kapczinski F. Increased oxidative stress and DNA damage in bipolar disorder: a twin-case report. Prog.Neuropsychopharmacol.Biol.Psychiatry. 2007;31:283–285. doi: 10.1016/j.pnpbp.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Friso S, Choi SW. Gene-nutrient interactions and DNA methylation. Journal of Nutrition. 2002;132:2382S–2387S. doi: 10.1093/jn/132.8.2382S. [DOI] [PubMed] [Google Scholar]
- Friso S, Choi SW, Dolnikowski GG, Selhub J. A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal.Chem. 2002a;74:4526–4531. doi: 10.1021/ac020050h. [DOI] [PubMed] [Google Scholar]
- Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R, Selhub J. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc.Natl.Acad.Sci.U.S.A. 2002b;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuso A, Seminara L, Cavallaro RA, D’Anselmi F, Scarpa S. S- adenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and beta-amyloid production. Mol.Cell Neurosci. 2005;28:195–204. doi: 10.1016/j.mcn.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Giordano V, Peluso G, Iannuccelli M, Benatti P, Nicolai R, Calvani M. Systemic and brain metabolic dysfunction as a new paradigm for approaching Alzheimer’s dementia. Neurochem.Res. 2007;32:555–567. doi: 10.1007/s11064-006-9125-8. [DOI] [PubMed] [Google Scholar]
- Greene LS. Asthma and oxidant stress: nutritional, environmental, and genetic risk factors. J Am.Coll.Nutr. 1995;14:317–324. doi: 10.1080/07315724.1995.10718516. [DOI] [PubMed] [Google Scholar]
- Guerra-Shinohara EM, Paiva AA, Rondo PH, Yamasaki K, Terzi CA, D’Almeida V. Relationship between total homocysteine and folate levels in pregnant women and their newborn babies according to maternal serum levels of vitamin B12. BJOG. 2002;109:784–791. doi: 10.1111/j.1471-0528.2002.01307.x. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Strange RC. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology. 2000;61:154–166. doi: 10.1159/000028396. [DOI] [PubMed] [Google Scholar]
- Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell.Mol.Life Sci. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs CA, Cleves MA, Melnyk S, Zhao W, James SJ. Congenital heart defects and abnormal maternal biomarkers of methionine and homocysteine metabolism. Am.J Clin.Nutr. 2005a;81:147–153. doi: 10.1093/ajcn/81.1.147. [DOI] [PubMed] [Google Scholar]
- Hobbs CA, Cleves MA, Zhao W, Melnyk S, James SJ. Congenital heart defects and maternal biomarkers of oxidative stress. Am.J.Clin.Nutr. 2005b;82:598–604. doi: 10.1093/ajcn.82.3.598. [DOI] [PubMed] [Google Scholar]
- Hogart A, Nagarajan RP, Patzel KA, Yasui DH, LaSalle JM. 15q11-13 GABAA receptor genes are normally biallelically expressed in brain yet are subject to epigenetic dysregulation in autism-spectrum disorders. Hum.Mol.Genet. 2007;16:691–703. doi: 10.1093/hmg/ddm014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingrosso D, D’angelo S, di Carlo E, Perna AF, Zappia V, Galletti P. Increased methyl esterification of altered aspartyl residues in erythrocyte membrane proteins in response to oxidative stress. Eur.J.Biochem. 2000;267:4397–4405. doi: 10.1046/j.1432-1327.2000.01485.x. [DOI] [PubMed] [Google Scholar]
- Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, Miller BJ, Henning SM, Swendseid ME. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. Journal of Nutrition. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- James SJ, Cutler P, Melnyk S, Jernigan S, Janak L, Gaylor DW, Neubrander JA. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am.J Clin.Nutr. 2004;80:1611–1617. doi: 10.1093/ajcn/80.6.1611. [DOI] [PubMed] [Google Scholar]
- James SJ, Melnyk S, Jernigan S, Cleves MA, Halsted CH, Wong DH, Cutler P, Bock K, Boris M, Bradstreet JJ, Baker SM, Gaylor DW. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. Am.J.Med.Genet.B Neuropsychiatr.Genet. 2006;141:947–956. doi: 10.1002/ajmg.b.30366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SJ, Melnyk S, Pogribna M, Pogribny IP, Caudill MA. Elevation in S- adenosylhomocysteine and DNA hypomethylation: Potential epigenetic mechanism for homocysteine-related pathology. Journal of Nutrition. 2002;132:2361S–2366S. doi: 10.1093/jn/132.8.2361S. [DOI] [PubMed] [Google Scholar]
- Kates WR, Burnette CP, Eliez S, Strunge LA, Kaplan D, Landa R, Reiss AL, Pearlson GD. Neuroanatomic variation in monozygotic twin pairs discordant for the narrow phenotype for autism. Am.J.Psychiatry. 2004;161:539–546. doi: 10.1176/appi.ajp.161.3.539. [DOI] [PubMed] [Google Scholar]
- Kilbourne AM, Brar JS, Drayer RA, Xu X, Post EP. Cardiovascular disease and metabolic risk factors in male patients with schizophrenia, schizoaffective disorder, and bipolar disorder. Psychosomatics. 2007;48:412–417. doi: 10.1176/appi.psy.48.5.412. [DOI] [PubMed] [Google Scholar]
- Kuratomi G, Iwamoto K, Bundo M, Kusumi I, Kato N, Iwata N, Ozaki N, Kato T. Aberrant DNA methylation associated with bipolar disorder identified from discordant monozygotic twins. Mol.Psychiatry. 2007 doi: 10.1038/sj.mp.4002001. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat.Rev.Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- Li Z, Dong T, Proschel C, Noble M. Chemically diverse toxicants converge on Fyn and c-Cbl to disrupt precursor cell function. PLoS Biol. 2007;5:e35. doi: 10.1371/journal.pbio.0050035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rangel E, Lewis ME. Loud and clear evidence for gene silencing by epigenetic mechanisms in autism and related neurodevelopmental disorders. Clin.Genet. 2006;69:21–22. doi: 10.1111/j.1399-0004.2006.00543a.x. [DOI] [PubMed] [Google Scholar]
- Lyons J, Rauh-Pfeiffer A, Yu YM, Lu XM, Zurakowski D, Tompkins RG, Ajami AM, Young VR, Castillo L. Blood glutathione synthesis rates in healthy adults receiving a sulfur amino acid-free diet. Proc.Natl.Acad.Sci U.S.A. 2000;97:5071–5076. doi: 10.1073/pnas.090083297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. Methylation and acetylation in nervous system development and neurodegenerative disorders. Ageing Res.Rev. 2003;2:329–342. doi: 10.1016/s1568-1637(03)00013-8. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Shea TB. Folate and homocysteine metabolism in neural plasticity and neurodegenerative disorders. Trends in Neurosciences. 2003;26:137–146. doi: 10.1016/S0166-2236(03)00032-8. [DOI] [PubMed] [Google Scholar]
- McKeever M, Molloy A, Weir DG, Young PB, Kennedy DG, Kennedy S, Scott JM. An abnormal methylation ratio induces hypomethylation in vitro in the brain of pig and man, but not in rat. Clin.Sci. 1995;88:73–79. doi: 10.1042/cs0880073. [DOI] [PubMed] [Google Scholar]
- Melnyk S, Pogribna M, Pogribny I, Hine RJ, James SJ. A new HPLC method for the simultaneous determination of oxidized and reduced plasma aminothiols using coulometric electrochemical detection. Journal of Nutritional Biochemistry. 1999;10:490–497. doi: 10.1016/s0955-2863(99)00033-9. [DOI] [PubMed] [Google Scholar]
- Melnyk S, Pogribna M, Pogribny IP, Yi P, James SJ. Measurement of plasma and intracellular S-adenosylmethionine and S-adenosylhomocysteine utilizing coulometric electrochemical detection: Alterations with plasma homocysteine and pyridoxal 5′-phosphate concentrations. Clinical Chemistry. 2000;46:265–272. [PubMed] [Google Scholar]
- Ming X, Stein TP, Brimacombe M, Johnson WG, Lambert GH, Wagner GC. Increased excretion of a lipid peroxidation biomarker in autism. Prostaglandins Leukot.Essent.Fatty Acids. 2005;73:379–384. doi: 10.1016/j.plefa.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Morrison JA, Jacobsen DW, Sprecher DL, Robinson K, Khoury P, Daniels SR. Serum glutathione in adolescent males predicts parental coronary heart disease. Circulation. 1999;100:2244–2247. doi: 10.1161/01.cir.100.22.2244. [DOI] [PubMed] [Google Scholar]
- Murphy MM, Fernandez-Ballart JD, Arija V, Scott JM, Molloy AM, Canals J. Maternal homocysteine at preconception is negatively correlated with cognitive achievement in children at 4 months and 6 years of age. Clinical Chemistry and Laboratory Medicine; Conference Proceedings, 6th International Conference on Homocysteine Metabolism; 2007.p. A23. [Google Scholar]
- Murphy MM, Scott JM, Arija V, Molloy AM, Fernandez-Ballart JD. Maternal homocysteine before conception and throughout pregnancy predicts fetal homocysteine and birth weight. Clin.Chem. 2004;50:1406–1412. doi: 10.1373/clinchem.2004.032904. [DOI] [PubMed] [Google Scholar]
- Nakayama A, Masaki S, Aoki E. Genetics and epigenetics in autism. Nihon Shinkei Seishin Yakurigaku Zasshi. 2006;26:209–212. [PubMed] [Google Scholar]
- Niculescu MD, Craciunescu CN, Zeisel SH. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 2006;20:43–49. doi: 10.1096/fj.05-4707com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu MD, Zeisel SH. Diet, methyl donors and DNA methylation: interactions between dietary folate, methionine and choline. J.Nutr. 2002;132:2333S–2335S. doi: 10.1093/jn/132.8.2333S. [DOI] [PubMed] [Google Scholar]
- Ono H, Sakamoto A, Sakura N. Plasma total glutathione concentrations in healthy pediatric and adult subjects. Clin.Chim.Acta. 2001;312:227–229. doi: 10.1016/s0009-8981(01)00596-4. [DOI] [PubMed] [Google Scholar]
- Pastore A, Federici G, Bertini E, Piemonte F. Analysis of glutathione: implication in redox and detoxification. Clin.Chim.Acta. 2003;333:19–39. doi: 10.1016/s0009-8981(03)00200-6. [DOI] [PubMed] [Google Scholar]
- Pennington K, Beasley CL, Dicker P, Fagan A, English J, Pariante CM, Wait R, Dunn MJ, Cotter DR. Prominent synaptic and metabolic abnormalities revealed by proteomic analysis of the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder. Mol.Psychiatry. 2007 doi: 10.1038/sj.mp.4002098. [DOI] [PubMed] [Google Scholar]
- Perna AF, Ingrosso D, Lombardi C, Acanfora F, Satta E, Cesare CM, Violetti E, Romano MM, De Santo NG. Possible mechanisms of homocysteine toxicity. Kidney Int. 2003;63:S137–S140. doi: 10.1046/j.1523-1755.63.s84.33.x. [DOI] [PubMed] [Google Scholar]
- Razin A. CpG methylation, chromatin structure and gene silencing - a three-way connection. EMBO Journal. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regland B, Johansson BV, Gottfries C-G. Homocysteinemia and schizophrenia as a case of methylation deficiency. J.Neural Transm. 1994;98:143–152. doi: 10.1007/BF01277017. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W. DNA methylation and mammalian epigenetics. Electrophoresis. 2001;22:2838–2843. doi: 10.1002/1522-2683(200108)22:14<2838::AID-ELPS2838>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Richardson B. DNA methylation and autoimmune disease. Clin.Immunol. 2003;109:72–79. doi: 10.1016/s1521-6616(03)00206-7. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- Samaco RC, Hogart A, LaSalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum.Mol.Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanen NC. Epigenetics of autism spectrum disorders. Hum.Mol.Genet. 2006;15(Spec No 2):R138–R150. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- Sogut S, Zoroglu SS, Ozyurt H, Ramazan YH, Ozugurlu F, Sivasli E, Yetkin O, Yanik M, Tutkun H, Savas HA, Tarakcioglu M, Akyol O. Changes in nitric oxide levels and antioxidant enzyme activities may have a role in the pathophysiological mechanisms involved in autism. Clin.Chim.Acta. 2003;331:111–117. doi: 10.1016/s0009-8981(03)00119-0. [DOI] [PubMed] [Google Scholar]
- Tycko B. DNA methylation in genomic imprinting. Mutat.Res.Rev.Mutat.Res. 1997;386:131–140. doi: 10.1016/s1383-5742(96)00049-x. [DOI] [PubMed] [Google Scholar]
- Ullegaddi R, Powers HJ, Gariballa SE. Antioxidant supplementation with or without B-group vitamins after acute ischemic stroke: a randomized controlled trial. JPEN J.Parenter.Enteral Nutr. 2006;30:108–114. doi: 10.1177/0148607106030002108. [DOI] [PubMed] [Google Scholar]
- Walker MC, Smith GN, Perkins SL, Keely EJ, Garner PR. Changes in homocysteine levels during normal pregnancy. American Journal of Obstetrics and Gynecology. 1999;180:660–664. doi: 10.1016/s0002-9378(99)70269-3. [DOI] [PubMed] [Google Scholar]
- Weaver IC, D’Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, Szyf M, Meaney MJ. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate- early genes. J.Neurosci. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- Yao JK, Leonard S, Reddy R. Altered glutathione redox state in schizophrenia. Dis.Markers. 2006a;22:83–93. doi: 10.1155/2006/248387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Walsh WJ, McGinnis WR, Pratico D. Altered vascular phenotype in autism: correlation with oxidative stress. Arch.Neurol. 2006b;63:1161–1164. doi: 10.1001/archneur.63.8.1161. [DOI] [PubMed] [Google Scholar]
- Yi P, Melnyk S, Pogribna M, Pogribny IP, Hines RJ, James SJ. Increase in plasma homocysteine associated with parallel increases in plasma S- adenosylhomocysteine and lymphocyte DNA hypomethylation. Journal of Biological Chemistry. 2000;275:29318–29323. doi: 10.1074/jbc.M002725200. [DOI] [PubMed] [Google Scholar]
- Zaidi SM, Al Qirim TM, Banu N. Effects of antioxidant vitamins on glutathione depletion and lipid peroxidation induced by restraint stress in the rat liver. Drugs R.D. 2005;6:157–165. doi: 10.2165/00126839-200506030-00004. [DOI] [PubMed] [Google Scholar]
- Zoroglu SS, Armutcu F, Ozen S, Gurel A, Sivasli E, Yetkin O, Meram I. Increased oxidative stress and altered activities of erythrocyte free radical scavenging enzymes in autism. Eur.Arch.Psychiatry Clin.Neurosci. 2004;254:143–147. doi: 10.1007/s00406-004-0456-7. [DOI] [PubMed] [Google Scholar]