Abstract
Detergents are commonly used for the extraction of hydrophobic proteins and must be removed for sensitive detection of peptides by mass spectrometry (MS). We demonstrate that ethyl acetate (EA) is able to extract octylglycoside (OG) from a protease digest without loss of peptides or interference with the MS peptide spectral profile. EA extraction was also found to reduce interference of SDS, NP-40 or Triton X-100 in the MS analysis.
Mass spectrometry (MS) is the preferred and dominant analytical tool for proteomic studies, including the identification of proteins and their post-translational modifications [1]. Protein sample preparation for MS analysis involves extraction and/or purification of proteins followed by site-specific enzymatic cleavage into small peptides. In-gel digestion of proteins separated by one dimensional or two dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) is the most commonly used procedure for protein separation. One disadvantage of in-gel digestion is low peptide yield. Furthermore, very large hydrophobic proteins are not efficiently resolved with SDS-PAGE [2]. The inclusion of low concentration of the saccharide-based detergent, octylglucoside (OG) or 5-cyclohexyl-1-pentyl-β-D-Maltoside, in the digestion buffer has been shown to increase peptide recovery from in-gel digestion [3, 4]. However, high concentrations of these detergents, interfere with the MS analysis [5]. Increased peptide recovery from in-gel protein digestion has also been achieved by using an expensive, acid-labile detergent that is degraded under acidic conditions into products that do not interfere with MS analysis [6].
In contrast to SDS-PAGE separation, protocols utilizing liquid phase separation result in high peptide yields and good resolution of large hydrophobic proteins and large protein complexes [2]. However, these procedures require the use of detergents such as Triton X-100, NP-40, or OG at concentrations of 0.5 – 1% to extract integral membrane proteins and to maintain the solubility of hydrophobic proteins throughout the protease digestion. The use of these detergents during protein purification also reduces non-specific protein-protein association and loss of protein due to adsorption to surfaces. OG has been described as MS compatible detergent and is among the commonly used detergents in protein extraction and isolation [3, 5]. Indeed, a dramatic improvement of selectivity in immunopurification of tyrosine phosphorylated peptides was shown by including 1% OG in the immunoprecipitation buffer [7]. However, 0.5–1% OG, Triton X-100 or NP-40 severely suppresses ionization in matrix-assisted laser desorption ionization-MS (MALDI-MS) [3] and decreases chromatographic resolution in liquid chromatography-MS (LC-MS). It is therefore necessary to remove these detergents prior to MS analysis [5]. This can be achieved by ion-exchange chromatography after protease digestion. However, this needs optimization for each digest, which results in an increase in volume and salt content and does not remove protein-bound detergent. Although the acid-labile detergent can be used with the liquid phase separation and digestion method [8], its high cost and the involvement of large-scale analyses severely limits its application.
In view of these limitations, we set out to find a simple method to remove OG after protease digestion by extracting the detergent with a water-immiscible organic solvent. The solubility of OG was tested against a variety of organic solvents. We found that OG is quite soluble in EA and used this solvent for further experiments. In addition to OG, both NP-40 and Triton X-100 are also soluble in EA, whereas ionic detergents like SDS (cationic) and dodecyl trimethylammonium bromide (anionic) are only sparingly soluble.
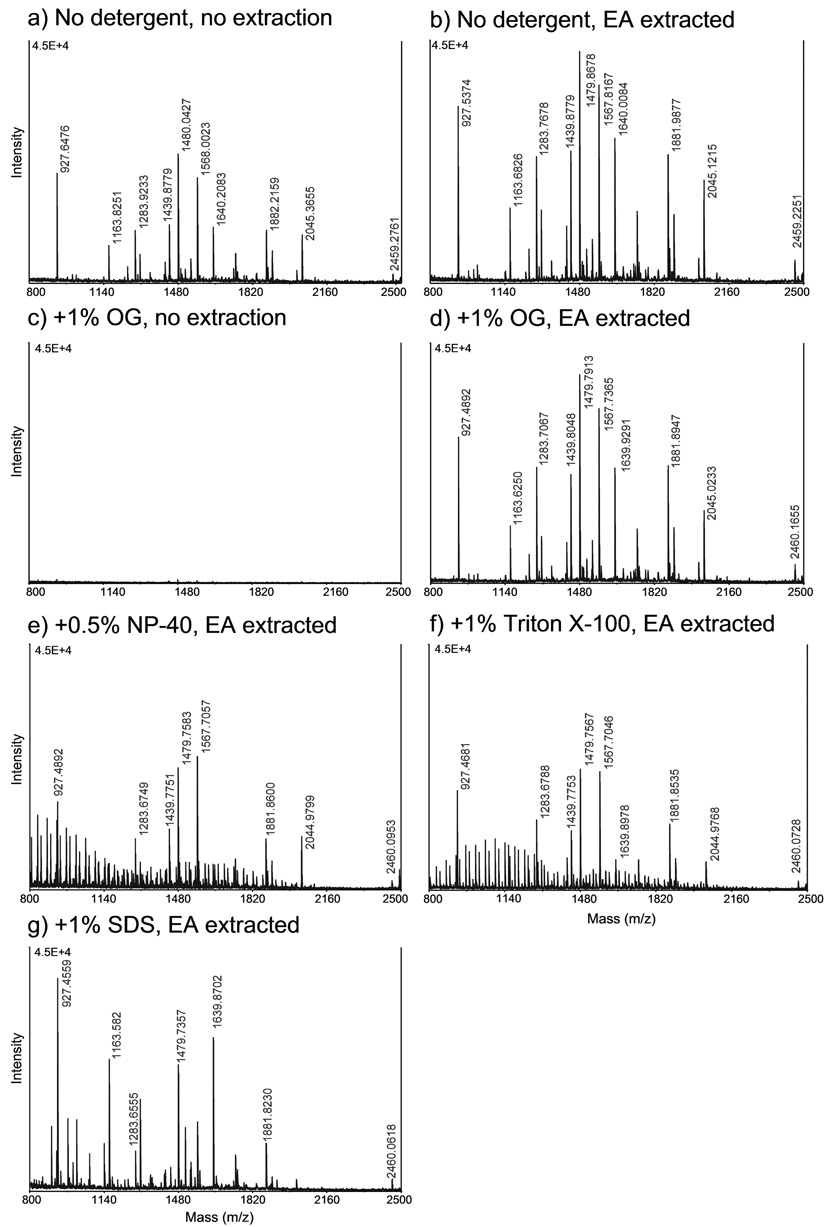
When a tryptic digest of bovine serum albumin (BSA) was extracted with 20 volumes of water saturated EA five times prior to MALDI-MS analysis (Supplementary Methods), there was an unexpected increase in peptide detection compared with unextracted tryptic digests (Fig. 1 a, b; Table 1, and Supplementary Table 1). This enhancing effect is mainly due to an increase in peptide signal from the EA-extracted sample that probably results from the removal by EA of unknown hydrophobic ionization-suppressing contaminants. Hence the EA extraction procedure is highly desirable even in the absence of detergent. The presence of 1% OG resulted in complete loss of peptide signals from the MALDI-MS spectrum (Fig. 1 c). This could be due to the blocking effect of the detergent on the C18 Zip tip and/or the suppression of ionization of the peptides by the detergent. EA extraction of the digest containing the same amount of OG eliminated all the interference of OG on the MALDI-MS analysis (Fig. 1 d, Table 1, and Supplementary Table 1).
Figure 1.
MALDI-MS spectra of tryptic digests of 25ng BSA, prepared with and without detergents, and with and without EA extraction as indicated (a – g). Spectra between 700 and 4000 m/z were acquired but only those between 800 and 2500 m/z are shown because peptide peaks from m/z 700 to 800 and 2500 to 4000 are not visible on the y-axis scale used and because expansion of the x-axis scale between 800 and 2500 m/z results in clear resolution of the major peptide peaks that are being compared.
Table 1.
Summary of the MS analysis of BSA digests prepared under the conditions indicated in Figure 1.
| Figure 1 panel | Detergent/EA Treatment | Total m/z matching BSA peptides ions* | Peptides common to a, b, & d |
|---|---|---|---|
| a) | No detergent, no EA | 30 | 25 |
| b) | No detergent, EA extracted | 37 | 25 |
| c) | 1% OG, no EA extraction | 0 | 0 |
| d) | 1% OG, EA extracted | 34 | 25 |
| e) | 0.5% NP-40, EA extracted | 26 | 17 |
| f) | 1% Triton X-100, EA extracted | 23 | 17 |
| g) | 1% SDS, EA extracted | 40 | 19 |
m/z between 700 and 4000
Data are derived from experiments depicted in Figure 1. The mass lists from the deisotoped peaks of the acquired m/z spectra were matched to BSA tryptic peptides with Expasy-FindPept (http://ca.expasy.org/tools/findpept.html). The number of predicted BSA tryptic peptides between 700 and 4000 m/z is 44.
EA extraction also removed most of the interference of 1% SDS, 0.5% NP-40 or 1% Triton X-100 from the tryptic digest and allowed detection of all the major BSA tryptic peptides (Fig. 1e–g; Table 1, Supplementary Table 1). However, the spectral profile in the extracted SDS peptide digest differed significantly from the detergent-free sample (Fig.1d, g). Signals from the larger peptides were reduced or lost after EA extraction, while the signals from the smaller peptides increased significantly, even revealing peptides in the lower m/z region of the MS spectrum that were not detected in the untreated or EA-treated samples (Supplementary Table 1). It is possible that SDS not only facilitates the extraction of some small hydrophobic contaminants, but also larger peptides into the EA layer. OG, NP-40 and Triton X-100 could be equally efficiently extracted by EA at pH 2.0 or at pH 8.5, whereas SDS extraction was only effective at pH 8.5 (data not shown). We also observed an increase in contaminants of low m/z peaks when we used EA that had been stored in a plastic tube. Hence, water-saturated EA should be prepared freshly in glass. A minimum of three EA extractions was needed to remove OG completely from tryptic digests and extended extractions, up to seven times, did not result in any significant loss of peptides from the BSA digest (Supplementary Figure 1). However, in the case of NP-40 or Triton X-100-containing BSA peptides (Fig. 1e, 1f) the residual interfering signal after EA extraction could not be further reduced even after an eight-fold of extraction (data not shown).
In the present study we have used BSA digest as an example. However, we have used the same EA extraction protocol in sample preparation for the LC-MS/MS identification of the membrane anti-phosphotryosine reactive proteins in buffer containing 0.4% OG and observed high quality m/z spectra with no detergent interference (data not shown). Our results demonstrate that EA extraction effectively removes high concentration of OG from protein digests without loss of peptides. EA extraction was also able to eliminate interference by SDS, but with some loss of larger peptides. Although EA extraction is unable to remove interference from NP-40 and Triton X-100 completely, their interference in MALDI-MS analysis was greatly minimized. Recently, Masuda, et al. [9] demonstrated that EA effectively extracted sodium deoxycholate from tryptic peptides and that no peptide was found in the EA phase. In conclusion, EA extraction results in a significant enhancement of peptide detection in the MS analysis of protease digests and especially in the case of proteins which require detergent in their preparation.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants CA26504 (E.R.S.), CA101150 (R.H.A.) and the Albert Einstein College of Medicine Cancer Center Grant 5P30-CA13330.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cravatt BF, Simon GM, Yates JR., III The Biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 2.Nagele E, Vollmer M, Horth P, Vad C. 2D-LC/MS techniques for the identification of proteins in highly complex mixtures. Expert Rev. Proteomics. 2004;1:37–46. doi: 10.1586/14789450.1.1.37. [DOI] [PubMed] [Google Scholar]
- 3.Katayama H, Nagasu T, Oda Y. Improvement of in-gel digestion protocol for peptide mass fingerprinting by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2001;15:1416–1421. doi: 10.1002/rcm.379. [DOI] [PubMed] [Google Scholar]
- 4.Katayama H, Tabata T, Ishihama Y, Sato T, Oda Y, Nagasu T. Efficient in-gel digestion procedure using 5-cyclohexyl-1-pentyl-β-D-Maltoside as an additive for gel-based membrane proteomics. Rapid Commun. Mass Spectrom. 2004;18:2388–2394. doi: 10.1002/rcm.1637. [DOI] [PubMed] [Google Scholar]
- 5.Zhang N, Li L. Effects of common surfactants on protein digestion and matrix-assisted laser desorption/ionization mass spectrometric analysis of the digested peptides using two-layer sample preparation. Rapid Commun. Mass Spectrom. 2004;18:889–896. doi: 10.1002/rcm.1423. [DOI] [PubMed] [Google Scholar]
- 6.Nomura E, Katsuta K, Ueda T, Toriyama M, Mori T, Inagaki N. Acid-labile surfactant improves in-sodium dodecyl sulfate polyacrylamide gel protein digestion for marrix-assisted laser desorption/ionization mass spectrometric peptide mapping. J Mass Spectrom. 2004;39:202–207. doi: 10.1002/jms.578. [DOI] [PubMed] [Google Scholar]
- 7.Chen EI, Cociorva D, Norris JL, Yates JR. Optimization of Mass Spectrometry-Compatible Surfactants for Shotgun Proteomics. J. Proteome Res. 2007;6:2529–2538. doi: 10.1021/pr060682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang G, Neubert TA. Use of detergents to increase selectivity of Immunoprecipitation of tyrosine phosphorylated peptides prior to identification by MALDI quadrupole-TOF MS. Proteomics. 2006;6:571–578. doi: 10.1002/pmic.200500267. [DOI] [PubMed] [Google Scholar]
- 9.Masuda T, Tomita M, Ishihama Y. Phase Transfer Surfactant-Aided Trypsin Digestion for Membrane Proteome Analysis. J. Proteome Res. 2008;7:731–740. doi: 10.1021/pr700658q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.