Abstract
Epidemiological studies have associated arsenic exposure with many types of human cancers. Arsenic has also been shown to act as a co-carcinogen even at low concentrations. However, the precise mechanism of its co-carcinogenic action is unknown. Recent studies indicate that arsenic can interfere with DNA repair processes. Poly (ADP-ribose) polymerase (PARP)-1 is a zinc-finger DNA repair protein, which can promptly sense DNA strand breaks and initiate DNA repair pathways. In the present study, we tested the hypothesis that low concentrations of arsenic could inhibit PAPR-1 activity and so exacerbate levels of ultraviolet radiation (UVR)-induced DNA strand breaks. HaCat cells were treated with arsenite and/or UVR, and then DNA strand breaks were assessed by comet assay. Low concentrations of arsenite (≤2 μM) alone did not induce significant DNA strand breaks, but greatly enhanced the DNA strand breaks induced by UVR. Further studies showed that 2 μM arsenite effectively inhibited PARP-1 activity. Zinc supplementation of arsenite-treated cells restored PARP-1 activity and significantly diminished the exacerbating effect of arsenite on UVR-induced DNA strand breaks. Importantly, neither arsenite treatment, nor zinc supplementation changed UVR-triggered reactive oxygen species (ROS) formation, suggesting their effects upon UVR-induced DNA strand breaks is not through a direct free radical mechanism. Combination treatments of arsenite with PARP-1 inhibitor 3-aminobenzamide or PARP-1 siRNA demonstrate that PARP-1 is the target of arsenite. Together, these findings show that arsenite at low concentration exacerbates UVR-induced DNA strand breaks by inhibiting PARP-1 activity, which may represent an important mechanism underlying the co-carcinogenicity of arsenic.
Keywords: arsenic, PARP-1, DNA damage repair, DNA strand break, comet assay, ROS, carcinogenesis
Introduction
Arsenic is a toxic metalloid, which is a naturally-occurring element found in water, soil, and air. High concentrations of arsenic in drinking water are found both within the United States (in the western and southwestern states and in Alaska), and in India, Mexico, Chile, Bangladesh and China (Kitchin, 2001; Huang et al., 2004; Hughes et al., 2007). Epidemiological studies have associated arsenic exposure not only with non-carcinogenic effects, but also many human cancers, including skin, lung, bladder, kidney and liver (Tchounwou et al., 2003; (IARC), 2004; Tapio and Grosche, 2006; Wang et al., 2007). An important mechanism of arsenic toxicity is the induction of DNA damage, a process involving reactive oxygen species (ROS) (Kessel et al., 2002; Kitchin and Ahmad, 2003; Shi et al., 2004b; Lantz and Hays, 2006; Valko et al., 2006). Our previous studies have demonstrated that arsenic can cause DNA damage via generating hydroxyl radical, superoxide anion, and hydrogen peroxide (Shi et al., 2004a; Ding et al., 2005). The International Agency for Research On Cancer (IARC) has classified arsenic as a human carcinogen ((IARC), 2004). U.S. Environmental Protection Agency (EPA) recently reduced the maximum contaminant level (MCL) standard for arsenic in drinking water from 50 μg/L to 10 μg/L (EPA, 2002). This reduction was because a growing number of laboratory studies, both in cell cultures and in experimental animals, have demonstrated biologic effects of arsenic at levels equivalent to or below the new 10 μg/L standard. Such effects, including endocrine disruption, activation of cell signaling pathways, alterations in cell cycle kinetics and proliferative response, are involved in carcinogenesis and other disease processes (Rossman, 2003; Andrew et al., 2006). Moreover, arsenic has been shown to potentiate the genotoxicity of other mutagen-carcinogens, including ultraviolet radiation (UVR). UV from sunlight is the most prominent carcinogen in our natural environment and the most important cause of skin cancers (de Gruijl, 1999; Guzman et al., 2003; Pi et al., 2005). It is estimated that in the United States 1.3 million new cases of skin cancer are diagnosed annually, making it by far the most prevalent cancer. Studies have found that low concentrations of arsenic, which are not mutagenic, can enhance the mutagenicity of UVR (Rossman et al., 2001; Rossman et al., 2002). Further investigations have attributed the cancer-enhancing effect of arsenic to decreased DNA repair (Hartwig et al., 1997; Hartwig et al., 2002; Andrew et al., 2006), but the mechanisms by which arsenic inhibits DNA repair are not clearly understood.
An important role in the regulation of DNA damage repair is played by the members of the poly (ADP-ribose) polymerase (PARP) family. Whereas 17 different PARP members have been found so far, PARP-1 activity accounts for about 90% of the total cellular poly(ADP-ribose) formation (Burkle et al., 2005; Schreiber et al., 2006). PARP-1 functions as a “nick sensor”. It is constitutively expressed in most tissues, but its activity is stimulated 500-fold by DNA with single-strand or double-strand breaks (SSBs and DSBs respectively). SSBs are the most common lesions induced by exogenous genotoxins (Caldecott, 2004). Moreover, SSBs encountered by a replication fork are most likely converted into DSBs (Strumberg et al., 2000; Kuzminov, 2001). DSB is one of the most toxic and mutagenic DNA lesions experienced in human cells: a single DSB can potentially lead to loss of more than 100 million base pairs of genetic information. Both SSBs and DSBs are closely involved in tumor initiation and progression (Helleday et al., 2007).
PARP-1 not only directly participates in the DNA repair, but also can affect a number of other DNA-repair and DNA-damage checkpoint proteins. When DNA is damaged (by ionizing radiation, alkylating agents, or oxidants), activated PARP-1 immediately transfers numerous ADP-ribosyl moieties from NAD+ to itself and other nuclear proteins. These include DNA methyltransferase 1, p53, p21, XPA, MSH6, DNA ligase III, XRCC1, DNA polymerase-ε, DNA-PKcs, Ku70, NF-κB, and telomerase, thus regulating DNA repair (Pleschke et al., 2000; Rossman et al., 2004). Hartwig, et al, found that very low concentrations (0.01-1 μM) of arsenic suppress PARP-1 activity induced by hydrogen peroxide in Hela S3 cells (Hartwig et al., 2003b). Our previous unpublished data has shown that 2 μM arsenic greatly inhibited PARP-1 activity induced by UVR in HaCat cells. At such a low concentration, arsenic itself did not induce significant oxidative DNA damage, which, could only be detected above background levels after exposure to concentrations of 10 μM or higher (Ding et al., 2005).
In the present study, we hypothesized that arsenic, at low concentration, exacerbates UVR-mediated DNA strand breaks by interfering with PAPR-1 activity. We found that 2 μM arsenite itself did not induce significant DNA strand breaks, but it greatly increased the DNA strand breaks induced by UVR. More importantly, we manipulated PARP-1 activity by using zinc, the PARP-1 inhibitor 3-aminobenzamide (3-AB), and PARP-1 siRNA: this demonstrated that arsenite exacerbated overall DNA strand breaks induced by UVR through inhibiting PARP-1 activity.
Materials and methods
Cell culture and treatments
The human keratinocyte cell line (HaCat) was generously provided by Dr. Mitch Denning (Loyola University Medical Center, Maywood, IL). HaCat cells were cultured in Dulbecco's Modified Eagle's Medium F:12 HAM (DMEM/F:12), supplemented with 10% newborn calf serum from Life Technologies/Gibco, four-fold final concentration of MEM amino acids, 2 mM L-glutamine and antibiotics (penicillin, 100 U/ml and streptomycin, 100 μg/ml) (Ding et al., 2005). The cells were cultured at 37 °C in 95% air/5% CO2 humidified incubator. When HaCat cells reached 60-70% confluence in 6-well plate, cells were placed into serum-free DMEM/F12 medium overnight prior to treatments. Sodium arsenite (As), zinc chloride (Zn) and 3-AB solutions were sterilized by passing through a 0.22 μm syringe filter. The working concentrations were diluted with serum-free DMEM/F12 medium. For all experiments involving incubation with arsenic and/or zinc (or 3-AB), HaCat cells were rinsed with PBS and placed into serum-free DMEM/F12 medium containing concentrations of arsenic and/or zinc (or 3-AB) as indicated in the figure legends. Serum-free conditions were used to eliminate potential effects of serum on the arsenite-stimulated responses. After 24 hours incubation, the medium was removed and the cells were rinsed three times with PBS. Then the cells were covered with a thin layer of PBS (0.5ml/well of 6-well plate) and placed on ice during UVR exposure. Cells were exposed to 1 J/cm2 solar-simulated light using an Oriel 1600 W Watt Solar Ultraviolet Simulator (Oriel Corp., Stratford, CT). This solar simulator produces a high intensity UVR beam in both the UVA (320-400 nm) and UVB (280-320nm) spectrum. After UVR exposure, PBS was removed and replaced with serum-free medium. Cells were incubated for another 6 hours before collected for further experimental procedures.
Comet assay
Cells grown in cell-culture plates were detached with 0.25% trypsin-EDTA and harvested by centrifugation. Cells were resuspended at 1 × 105 cells/ml in ice cold PBS (Ca++ and Mg++ free). Then the cells (1×105ml) were combined with molten LMAgarose (Trevigen, Gaithersburg, MD) (at 37 °C) at a ratio of 1: 10 (v/v), and 40 μl of the mixture was immediately pipetted onto each well of a CometSlide™ (20 well slide) (Trevigen). The slides were placed at 4 °C in the dark in refrigerator for 30 min. A 0.5 mm clear ring appears at edge area of each well. The slides were immersed in prechilled Lysis Solution (Trevigen) and left at 4 °C in a refrigerator for 50 min. Then the slides were immersed in freshly prepared Alkaline Solution (300 mM NaOH and 1 mM EDTA, pH>13) and left at room temperature in the dark for 30 min. The slides were transferred to a horizontal electrophoresis apparatus. They were placed flat on a gel tray and aligned equidistant from the electrodes. The Alkaline Solution was carefully poured until the solution level just covered samples. The voltage was set to about 1 Volt/cm and buffer was added or removed until the current was approximately 300 mA and electrophoresis was performed for 35 min. The slides were rinsed by dipping several times in distilled H2O and then immersed in 70% ethanol for 5 min and then air dried. 50 μl of diluted SYBR Green I (1:10 000 SYBR Green in TE buffer) was placed onto each circle of dried agarose. Then the slides were viewed by an Olympus BH2-RFCA fluorescence microscope (Olympus, Melville, NY) (SYBR Green I's maximum excitation and emission are respectively 494nm/521nm). Images were taken by Omegafire digital camera with MagnaFire 2.1 software (Optronix, Goleta, CA) and analyzed using the Comet Assay Software Project (CASP). The results were presented as the mean comet-tail moment. The comet-tail moment is defined as the product of the fraction of cellular DNA in the comet tail and the tail length. A higher comet-tail moment value represents a greater number of cellular DNA strand breaks (Lockett et al., 2006; Ueno et al., 2007). 50 cells on each slide and at least three slides in each group were analyzed.
Detection of cellular poly (ADP-ribosyl)ation level
The level of poly (ADP-ribosyl)ation (PAR) was detected in HaCat cells according to Hartwig (Hartwig et al., 2003b) with some modifications. Coverslips with cells were rinsed with ice-cold PBS and fixed in ice-cold 10% trichloroacetic acid (TCA) for 20 min followed by successive 10 min washing in 75%, 90%, and 100% ethanol (−20 °C). Then the coverslips were air-dried, rehydrated in PBS and incubated in blocking reagent (1% BSA in PBS) at 37 °C for 1 hr. The coverslips were then incubated with mouse monoclonal antibody raised against poly(ADP-ribose) (Axxora, LLC. San Diego, CA) (diluted to 10 μg/ml in blocking reagent) in a humid chamber at 37 °C for 1 hr. The secondary, FITC-conjugated anti-mouse antibody (Chemicon, Temecula, CA) (diluted 1:200 in blocking reagent) was applied and the samples were incubated in a dark humid chamber at 37 °C for 1 hr. Then the coverslips were mounted on microscope slides using Vectashield mounting medium containing 2 μg/ml DAPI (Vector Labs, Burlingame, CA) to visualize nuclei. Images were obtained using an Olympus BH2-RFCA fluorescence microscope and Omegafire digital camera with MagnaFire 2.1 software.
Measurement of reactive oxygen species
Intracellular ROS production was assessed using dihydroethedium (DHE, Molecular Probes, Carlsbad, CA) as previously (Cooper et al., 2007). HaCat cells were cultured on plastic coverslips (NUNC, Rochester, NY) in 24-well plates. When the cells reached 70% confluence, they were incubated with arsenic (2 μM) and/or zinc (2 μM) for 24 hrs. Cells were exposed to 1 J/cm2 UV. 30 min after exposure, DHE (5 μM) was added as a fluorescent indicator of ROS and incubated for another 30 min in cell culture incubator. Then coverslips were washed three times with PBS, fixed with 4% paraformaldehyde, and mounted to glass slides with Vecta-Shield (Vector Labs). Images were collected with an Olympus IX70 fluorescence microscope fitted with an Olympus America camera and MagnaFire2.1 software. Relative fluorescence intensity was quantified by measuring pixel intensity along a line drawn through the center of a cluster cells using Metamorph software (version 6.3r6). A minimum of 3 independent samples were analyzed per treatment. The upper 95% of each measurement was pooled and averaged and the resulting data analyzed for significance.
PARP-1 knockdown by transfection of PARP-1 siRNA
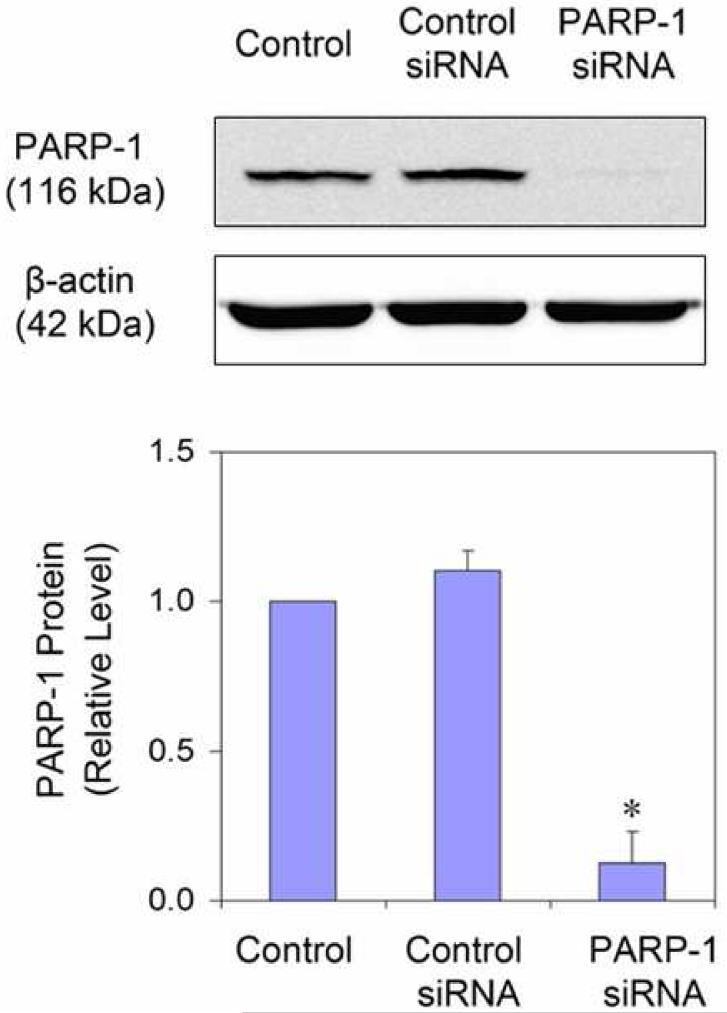
Transfection of siRNA was performed according to the procedure described by Liu (Liu et al., 2006). Briefly, SMARTpool siRNA specific for human PARP-1 sequence (GenBank accession number: NM_001618) was obtained from Dharmacon Research, Inc. (Lafayette, CO). SiGLO RISC-Free siRNA (Dharmacon) was used as a control siRNA. HaCaT cells were seeded at a density of 2 × 105 cells/well in 6-well plate the day before transfection in DMEM/F12 containing 10% FBS without antibiotics. Transfection of siRNAs was carried out using DharmaFECT transfection reagent (Dharmacon). DharmaFECT reagent was diluted 1:50 in serum free DMEM/F12 and incubated at room temperature for 5 min. In parallel, 2 μM siRNA in 1×siRNA buffer (Dharmacon) was diluted 1:1 in serum-free DMEM/F12. The two mixtures were combined and incubated for 20 min at room temperature prior to addition to cells with a final siRNA concentration of 100 nM. After 24 hrs, the medium was replaced with a complete growth medium. After another 48 hrs, cells were treated with arsenic and/or UVR, as indicated in the figure legends. Specific silencing was confirmed by Western Blot analysis.
Western Blot analysis of PARP-1 knockdown
Cells in 6-well plate were washed twice with ice-cold PBS and harvested. 50 μl Chaps Cell Extract Buffer (Cell Signaling, Danvers, MA) supplemented with complete proteinase inhibitor cocktail was added and then samples were lysed on ice for 30 min. After being frozen and thawed three times, the extracts were clarified by centrifugation at 14,000 g for 15 min at 4 °C. Protein concentrations were determined by the Coomassie Plus Protein assay (Pierce, Rockford, IL). Cell lysate (30 μg of protein) was resolved in an 8% SDS-polyacrylamide gel and transferred onto nitrocellulose membrane (Bio-Rad, Hercules, CA), and incubated for 1 hr in TBST (10 mM Tris, pH 8.0, 150 mM NaCl and 0.1% Tween 20) containing 5% non-fat milk at room temperature. The membrane was then incubated with the rabbit polyclonal anti-PARP-1 antibody (1:500, Santa Cruz, CA) overnight at 4 °C. After washing with TBST, the membrane was incubated for 1 hr with horseradish peroxidase conjugated secondary antibody (1:1000, Cell Signaling, Danvers, MA), and signal was detected using the SuperSignal West Pico chemiluminescent kit (Pierce) on a Kodak Image Station 4000MM following the manufacturer's instructions. To control sample loading and protein transfer, the membrane was stripped and re-probed to detect β-actin (1:1000, Santa Cruz). The intensities were quantified by KODAK Molecular Imaging Software version 4.0. The PARP-1 protein levels were normalized to β-actin as compared with the untreated control (set to 1).
Data analysis
Data were presented as means ± S.D. Statistical analysis was performed using ANOVA and Student's t-test. A value of P < 0.05 was considered statistically significant.
Results
Low concentration of arsenite increases DNA strand breaks induced by UVR
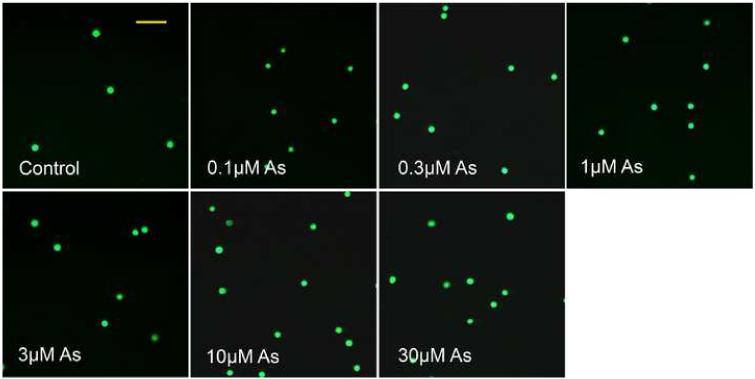
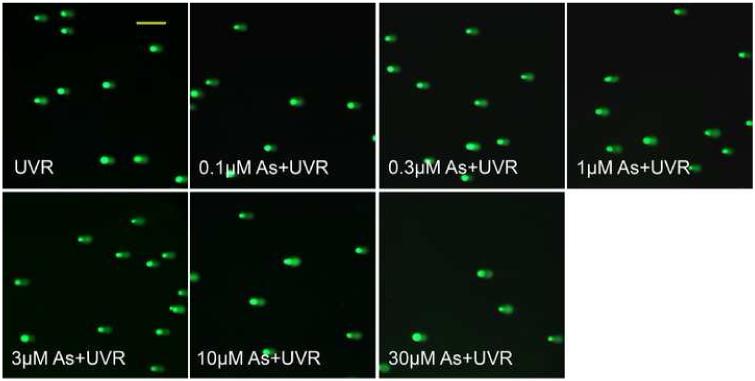
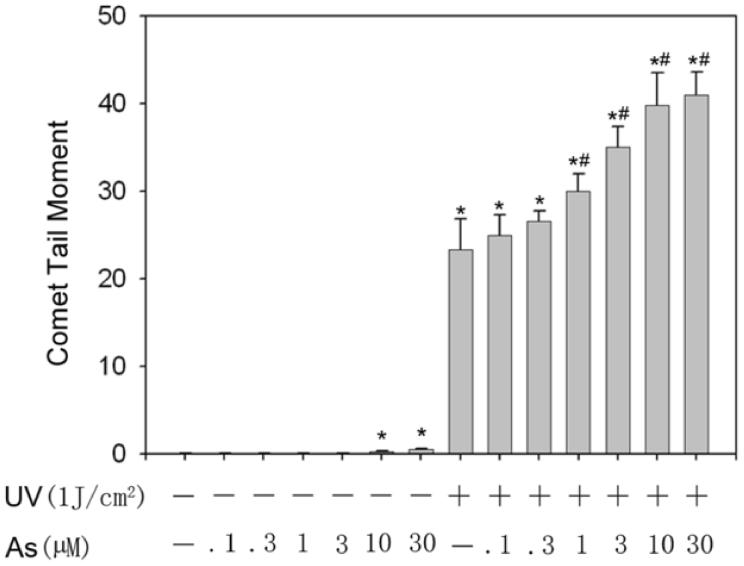
UVR is the most prominent and ubiquitous physical carcinogen in our natural environment (de Gruijl, 1999). UVR can induce many kinds of DNA damage, such as 8-OHdG, SSBs, DSBs, base loss, cyclobutane-pyrimidine dimers (CDPs) and pyrimidine-pyrimidone (6-4) photoproducts ((6-4)-PP). The comet assay is a sensitive and rapid method for detection of both SSBs and DSBs in individual cells (Fairbairn et al., 1995). In the present study, the total DNA strand breaks (TSBs =SSBs+DSBs) were detected by alkaline comet assay. As shown in Figure 1, DNA strand breaks produced by arsenite treatment alone were detectable after exposure to 10 μM or higher concentrations of arsenite, but not at 3 μM or lower concentrations. In contrast, UVR alone greatly increased DNA strand breaks. Most noticeably, pre-incubation of HaCat cells with arsenite concentrations as low as 1 μM increased UVR-induced TSB in a dose-dependent manner (Figure 1B and 1C). These results suggest that arsenite itself, at high concentrations (10 μM or higher), induces DNA strand breaks in HaCat cells. More importantly, arsenite exacerbates UVR-induced DNA strand breaks at much lower concentration, i.e., 1 μM under our experimental conditions.
Fig. 1.
DNA strand breaks induced by different concentrations of arsenite with or without UVR. (A)(B). HaCat cells were incubated with different concentrations of arsenite (0.1, 0.3, 1, 3, 10, 30 μM) for 24 hrs. Then the indicated groups were exposed to UVR (1J/cm2). The DNA strand breaks were measured by comet assay 6 hrs after UVR. Images were obtained using an Olympus BH2-RFCA fluorescence microscope and Omegafire digital camera with MagnaFire 2.1 software. Scale bar = 200 μm. (C). The results from (A) and (B) were quantified by measuring the comet-tail moment using CASP software. The experiment was repeated at least 3 times. Data were presented as means. ± S.D. * p<0.05 vs Control, # p<0.05 vs UVR.
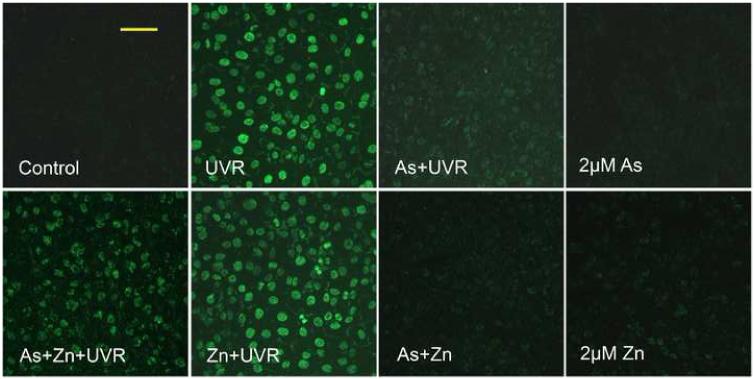
Low concentration of arsenite inhibits PARP-1 activity induced by UVR
PARP-1 is activated by DNA strand breaks and is directly involved in their repair (Miwa and Masutani, 2007). Our data above (Figure 1) confirmed the production of DNA strand breaks induced by UVR. We next determined whether low concentrations of arsenite could inhibit UVR-induced PARP-1 activation. Activated PARP-1 consumes NAD+ and produces PAR. Therefore, PARP-1 activity can be measured by detecting the level of PAR (Kupper et al., 1990; Hartwig et al., 2003b). As shown in Figure 2, the control (untreated) cells did not show any visible PARP-1 activity. After the cells were treated with UVR, PARP-1 was activated and the fluorescence intensity was strong (Figure 2, UVR). Pre-incubating the cells with 2 μM arsenite effectively abolished UVR-stimulated fluorescent PAR production, suggesting that arsenite inhibited the activity of PARP-1 (Figure 2, As+UVR). PARP-1 is a protein with two zinc finger domains that are essential for DNA binding (Ikejima et al., 1990). Literature reports suggest that arsenite could replace the zinc in the zinc finger (Hartwig et al., 2003a; Kitchin and Wallace, 2005). We therefore speculated that arsenite may interfere the zinc finger domains of PARP-1 by replacing zinc. If this is the case, zinc supplementation might be able to reverse the process, resulting in the revival of PARP-1 activity. To test this possibility, we co-incubated cells with 2 μM arsenite and 2 μM zinc before UVR exposure. Indeed, zinc supplementation partially restored PARP-1 activity (Figure 2, As+Zn+UVR). These results demonstrate that arsenite at low concentration (2 μM)can effectively inhibit UVR-stimulated PARP-1 activity through interfering with the function of its zinc finger domains. It is important to note that PARP-1 activity was not detectable in the arsenic alone treated cells, even with zinc supplementation (Figure 2, As+Zn). The most plausible explanation is that 2 μM arsenic did not cause significant DNA strand breaks, so PARP-1 activity was below the detection limit.
Fig. 2.
Effect of arsenite and/or zinc on PARP-1 activity induced by UVR. HaCat cells were incubated with 2 μM arsenite with or without 2 μM zinc for 24 hrs before UVR (1J/cm2). The PARP-1 activity was detected by measuring the production of PAR using immunofluorescence. Images were obtained using an Olympus BH2-RFCA fluorescence microscope and Omegafire digital camera with MagnaFire 2.1 software. Scale bar = 100 μm.
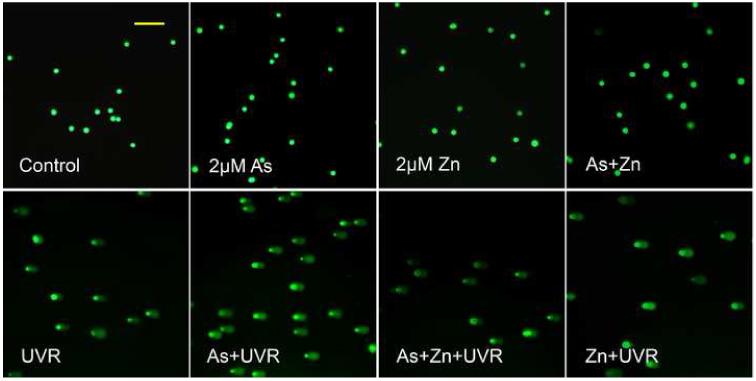
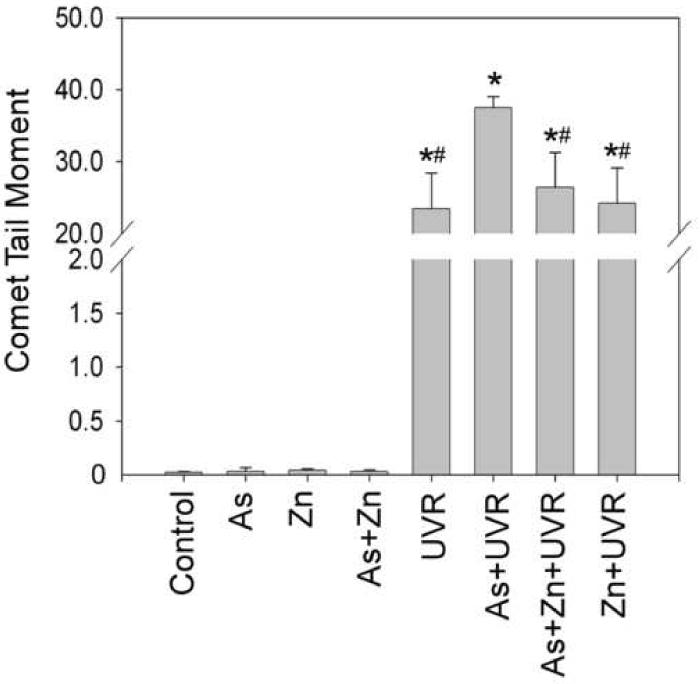
Protection of zinc against arsenite-exacerbated DNA strand breaks induced by UVR
Our results in Figure 2 showed that arsenite inhibition of PARP-1 activity could be reversed by zinc. We next examined whether the protection of PARP-1 activity by zinc is functional in terms of DNA strand break repair. The cells were co-treated with arsenite and/or zinc before UVR. DNA strand breaks were determined by comet assay. As shown in Figure 3, 2 μM arsenite and/or 2 μM zinc did not induce DNA strand breaks, whereas UVR alone did. Pre-treating cells with 2 μM arsenite significantly exacerbated the UVR-induced DNA strand break level. Consistent with its protective effect on PARP-1 activity, zinc supplementation effectively prevented the exacerbating effect of arsenite on UVR-induced DNA strand breaks. These results suggest that inhibition of PARP-1 by arsenite may be an important mechanism underlying its exacerbating effect on UVR-induced DNA strand breaks.
Fig. 3.
Effect of zinc on arsenite-exacerbated DNA strand breaks induced by UVR. (A). HaCat cells were incubated with 2 μM arsenite with or without 2 μM zinc for 24 hrs before UVR (1J/cm2). DNA strand breaks were measured by comet assay 6 hrs after UVR. Images were obtained using an Olympus BH2-RFCA fluorescence microscope and Omegafire digital camera with MagnaFire 2.1 software. Scale bar = 200 μm. (B). The above images were quantified by measuring the comet-tail moment using CASP software. The experiment was repeated at least 3 times. Data were presented as means ± S.D. * p<0.05 vs Control, # p<0.05 vs As+UVR.
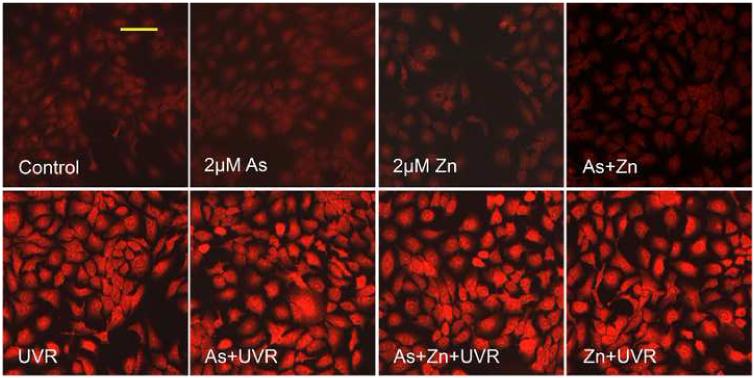
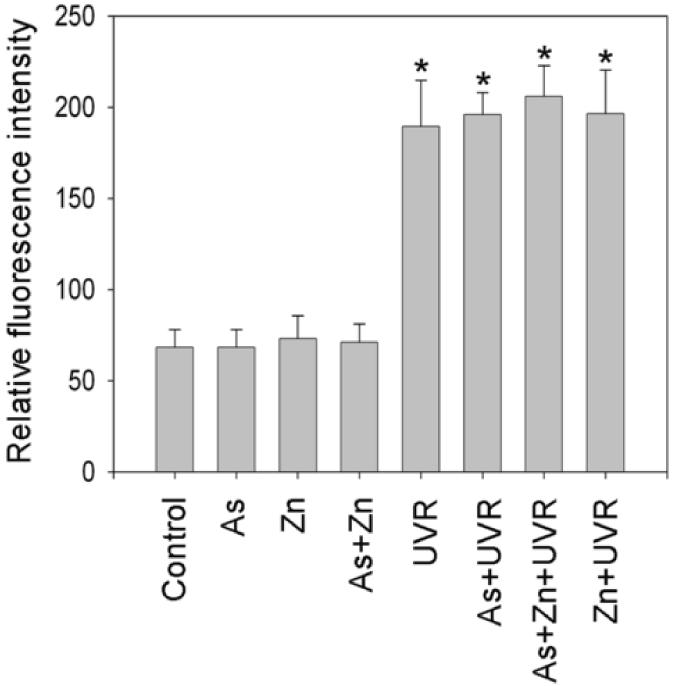
The effects of arsenite or zinc on UVR-induced DNA strand breaks is not through altering ROS generation
Zinc has been reported to possess antioxidant properties (Powell, 2000; Rostan et al., 2002), while arsenite is well-known oxidative stressor (Valko et al., 2005; Lantz and Hays, 2006). There is therefore a possibility that they affect UVR-induced DNA strand breaks directly by altering ROS generation, providing an alternative explanation for our observed results in Figure 1 and 3. DHE staining was used to measure cellular ROS generation. As shown in Figure 4A, there was no significant increase of ROS in the cells treated with 2 μM arsenite and/or zinc, whereas UVR greatly increased ROS generation. However, pre-treatment with arsenite and/or zinc did not cause any significant difference in the UVR-induced ROS generation. Fluorescence intensity was quantified using Metamorph software, and the quantitative fluorescence intensities were shown in Figure 4B. These results demonstrated that arsenite or zinc did not affect UVR-induced ROS generation. Thus, the increased DNA strand breaks by arsenite (Figure 1) or the decreased DNA strand breaks by zinc (Figure 3) were not due to increased ROS production.
Fig. 4.
Effect of arsenite or zinc on the generation of ROS induced by UVR. (A). HaCat cells were incubated with 2 μM arsenite and/or 2 μM zinc for 24 hrs before UVR (1J/cm2). DHE (5 μM) was added as a fluorescent indicator of ROS 30 min after UVR and incubated for another 30 min. Images were obtained using an Olympus BH2-RFCA fluorescence microscope and Omegafire digital camera with MagnaFire 2.1 software. Scale bar = 100 μm. (B). Relative fluorescence intensity was quantified using Metamorph software. The experiment was repeated at least 3 times. The relative level of ROS were presented as means ± S.D. * p<0.05 vs Control.
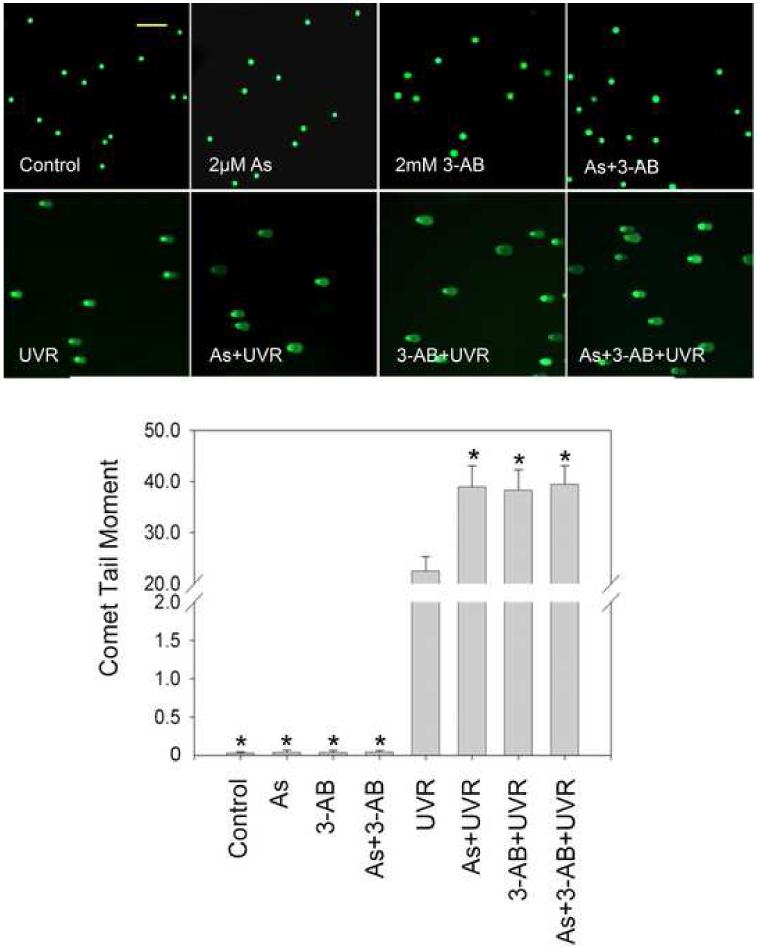
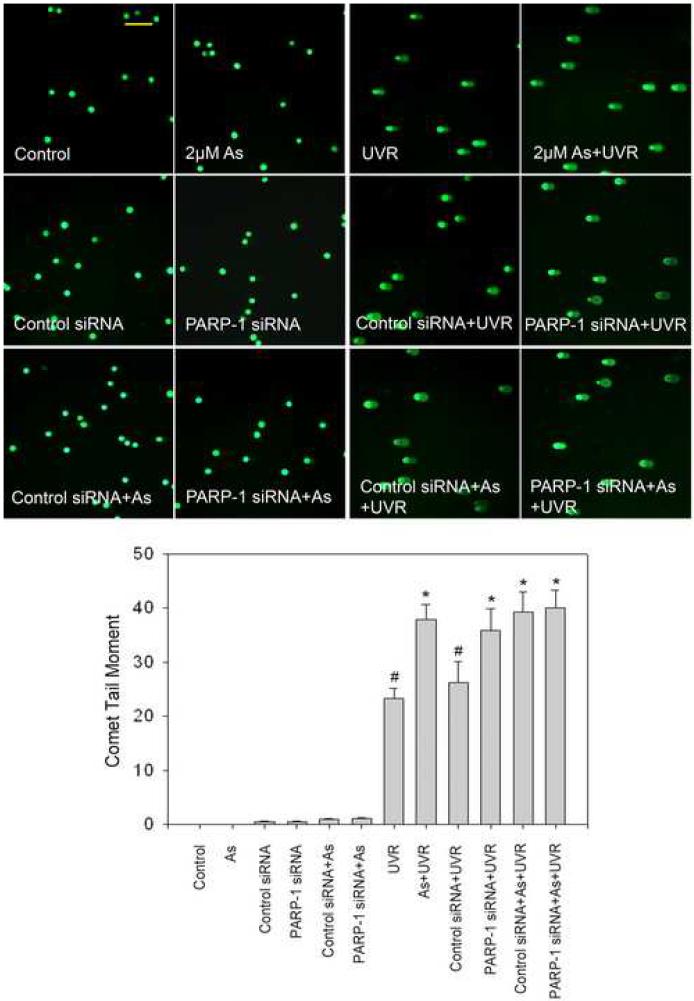
PARP-1 inhibitor 3-AB or PARP-1 siRNA exacerbates UVR-induced DNA strand breaks
Findings from the above experiments suggest that arsenite can inhibit the activity of PARP-1, which leads to decreased repair of DNA strand break, ultimately resulting in an increase in the observed DNA strand break level. In order to further verify this hypothesis PARP-1 activity was modulated by the inhibitor 3-AB or PARP-1 siRNA. PARP-1 siRNA decreased the protein level by nearly 88% (Figure 5A), demonstrating efficient knockdown. Pre-treatment of 3-AB or PARP-1 siRNA greatly increased UVR-induced DNA strand break formation, confirming PARP-1 is involved in the repair of DNA strand breaks (Figure 5B and 5C). Moreover, 3-AB or PARP-1 siRNA did not cause any further increase of UVR-induced DNA strand breaks in the presence of 2 μM arsenite (Figure 5B and 5C). These results suggest that arsenite, 3-AB, and PARP-1 siRNA were acting on the same target, PARP-1 protein.
Fig. 5.
Effect of PARP-1 inhibition on the arsenite-exacerbated DNA strand breaks induced by UVR. (A). Effect of PARP-1 siRNA on PARP-1 protein level. HaCat cells were incubated with PARP-1 siRNA or control siRNA for 24 hrs. Then the medium was replaced with complete growth medium. After another 48 hrs, the cells were collected to measuring the PARP-1 protein by Western-Blot. The experiment was repeated at least 3 times. Data were presented as means ± S.D. * p<0.05 vs control group. (B). Effect of PARP-1 inhibitor (3-AB) on the arsenite-exacerbated DNA strand breaks induced by UVR. HaCat cells were incubated with 2 μM arsenite and/or 2 mM 3-AB for 24 hrs before UVR (1J/cm2). DNA strand breaks were measured by comet assay 6 hrs after UVR. The experiment was repeated at least 3 times. The results were quantified by measuring the comet-tail moment using CASP software. Data were presented as means ± S.D. * p<0.05 vs UVR. (C). Effect of PARP-1 siRNA on the arsenite-exacerbated DNA strand breaks induced by UVR. After PARP-1 was knockdowned by siRNA described as (A), the cells were incubated with 2 μM arsenite for 24 hrs before UVR (1J/cm2). DNA strand breaks were measured by comet assay 6 hrs after UVR. The experiment was repeated at least 3 times. The results were quantified by measuring the comet-tail moment using CASP software. Data were presented as means ± S.D. * p<0.05 vs UVR, # p<0.05 vs As+UVR. Scale bar = 200 μm.
Discussion
Arsenic has been well documented to stimulate ROS generation in many cell types at high concentrations (Kessel et al., 2002; Kitchin and Ahmad, 2003). Oxidative DNA damage by arsenite-derived ROS has long been considered as one of the most important mechanisms underlying arsenic carcinogenicity (Kessel et al., 2002; Shi et al., 2004b; Valko et al., 2006). However, most human exposure does not involve high levels, but rather are to low concentrations of arsenic. Accumulating evidence indicates that even a low concentration of arsenic is closely associated with carcinogenesis, by acting as a co-carcinogen or co-promoter (Rossman et al., 2002; Rossman et al., 2004; Salnikow and Zhitkovich, 2008). For example, recent studies showed that oral arsenic greatly enhances the incidence and progression of skin malignancies induced by UVR (Germolec et al., 1998; Rossman et al., 2001; Pi et al., 2005). In the present study, using the human keratinocyte HaCat cell line, treatment with arsenite alone did not produce significant amount of DNA strand breaks at arsenite concentrations of 3 μM or less. It is only when treated with arsenite at concentrations of 10 μM or more that the DNA strand breaks became detectable (Figure 1C). Even then, the total amount of DNA strand breaks, as measured by the tail moment of the comet assay, was much less than that induced by UVR (Figure 1C). In contrast, at low concentration of 1μM, arsenite significantly increased the DNA strand breaks induced by UVR, even though 1μM arsenite did not induce detectable DNA strand breaks (Figure 1). Thus a much lower concentration of arsenite is needed to enhance UVR-induced DNA strand breaks than to produce similar damage alone. These findings imply that arsenite is a much more effective co-carcinogen to UVR (and perhaps other oxidants as well) than a carcinogen by itself.
Although the precise mechanism underlying the co-carcinogenicity of arsenic remains unknown, many studies suggest that arsenic may interfere with DNA repair processes. Hartwig et al found that low concentration (2.5 μM) of arsenite reduced the efficiency of nucleotide excision repair, and incision frequency after UVR exposure (Hartwig et al., 1997). Andrew et al found arsenite exposure is associated with decreased DNA repair by inhibiting excision repair cross-complement 1 (ERCC1) (Andrew et al., 2003; Andrew et al., 2006). PARP-1 is the earliest DNA repair protein activated by DNA strand breaks and initiates repair pathways. Previous studies have found that arsenite could inhibit PARP-1 activity (Yager and Wiencke, 1997; Hartwig et al., 2003b). In the present study, PARP-1 was activated by UVR. This activation could be effectively inhibited by low concentration arsenite (2 μM) (Figure 2). PARP-1 is a highly important zinc-finger DNA repair protein. This enzyme possesses two zinc fingers within its DNA-binding domain through which it binds tightly to DNA SSBs or DSBs, arising either as primary DNA damage or as DNA repair intermediates. Zinc finger motifs are protein domains containing typically 30-40 amino acid residues, whose structures are maintained by coordination of Zn(II) ions to side chain donors of histidine and/or cysteine residues in a tetrahedral geometry (Papworth et al., 2006; Witkiewicz-Kucharczyk and Bal, 2006). Zinc fingers in DNA repair proteins are suggested as sensitive intracellular target of arsenite (Hartwig, 2001; Hartwig et al., 2003a). The proposed modes of zinc finger damage by toxic metals include isostructural substitution, substitution with altered geometry, mixed complex formation, and catalysis of thiol oxidation (Virag, 2005). So we reasoned that zinc supplementation might be able to reverse the process, resulting in the restoration of PARP-1 activity. As expected, zinc treatment partially recovered PARP-1 activity (Figure 2), suggesting that arsenite may interfere with the zinc finger motifs of the PARP-1 and thus inhibit its activity. As zinc could retrieve the arsenite-inhibited PARP-1 activity, it is reasonable to deduce that zinc supplementation has protection on arsenite-exacerbated DNA strand breaks induced by UVR. This hypothesis was confirmed by the effective protection of zinc on arsenite-exacerbated DNA strand breaks by UVR (Figure 3), suggesting that inhibited PARP-1 activity by low concentration arsenite contributed to the exacerbated DNA strand breaks.
Zinc possesses antioxidant properties (Powell, 2000; Rostan et al., 2002), while arsenite is a well-known oxidative stressor (Valko et al., 2005; Lantz and Hays, 2006). It was therefore possible that arsenic or zinc may affect UVR-induced DNA strand breaks by altering ROS generation. Our finding that low concentration of arsenite or zinc supplementation did not change UVR-induced ROS generation rules out this possibility. To further confirm the important role of PARP-1 in repairing UVR-induced DNA strand breaks, we tested the effects of PARP-1 inhibitor 3-AB and PARP-1 siRNA on DNA strand break formation. Our results clearly showed that inhibition of PARP-1 by 3-AB or siRNA greatly increased the DNA strand breaks induced by UVR. The finding that 3-AB or PARP-1 siRNA did not further increase UVR-induced DNA strand breaks in the presence of 2 μM arsenite indirectly supports that PARP-1 is an important target of arsenite, as 2 μM arsenite had already inhibited UVR-triggered PARP-1 activation.
In conclusion, the results presented demonstrate that arsenite at low concentrations could greatly exacerbate UVR-induced DNA strand breaks through inhibiting PARP-1 mediated DNA repairing process. This may represent an important mechanism underlying the co-carcinogenicity of arsenic.
Acknowledgements
This study was funded by a Department of Health and Human Services grant from the U.S. National Institutes of Health R01 ES012938 and R01 ES015826. Support was also provided by the UNM Cancer Research and Treatment Center P30 CA118100 and the UNM NIEHS Center P30 ES-012072. We would like to thank Ms. Leslie Lund for her help with the comet assay.
Abbreviations
- As
sodium arsenite
- UVR
ultraviolet radiation
- PARP-1
poly(ADP-ribose) polymerase-1
- SSB
single strand break
- DSB
double strand break
- ROS
reactive oxygen species
- 8-OHdG
8-hydroxyl-2′-deoxyguanine
- BER
base excision repair
- NER
nucleotide excision repair
- DHE
dihydroethedium
- BSA
bovine serum albumin
- PBS
phosphate-buffered saline
- siRNA
small interfering RNA
- Zn
zinc chloride
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- (IARC), I. A. f. R. O. C. Arsenic in drinking-water. IARC Monogr Eval Carcinog Risks Hum. 2004;84:39–267. [PMC free article] [PubMed] [Google Scholar]
- Andrew AS, Burgess JL, Meza MM, Demidenko E, Waugh MG, Hamilton JW, Karagas MR. Arsenic exposure is associated with decreased DNA repair in vitro and in individuals exposed to drinking water arsenic. Environ Health Perspect. 2006;114:1193–1198. doi: 10.1289/ehp.9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew AS, Karagas MR, Hamilton JW. Decreased DNA repair gene expression among individuals exposed to arsenic in United States drinking water. Int J Cancer. 2003;104:263–268. doi: 10.1002/ijc.10968. [DOI] [PubMed] [Google Scholar]
- Burkle A, Diefenbach J, Brabeck C, Beneke S. Ageing and PARP. Pharmacol Res. 2005;52:93–99. doi: 10.1016/j.phrs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Caldecott KW. DNA single-strand breaks and neurodegeneration. DNA Repair (Amst) 2004;3:875–882. doi: 10.1016/j.dnarep.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Cooper KL, Liu KJ, Hudson LG. Contributions of reactive oxygen species and mitogen-activated protein kinase signaling in arsenite-stimulated hemeoxygenase-1 production. Toxicol Appl Pharmacol. 2007;218:119–127. doi: 10.1016/j.taap.2006.09.020. [DOI] [PubMed] [Google Scholar]
- de Gruijl FR. Skin cancer and solar UV radiation. Eur J Cancer. 1999;35:2003–2009. doi: 10.1016/s0959-8049(99)00283-x. [DOI] [PubMed] [Google Scholar]
- Ding W, Hudson LG, Liu KJ. Inorganic arsenic compounds cause oxidative damage to DNA and protein by inducing ROS and RNS generation in human keratinocytes. Mol Cell Biochem. 2005;279:105–112. doi: 10.1007/s11010-005-8227-y. [DOI] [PubMed] [Google Scholar]
- EPA, U. S. National primary drinking water regulations: Long Term 1 Enhanced Surface Water Treatment Rule. Final rule. Fed Regist. 2002;67:1811–1844. [PubMed] [Google Scholar]
- Fairbairn DW, Olive PL, O'Neill KL. The comet assay: a comprehensive review. Mutat Res. 1995;339:37–59. doi: 10.1016/0165-1110(94)00013-3. [DOI] [PubMed] [Google Scholar]
- Germolec DR, Spalding J, Yu HS, Chen GS, Simeonova PP, Humble MC, Bruccoleri A, Boorman GA, Foley JF, Yoshida T, Luster MI. Arsenic enhancement of skin neoplasia by chronic stimulation of growth factors. Am J Pathol. 1998;153:1775–1785. doi: 10.1016/S0002-9440(10)65692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman E, Langowski JL, Owen-Schaub L. Mad dogs, Englishmen and apoptosis: the role of cell death in UV-induced skin cancer. Apoptosis. 2003;8:315–325. doi: 10.1023/a:1024112231953. [DOI] [PubMed] [Google Scholar]
- Hartwig A. Zinc finger proteins as potential targets for toxic metal ions: differential effects on structure and function. Antioxid Redox Signal. 2001;3:625–634. doi: 10.1089/15230860152542970. [DOI] [PubMed] [Google Scholar]
- Hartwig A, Asmuss M, Ehleben I, Herzer U, Kostelac D, Pelzer A, Schwerdtle T, Burkle A. Interference by toxic metal ions with DNA repair processes and cell cycle control: molecular mechanisms. Environ Health Perspect. 2002;110(Suppl 5):797–799. doi: 10.1289/ehp.02110s5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig A, Blessing H, Schwerdtle T, Walter I. Modulation of DNA repair processes by arsenic and selenium compounds. Toxicology. 2003a;193:161–169. doi: 10.1016/j.tox.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Hartwig A, Groblinghoff UD, Beyersmann D, Natarajan AT, Filon R, Mullenders LH. Interaction of arsenic(III) with nucleotide excision repair in UV-irradiated human fibroblasts. Carcinogenesis. 1997;18:399–405. doi: 10.1093/carcin/18.2.399. [DOI] [PubMed] [Google Scholar]
- Hartwig A, Pelzer A, Asmuss M, Burkle A. Very low concentrations of arsenite suppress poly(ADP-ribosyl)ation in mammalian cells. Int J Cancer. 2003b;104:1–6. doi: 10.1002/ijc.10911. [DOI] [PubMed] [Google Scholar]
- Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Huang C, Ke Q, Costa M, Shi X. Molecular mechanisms of arsenic carcinogenesis. Mol Cell Biochem. 2004;255:57–66. doi: 10.1023/b:mcbi.0000007261.04684.78. [DOI] [PubMed] [Google Scholar]
- Hughes MF, Kenyon EM, Kitchin KT. Research approaches to address uncertainties in the risk assessment of arsenic in drinking water. Toxicol Appl Pharmacol. 2007;222:399–404. doi: 10.1016/j.taap.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Ikejima M, Noguchi S, Yamashita R, Ogura T, Sugimura T, Gill DM, Miwa M. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. J Biol Chem. 1990;265:21907–21913. [PubMed] [Google Scholar]
- Kessel M, Liu SX, Xu A, Santella R, Hei TK. Arsenic induces oxidative DNA damage in mammalian cells. Mol Cell Biochem. 2002;234-235:301–308. [PubMed] [Google Scholar]
- Kitchin KT. Recent advances in arsenic carcinogenesis: modes of action, animal model systems, and methylated arsenic metabolites. Toxicol Appl Pharmacol. 2001;172:249–261. doi: 10.1006/taap.2001.9157. [DOI] [PubMed] [Google Scholar]
- Kitchin KT, Ahmad S. Oxidative stress as a possible mode of action for arsenic carcinogenesis. Toxicol Lett. 2003;137:3–13. doi: 10.1016/s0378-4274(02)00376-4. [DOI] [PubMed] [Google Scholar]
- Kitchin KT, Wallace K. Arsenite binding to synthetic peptides based on the Zn finger region and the estrogen binding region of the human estrogen receptor-alpha. Toxicol Appl Pharmacol. 2005;206:66–72. doi: 10.1016/j.taap.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Kupper JH, de Murcia G, Burkle A. Inhibition of poly(ADP-ribosyl)ation by overexpressing the poly(ADP-ribose) polymerase DNA-binding domain in mammalian cells. J Biol Chem. 1990;265:18721–18724. [PubMed] [Google Scholar]
- Kuzminov A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc Natl Acad Sci U S A. 2001;98:8241–8246. doi: 10.1073/pnas.131009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz RC, Hays AM. Role of oxidative stress in arsenic-induced toxicity. Drug Metab Rev. 2006;38:791–804. doi: 10.1080/03602530600980108. [DOI] [PubMed] [Google Scholar]
- Liu W, Rosenberg GA, Liu KJ. AUF-1 mediates inhibition by nitric oxide of lipopolysaccharide-induced matrix metalloproteinase-9 expression in cultured astrocytes. J Neurosci Res. 2006;84:360–369. doi: 10.1002/jnr.20895. [DOI] [PubMed] [Google Scholar]
- Lockett KL, Hall MC, Clark PE, Chuang SC, Robinson B, Lin HY, Su LJ, Hu JJ. DNA damage levels in prostate cancer cases and controls. Carcinogenesis. 2006;27:1187–1193. doi: 10.1093/carcin/bgi288. [DOI] [PubMed] [Google Scholar]
- Miwa M, Masutani M. PolyADP-ribosylation and cancer. Cancer Sci. 2007;98:1528–1535. doi: 10.1111/j.1349-7006.2007.00567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papworth M, Kolasinska P, Minczuk M. Designer zinc-finger proteins and their applications. Gene. 2006;366:27–38. doi: 10.1016/j.gene.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Pi J, He Y, Bortner C, Huang J, Liu J, Zhou T, Qu W, North SL, Kasprzak KS, Diwan BA, Chignell CF, Waalkes MP. Low level, long-term inorganic arsenite exposure causes generalized resistance to apoptosis in cultured human keratinocytes: potential role in skin co-carcinogenesis. Int J Cancer. 2005;116:20–26. doi: 10.1002/ijc.20990. [DOI] [PubMed] [Google Scholar]
- Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- Powell SR. The antioxidant properties of zinc. J Nutr. 2000;130:1447S–1454S. doi: 10.1093/jn/130.5.1447S. [DOI] [PubMed] [Google Scholar]
- Rossman TG. Mechanism of arsenic carcinogenesis: an integrated approach. Mutat Res. 2003;533:37–65. doi: 10.1016/j.mrfmmm.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Rossman TG, Uddin AN, Burns FJ. Evidence that arsenite acts as a cocarcinogen in skin cancer. Toxicol Appl Pharmacol. 2004;198:394–404. doi: 10.1016/j.taap.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Rossman TG, Uddin AN, Burns FJ, Bosland MC. Arsenite is a cocarcinogen with solar ultraviolet radiation for mouse skin: an animal model for arsenic carcinogenesis. Toxicol Appl Pharmacol. 2001;176:64–71. doi: 10.1006/taap.2001.9277. [DOI] [PubMed] [Google Scholar]
- Rossman TG, Uddin AN, Burns FJ, Bosland MC. Arsenite cocarcinogenesis: an animal model derived from genetic toxicology studies. Environ Health Perspect. 2002;110(Suppl 5):749–752. doi: 10.1289/ehp.02110s5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostan EF, DeBuys HV, Madey DL, Pinnell SR. Evidence supporting zinc as an important antioxidant for skin. Int J Dermatol. 2002;41:606–611. doi: 10.1046/j.1365-4362.2002.01567.x. [DOI] [PubMed] [Google Scholar]
- Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem Res Toxicol. 2008;21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Shi H, Hudson LG, Ding W, Wang S, Cooper KL, Liu S, Chen Y, Shi X, Liu KJ. Arsenite causes DNA damage in keratinocytes via generation of hydroxyl radicals. Chem Res Toxicol. 2004a;17:871–878. doi: 10.1021/tx049939e. [DOI] [PubMed] [Google Scholar]
- Shi H, Shi X, Liu KJ. Oxidative mechanism of arsenic toxicity and carcinogenesis. Mol Cell Biochem. 2004b;255:67–78. doi: 10.1023/b:mcbi.0000007262.26044.e8. [DOI] [PubMed] [Google Scholar]
- Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol Cell Biol. 2000;20:3977–3987. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapio S, Grosche B. Arsenic in the aetiology of cancer. Mutat Res. 2006;612:215–246. doi: 10.1016/j.mrrev.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Tchounwou PB, Patlolla AK, Centeno JA. Carcinogenic and systemic health effects associated with arsenic exposure--a critical review. Toxicol Pathol. 2003;31:575–588. doi: 10.1080/01926230390242007. [DOI] [PubMed] [Google Scholar]
- Ueno S, Kashimoto T, Susa N, Natsume H, Toya M, Ito N, Takeda-Homma S, Nishimura Y, Sasaki YF, Sugiyama M. Assessment of DNA damage in multiple organs of mice after whole body X-irradiation using the comet assay. Mutat Res. 2007;634:135–145. doi: 10.1016/j.mrgentox.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Virag L. Structure and function of poly(ADP-ribose) polymerase-1: role in oxidative stress-related pathologies. Curr Vasc Pharmacol. 2005;3:209–214. doi: 10.2174/1570161054368625. [DOI] [PubMed] [Google Scholar]
- Wang CH, Hsiao CK, Chen CL, Hsu LI, Chiou HY, Chen SY, Hsueh YM, Wu MM, Chen CJ. A review of the epidemiologic literature on the role of environmental arsenic exposure and cardiovascular diseases. Toxicol Appl Pharmacol. 2007;222:315–326. doi: 10.1016/j.taap.2006.12.022. [DOI] [PubMed] [Google Scholar]
- Witkiewicz-Kucharczyk A, Bal W. Damage of zinc fingers in DNA repair proteins, a novel molecular mechanism in carcinogenesis. Toxicol Lett. 2006;162:29–42. doi: 10.1016/j.toxlet.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Yager JW, Wiencke JK. Inhibition of poly(ADP-ribose) polymerase by arsenite. Mutat Res. 1997;386:345–351. doi: 10.1016/s1383-5742(97)00011-2. [DOI] [PubMed] [Google Scholar]