Abstract
The GUCA1A gene encodes a guanylate cyclase-activating protein (GCAP1) that is involved in regulation of phototransduction in the vertebrate retina. We discovered a novel C312A transversion in exon 2 of the human GUCA1A gene, replacing Asn-104 (N104) in GCAP1 with Lys (K), in two affected members of a family with dominant cone dystrophy. The mutation N104K is located in the third EF hand motif (EF3) shown previously to be instrumental in converting Ca2+-free GCAP1 to a GC inhibitor in the Ca2+-bound form. In one patient, rod ERGs were fairly stable over a 12-year-period whereas 30 Hz flicker ERG and single-flash cone ERGs declined. In both patients, double flash ERGs showed that rod recovery from an intense test flash was significantly delayed. The EC50 for GC stimulation shifted from ~250 nM in wild-type GCAP1 to ~800 nM in the GCAP1(N104K) mutant suggesting inability of the mutant to assume an inactive form under physiological conditions. The replacement of N104 by K in GCAP1 is the first naturally occurring mutation identified in the EF3 loop. The rod recovery delays observed in double-flash ERG of affected patients suggest a novel dominant-negative effect that slows GC stimulation.
Introduction
Photoreceptors, using a process termed phototransduction, receive light and generate an electrical impulse that is sent to the visual cortex (Ridge et al., 2003; Burns & Arshavsky, 2005; Stephen et al., 2008). Key events in phototransduction are the hydrolysis of cGMP, the secondary messenger of phototransduction, and closure of cGMP-gated cation channels (Polans et al., 1996). Channel closure triggers a change in free [Ca2+] (Pugh, Jr. et al., 1999; Nakatani et al., 2002) and re-synthesis of cGMP by membrane-associated guanylate cyclases (GCs) present in rod and cone outer segments (Baehr et al., 2007). Changes in cytoplasmic [Ca2+] are monitored by GC-activating proteins (GCAPs) which are Ca2+-binding proteins of the calmodulin superfamily (Palczewski et al., 2004). Conformations of GCAPs change in response to Ca2+ binding. In dark-adapted outer segments, free Ca2+ is controlled by a light-insensitive NCKX and the cGMP-gated channel, and adjusts to about 250–600 nM (Woodruff et al., 2007). Under these conditions, high affinity Ca2+-binding sites (EF hands) on GCAPs are saturated, GCAPs are inactive and the GCs display a low basal activity (Gorczyca et al., 1994a). In response to light, free [Ca2+] drops to less than 50 nM, Ca2+ dissociates from Ca2+-binding sites, and GCAPs convert into activators accelerating cGMP synthesis roughly 8–10 fold.
GCAPs are N-myristoylated neuronal Ca2+ sensors in which the acyl side chain is buried in the Ca2+-bound as well as the Ca2+-free state (Stephen et al., 2007). A hallmark of the GCAP structure are high affinity Ca2+-binding sites termed EF hands (Persechini et al., 1989; Falke et al., 1994; Gifford et al., 2007) consisting of a helix-loop-helix secondary structure that is able to chelate Ca2+ ions. EF hands also have affinity for Mg2+ ions but the interaction is several orders of magnitude weaker (Gifford et al., 2007). In the canonical EF hand, the loop consists of 12 amino acids rich in acidic residues providing oxygen ligands for Ca2+ coordination. The loop is flanked by hydrophobic residues (I, L, Y, W). GCAPs have two pairs of EF hands, one each in the N-terminal and C-terminal half of the molecule. The first EF hand in the N-terminal region is nonfunctional, as Ca2+ coordination is prevented by lack of acidic side chains providing oxygen for Ca2+ coordination (Palczewski et al., 2004). The N-terminal region including EF1 has a key role in interaction with the target protein GC (Krylov et al., 1999; Li et al., 2001; Ermilov et al., 2001). EF hands 2–4 are fully functional, canonical EF-hand Ca2+-binding sites. Their individual roles have been explored mostly by site-directed mutagenesis, and recording of conformational changes in the absence and presence of Ca2+ and/or Mg2+ (Otto-Bruc et al., 1997; Rudnicka-Nawrot et al., 1998; Sokal et al., 1999; Peshenko & Dizhoor, 2007).
Pathogenic mutations of residues flanking EF3 and EF4, as well as several residues of the EF4 loop of GCAP1, are associated with autosomal dominant cone dystrophy (adCD) or autosomal dominant cone-rod dystrophy (adCRD) (for review: (Baehr & Palczewski, 2007)). Four mutations are known to affect Ca2+-binding of EF hands (Y99C, I143NT, L151F, E155G) (Sokal et al., 1998; Dizhoor et al., 1998; Wilkie et al., 2001; Nishiguchi et al., 2004; Jiang et al., 2005; Sokal et al., 2005). The Y99C mutation is located adjacent to EF3 (Payne et al., 1998) and I143NT adjacent to the EF4-hand motif (Nishiguchi et al., 2004). Two mutations (E155G, L151F) are located in EF4. These mutations alter the dissociation constant and coordination of Ca2+ to the mutant loop, and change the Ca2+ sensitivity of GCAP1. As a result, mutant GCAPs are not fully inactivated at dark Ca2+ levels, leading to the persistent stimulation of GC1 in the dark, elevated cGMP and Ca2+ levels, and cell death. To date, no naturally-occurring mutations affecting one of the 12 amino acids of the Ca2+-binding loop of EF3 had been identified. Here, we investigated a single family with dominant cone dystrophy and identified a novel mutation in the GUCA1A gene in which Asn 104 (N104), located in EF3, is replaced by lysine. It is predicted that the N104K substitution will dramatically affect the ability of the mutant GCAP1 to inhibit GC1 in dark-adapted cone photoreceptors.
Methods
Patients and Mutation Identification
This study was approved by each Institutional Review Board of the University of Utah Hospitals and Clinics, and the University of Texas Southwestern Medical Center-Dallas. All subjects provided informed consent prior to participation. Some subjects underwent complete ophthalmologic examination including visual acuity measurements and fundus examinations. Blood samples from two family members (#8279 and #4940, Figure 1) were collected under consignment and genomic DNA was extracted using the Puregene DNA isolation kit. Each of the four exons of GCAP1 was amplified by PCR using flanking intron-specific primers (GCAP1_exon1F: 5’-GGCCTGTCCATCTCAGACGT; GCAP1_ exon1R: 5’-CCCCAGCTGGTCAGGCTTCCAG; GCAP1_ exon2F: 5’-GCCTGAGGCTGGAGTGAGCG; GCAP1_ exon2R: 5’-CTAACCCTGGGCTCTCAGTTCC; GCAP1_exon3F: 5’-CCTGAGATAGGATAAGGATGG; GCAP1_exon3R: 5’-ACCCCACATCCATGGTGACC; GCAP1_exon4F: 5’-CTGGACTGCAGAAATGAACACCCTC; GCAP1_exon4R: 5’-GGCGAGCTAAGCCTCTGAGTTC) and screened for mutations by denaturing high performance liquid chromatography (DHPLC; WAVE® System, Transgenomic, Omaha, NE). Sequence alterations were identified by direct sequencing with a CEQ Dye Terminator Cycle Sequencing Kit on Beckman-Coulter CEQ 8000 Genetic Analysis System, according to the manufacturer’s instructions and using established methods (Yang et al., 2005; Yang et al., 2006).
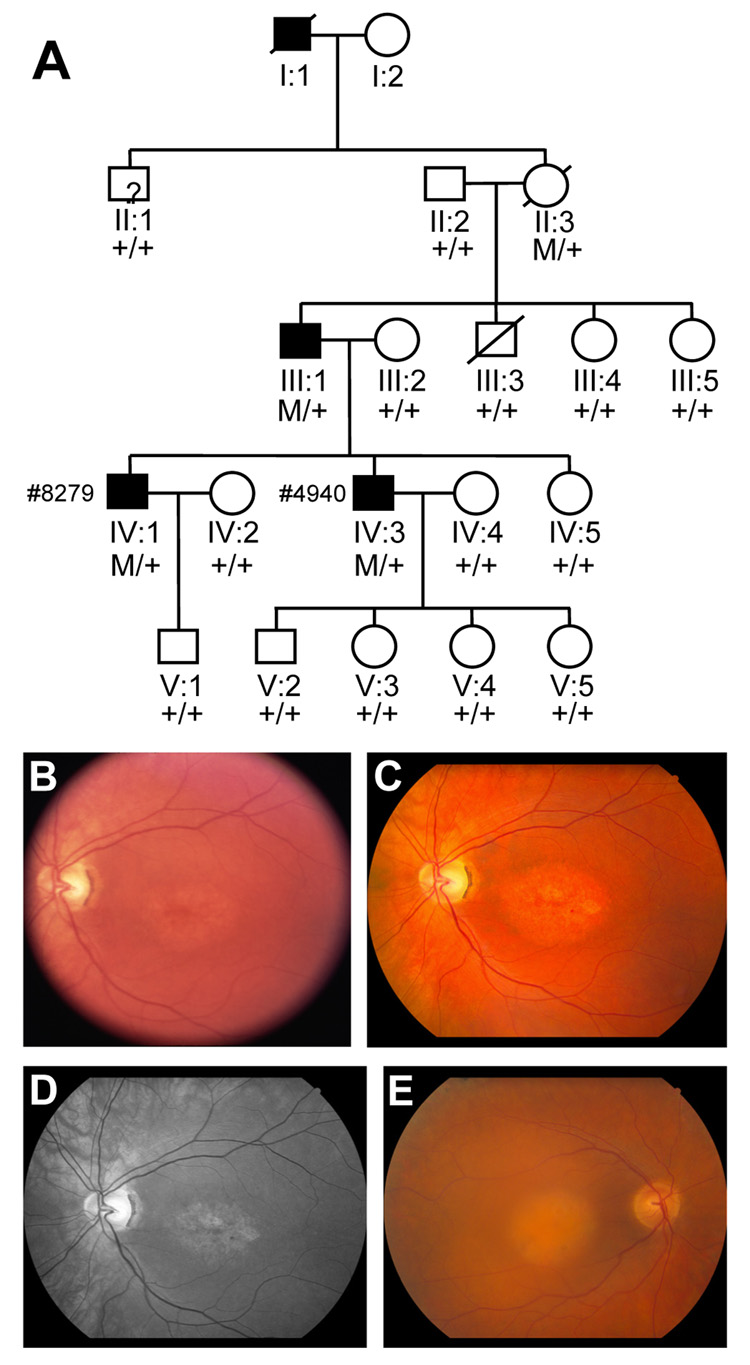
Figure 1.
A. Pedigree of the study family with autosomal dominant cone dystrophy: Individuals are identified by pedigree number. Squares indicate males; circle, females; slashed symbols, deceased; solid symbols, affected; open symbols, unaffected; +/+, two copies of wild type; M/+, one copy of wild type, one copy of mutant. B–D. Fundus photos of the posterior pole of the left eye of patient # 4940 at two separate visits, seven years apart. In June of 2000 a Topcon 50 degree camera was used with Kodak Ektachrome 100 film and later digitized (B). In April of 2007, a Canon camera with EMI digital capturing system was used to take color posterior pole at 60 degrees (C) and a “red-free” image at 40 degrees for better contrast of the macular changes (D). E. Fundus photo of the posterior pole in patient # 8279 taken with Canon camera.
Electroretinography
Full-field ERGs were obtained following ISCEV standards (Marmor et al., 2004). Following pupil dilation (1% cyclopentolate hydrochloride and 2.5% phenylephrine hydrochloride) and 45 min dark-adaptation, ERGs were recorded as detailed (Birch & Fish, 1987). PC-based custom software was used for stimulus control and timing, data acquisition, averaging, and analysis. Inactivation kinetics were probed with the “paired-flash” paradigm (Birch et al., 1995), using a test flash of 2.4 log sc td-s. The paired-flash ERG method involves the presentation of a bright probe flash at a defined time after the test flash, and determination of the prevailing response to the test stimulus by analysis of the probe flash response. Underlying the method is the notion that the intense probe flash rapidly drives the rods to saturation and that the amplitude of the probe-generated ERG a-wave titrates the prevailing circulating current. The peak amplitude of the probe response as referred to the pre-probe baseline will approximate the circulating current at time t. From this determination of the probe response amplitude, and from the maximal amplitude exhibited by the probe-alone response, one obtains by subtraction an amplitude A that represents the rod response to the test flash at time t. That is,
where Am(t) is the probe response amplitude determined in the paired-flash trial, Amo is the amplitude of the probe-alone response, and A(t) is the amplitude at time t of the derived response to the test flash. We determined the entire time course of the response to the 2.4 log sc td-s flash in two patients (#4940; IV-3 and #8279; IV-1).
Expression and purification of recombinant human GCAP1s
Human GCAP1 coding sequence was amplified by PCR with primers including a 6-histidine tag just before the stop codon, two flanking restriction sites XbaI and XhoI, and then cloned into pFastBac-1. To generate a N104K point mutation, a site-directed mutagenesis (QuikChange® Site-Directed Mutagenesis Kit, Stratagene) was performed in this hGCAP1 pFastBac construct by using the following sense and antisense primer: 5’-GATGTAGATGGCAAAGGCTGCATTGACCGCG, 5’ –CGCGGTCAATGCAGCCTTTGCCATCTACATC. hGCAP1 wild type and N104K mutant plasmids were transformed into DH10Bac competent cells to generate the recombinant bacmids, and then the bacmids were transfected into High five insect cells to produce recombinant baculovirus and hGCAP1 proteins (Bac-to-Bac® Baculovirus Expression System, Invitrogen). Expression of the recombinant hGCAP1s was confirmed by Western blot analysis with GCAP1 antibody UW101. Recombinant hGCAP1 proteins were purified with Ni columns (Ni-NTA Columns, Qiagen). The purified proteins were dialyzed twice for 8–12 h against 2 L of protein buffer with glycerol (50 mM HEPES, 90 mM KCl, 10 mM NaCl, 50% v/v glycerol, pH 7.4) at 4°C. Dialysis of each 1 ml aliquot of GCAP preparat ion was performed using a 10 kDa MWCO Slide-A-Lyzer cassette (Pierce) in a cold room. The purity and concentration of hGCAP1 proteins were estimated by SDS-PAGE with Coomassie blue staining and Bradford’s protein assay (Bio-Rad).
SDS-PAGE and limited proteolysis of hGCAP1s
SDS-PAGE in the presence and absence of Ca2+ (1mM Ca2+ or 1mM EGTA) and limited proteolysis of purified GCAP1 and GCAP1 mutants were carried out as described (Rudnicka-Nawrot et al., 1998; Nishiguchi et al., 2004). For limited proteolysis, the recombinant hGCAP1 wild type or mutant protein in the present or absence of Ca2+ (1mM Ca2+ or 1mM EGTA) was incubated with trypsin (at a mass ratio of 100:1) at 30°C. Proteolysis of hGCAP1 protein was stopped at various times with trypsin inhibitor (0, 5, 10, 20, 40, and 60 min), and the products were analyzed by SDS-PAGE with Coomassie staining.
Recombinant mouse Guanylate Cyclase 1 and preparation of washed cell membranes
HEK293 cells stably overexpressing mouse GC1 were plated at 15·× 106 cells /150 cm2 dish, grown for 48 h, washed twice with 5 ml of protein buffer (50 mM HEPES, 90 mM KCl, 10 mM NaCl, pH 7.4) with 1 µM leupeptin, and detached by scraping in protein buffe. The cells were then transferred to 15 ml conical tubes, pelleted by centrifugation and after removal of supernatant, snap frozen in liquid N2. To deplete the soluble proteins, each cell pellet was resuspended in 2 ml of water with 1 µM leupeptin and sonicated for 5 s at power level 3 (Branson Sonifier 150) followed by 20 min centrifugation at 16,000g. The cell membrane pellet was resuspended in 150 µl protein buffer with 1 µM leupeptin and immediately used for GC assay.
Guanylate Cyclase Assay
The GC activity was assayed as described (Gorczyca et al., 1994a; Gorczyca et al., 1994b) with minor modifications. Briefly, each 50 µl reaction mix contained 10 µl of 5X assay buffer (150 mM HEPES, 270 mM KCl, 30 mM NaCl, 50 mM MgCl2, 2.5 mM EGTA), 10 µl of 5 mM IBMX (3-Isobuyl-1-methyl-xanthine, Sigma), 10 µl of washed cell membranes in protein buffer (50 mM HEPES, 90 mM KCl, 10 mM NaCl, pH 7.4), 10 µl of 25 µM wildtype or mutant(N104K) GCAP in protein buffer with 50% v/v glycerol, 5 µl of CaCl2 (concentration ranging from 0.34 – 6 mM to obtain free Ca2+ concentration as calculated by computer program WEBMAXC STANDARD v. 5/21/2007 (Patton et al., 2004)), and 5 µl of nucleotide mix (10 mM GTP, 0.5 µCi α-33P-GTP). The reaction was initiated by addition of nucleotides, incubated at 30°C for 10 min and stopped by addition of 15 µl of 0.4M HCl. Cell membranes were removed by centrifugation at 16,000gfor 4 min; the supernatant (40 µl) was transferred to 150 mg of neutral alumina (ICN Biomedicals) in 0.5 ml of 200 mM Tris, 50 mM EDTA, pH 7.4 and vortexed for 8 min. Following centrifugation at 16,000g for 4 min, the supernatant (0.3 ml) was mixed with 3 ml of scintillation cocktail and radioactivity was measured by a scintillation counter.
Results
Human patients
The proband (IV-3; #4940) was first seen at the Retina Foundation of the Southwest in 1995 at age 33. His acuity was OD 20/200 and OS 20/80. His refraction was OD: −2.00 +3.00 * 095; OS: −2.00 +3.00 * 082. He was diagnosed with dominant cone dystrophy by the referring retinal specialist. The pedigree (Fig. 1A), fundus appearance (Fig. 1B), and full-field ERG (Fig. 2) were all consistent with the diagnosis. On successive visits, his acuity dropped to OD: 20/320, OS:20/250 in 2000, OD: 20/400, OS:20/400 in 2004 and OD: 20/500, OS:20/500 in 2007. Humphrey visual fields showed a dense central scotoma which enlarged over time with near normal sensitivity in the peripheral retina. Visual thresholds 7 degrees below fixation following 45 min of dark adaptation were elevated 0.1 log unit in 1995, within normal limits in 2000 and 2004 and elevated 0.1 log unit in 2007.
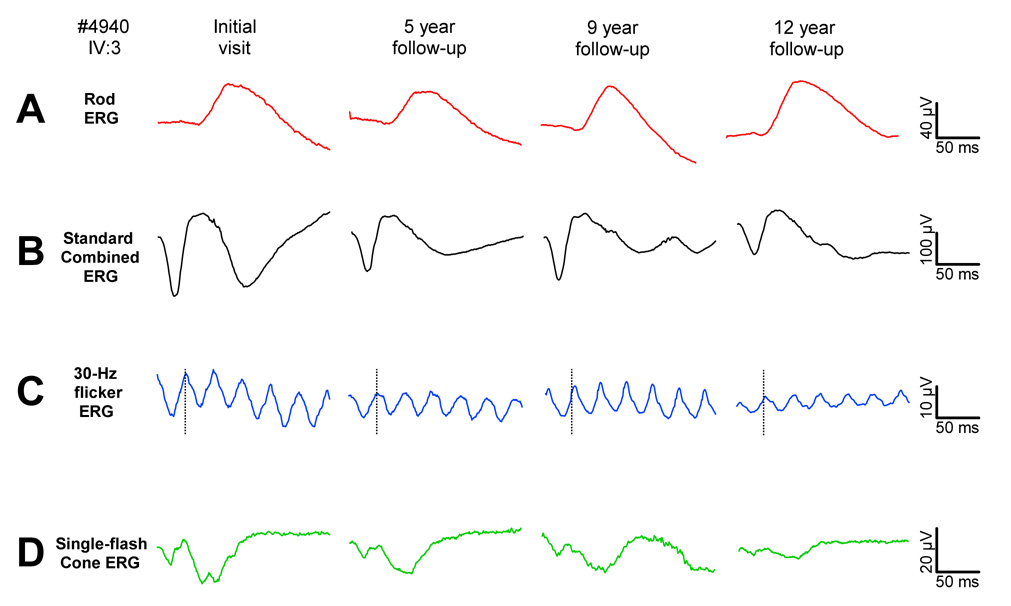
Figure 2.
Standard ISCEV (International Society for Clinical Electrophysiology of Vision) protocol ERGs obtained over a 12-year-period of follow-up in patient # 4940. A, The ISCEV standard rod response. B. The standard combined (rod and cone) ERG response. C. The cone response to a 30 Hz flickering stimulus. D. The cone response presented in the presence of a rod-saturating background. Typical of patients with CD, rod responses remain fairly stable over the follow-up period, while cone responses show considerable progression.
The proband’s brother (IV-1; #8279) was first seen at age 47. His acuity was OD 20/320 and OS 20/200. His refraction was OD: −5.00 +3.75 * 113; OS: −3.50 +2.00 * 077. Humphrey visual fields showed a dense central scotoma with near normal sensitivity in the peripheral retina. Visual thresholds 7 degrees below fixation following 45 min of dark-adaptation were within the normal range.
Electroretinography
Full-field ERGs from the proband are shown in Fig. 2. Rod responses to the ISCEV standard stimulus (Fig. 2A) were reduced 40% in amplitude compared to the lower limit of normal, and borderline delayed in b-wave implicit time (89.6 ms vs. upper limit of normal of 88.2 ms). Similarly, mixed rod and cone combined ERGs were reduced 40% in amplitude (compared to the lower limit of normal (Fig. 2B). Cone responses to the 30 Hz flicker (Fig. 2C) were reduced 65% in amplitude compared to the lower limit of normal. With a b-wave implicit time of 34.4 ms, these responses were only slightly delayed relative to the upper limit of normal (31.5 ms). Similarly, the single-flash cone ERG (Fig. 2D) was 70% reduced in amplitude and borderline normal in b-wave implicit time. Over 12 years of follow-up, rod responses fluctuated somewhat in amplitude but showed no clear trend toward progression. Cone responses, on the other hand, declined progressively over the 12 years, but at a slow rate of 0.66 mV (5%) per year.
The brother of the proband (IV-1, #8279) was tested once at age 47 yrs. His ISCEV standard rod response was reduced in amplitude by 76%, while cone responses to 30 Hz flicker and single flashes were reduced in amplitude by greater than 99% (results not shown). Both rod and cone responses were borderline delayed in b-wave implicit time.
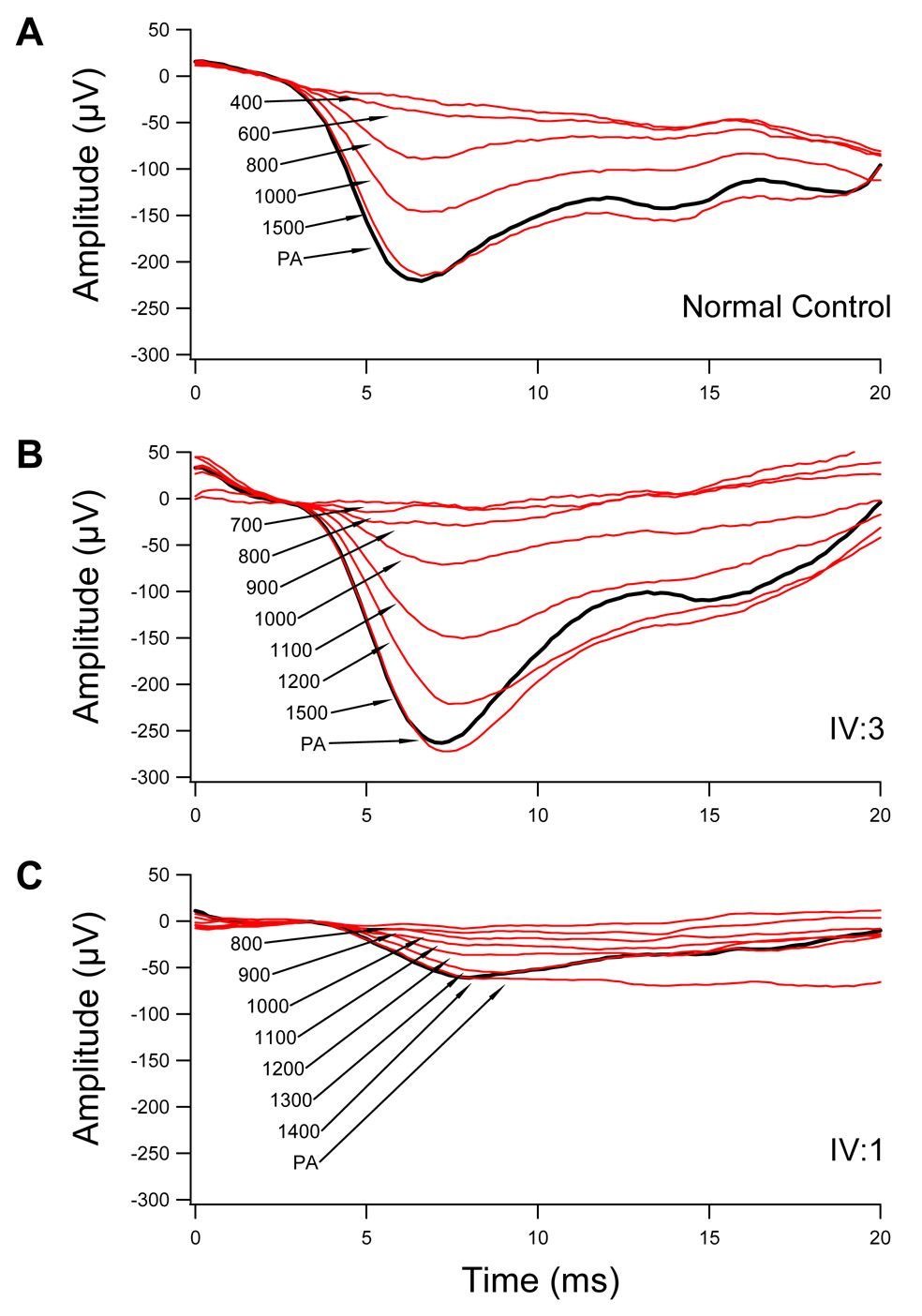
Results from the paired-flash paradigm are shown in Fig. 3 for a representative normal subject (3A), the probands (3B), and his brother (3C). The trace marked PA represents the first 20 ms of the maximal, saturated rod response to a probe flash of 4.3 log sc td-s (log scotopic troland-seconds) presented singly to the dark-adapted eye. When presented shortly after a just-saturating test flash of 2.4 log sc td-s, no response to the probe is detected as the photoresponse is still saturated. The a-wave first becomes evident at about 600 ms in the normal subject, and about 900 ms in the patients. As the interval between paired flashes lengthens, the response to the probe grows in amplitude.
Figure 3.
A. Paired-flash method in a normal control. Cone responses are computer-subtracted to isolate the rod photoresponse. When presented singly, the probe alone (PA) produces a maximal, saturated response from the rod photoreceptors. When presented shortly after a just-saturating test flash of 2.4 log sc td-s, the probe elicits no response as the photoresponse is still saturated. As the interval between paired flashes lengthens, the response to the probe becomes evident and progressively grows in amplitude. With an interval of approximately 1500 ms, the response has fully recovered; i.e., inactivation of the photoresponse is complete. B. Paired-flash responses from patient IV:3. C. Paired-flash responses from patient IV:1
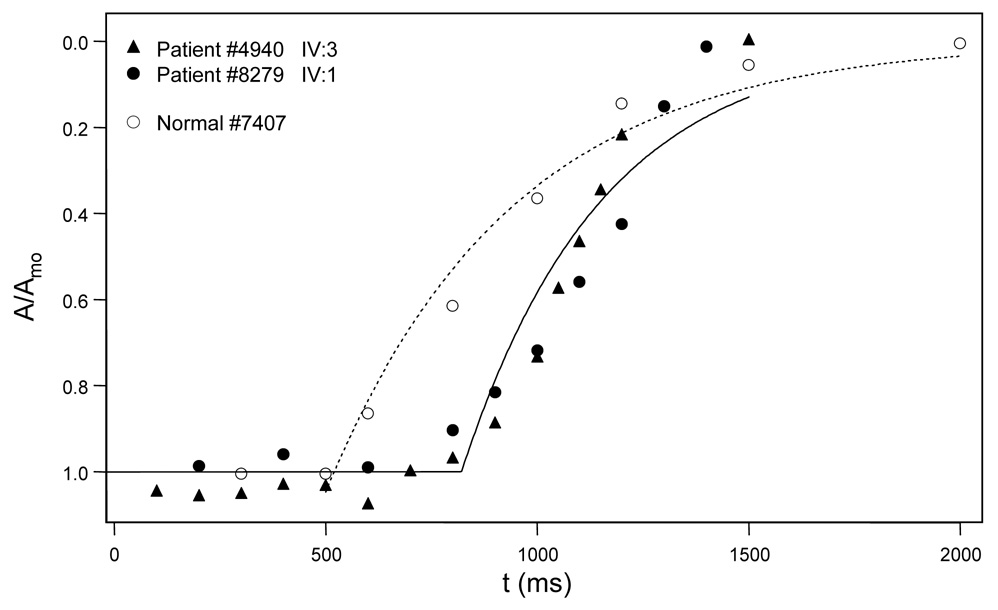
Full recovery functions for the proband (#4940), his brother (#8279) and the representative normal subject are shown in Fig. 4. Relative amplitude [the ratio of A to maximum saturated amplitude (Amo)] is plotted as a function of the time (t) between the test flash and the probe flash. The best-fit curves are exponential recovery functions: A/Amo = exp[-(t-Tsat)/ τ], where Tsat is the period of rod saturation that precedes recovery and t is a recovery time constant. Compared to the representative normal subject (Tsat = 520 ms) and a mean normal of 490.1 ± 111.2 ms (Kozma et al., 2005), both the proband (889 ms) and his brother (802 ms) showed large delays.
Figure 4.
Recovery functions derived from responses shown in Fig. 3A (patient IV:3, filled circles), Fig. 3B (patient IV:1, triangles), and Fig. 3C (normal control, open circles). Open circles (control), the amplitude of each photoresponse is measured at a fixed time following the probe flash. Relative amplitude is the ratio of A to maximum saturated amplitude (Amo). The dotted curve is an exponential recovery function: A/Amo = exp[-(t-Tsat)/τ], where Tsat is the period of rod saturation that preceded recovery and t is a recovery time constant. Filled circles, recovery function from patient IV:3. Filled triangles, recovery function from patient IV:1. Note the delay in Tsat for each patient (#4940 = 889 ms; #8279 = 802 ms) compared to the normal control (Tsat = 520 ms). Mean normal (based on 10 normal subjects) for Tsat is 490.1 ms, with sd = 111.2 ms.
Identification of the mutation in the GUCA1A gene
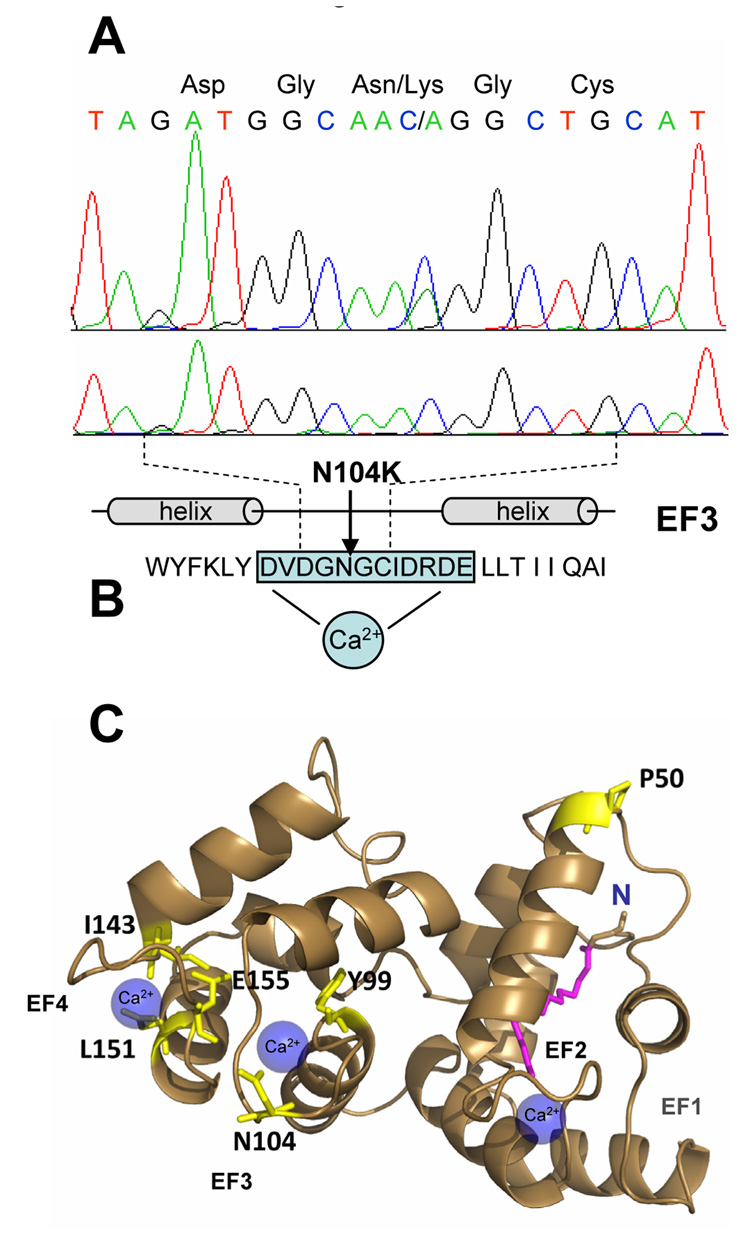
Mutation screening by DHPLC (denaturing high pressure liquid chromatography) and direct sequencing of amplified GUCA1A exons of patient IV:3 identified a novel C312A transversion in exon 2 (Fig. 5A). This mutation replaced N104 in GCAP1 with Lys resulting in a novel N104K change in the GCAP1 amino acid sequences in the affected individual. No mutation was identified in exon 2 of over 200 normal controls. The N104K mutation occurs within the third EF-hand domain of GCAP1 and represents the first naturally occurring mutation in this 12-amino-acid-loop (Fig. 5B). A mutation of a residue flanking EF3 (Y99C) disrupts the N-terminal helix of the helix-loop-helix conformation of EF3, severely affecting Ca2+ binding at this site. Another pathogenic mutation of a flanking hydrophobic residue (I143NT) (Nishiguchi et al., 2004) was observed in EF4, emphasizing the importance of an intact N-terminal helix for Ca2+ binding. Other mutations linked to cone dystrophy (E155G, and L151F) affect exclusively Ca2+ coordination in EF4 (Wilkie et al., 2001; Jiang et al., 2005; Sokal et al., 2005) (Fig. 5B).
Figure 5.
A. DNA sequences of the affected individual IV:3 (top) and a normal control (bottom) showing a C to A transversion in exon 2 of GUCA1A, resulting in a Asp to Lys change (N104K) in the EF3 hand of GCAP1. B. Location of N104 in EF3. Binding of Ca2+ to this loop contributes in switching GCAP1 from the inhibited to the activated state. C. The structure of myristoylated chicken GCAP1 showing the arrangement of the four EF hands and the position of mutant amino acids homologous to the human sequence. The structure was drawn with Pymol software based on the 2.0 A resolution crystal structure (Stephen et al., 2007). Amino acids mutated in autosomal dominant cone dystrophy are indicated in red. The Y99C mutation is located adjacent to the EF3-hand motif (Payne et al., 1998) and three mutations (I143NT, E155G, L151F) have been described in EF4-hand motif (Wilkie et al., 2001; Nishiguchi et al., 2004; Jiang et al., 2005; Sokal et al., 2005). A fifth mutation (P50L) is located between EF1 and EF2, and does not affect Ca2+ binding (Newbold et al., 2001). The N-terminal myristoyl side chain, buried in the N-terminal domain, is shown in purple in close proximity to the C-terminal helix.
Ca2+-dependent conformational changes in wild-type and mutant GCAP1
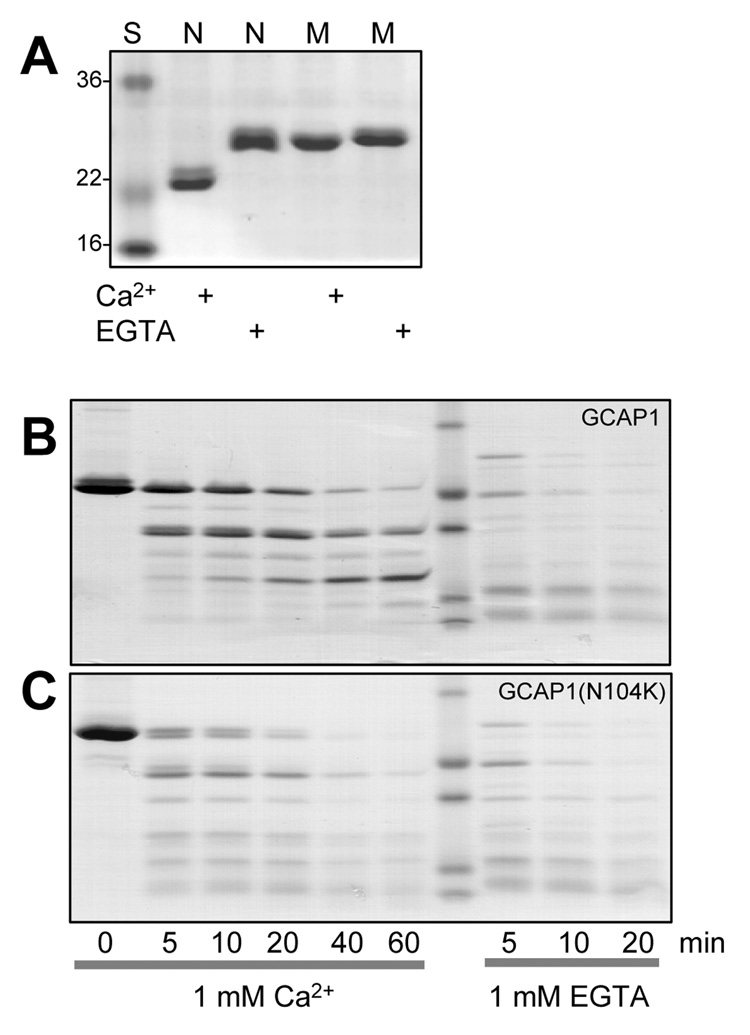
N104, the 5th amino acid residue of the EF3 loop, provides a key ligand for Ca2+ coordination. The N104K mutation is predicted to severely weaken Ca2+-binding in EF3 since it introduces a positively charged amino acid destroying Ca2+ coordination of the amide side chain of Asn. To verify the effects of N104K on Ca2+-binding, we analyzed recombinant GCAP1(N104K) by two biochemical methods, mobility shift in denaturing polyacrylamide gels in the presence and absence of Ca2+, and susceptibility to proteolysis in the Ca2+-free state. Ca2+-binding proteins of the calmodulin superfamily undergo characteristic conformational changes and shifts in mobility in SDS gels, depending on the state of Ca2+ coordination. In its Ca2+-free form, GCAP1 migrates slower than the Ca2+-bound form most likely based on conformational changes. As shown in Fig. 6A, wildtype Ca2+-free GCAP1 displays an apparent mobility of nearly 30 kDa while Ca2+-bound GCAP1 migrates like a 23 kDa protein, corresponding to its calculated molecular mass. Recombinant GCAP1(N104K) showed a Ca2+-shift very similar to the Ca2+-free form regardless of high or low [Ca2+], an effect that is in contrast to GCAP1(Y99C) whose Ca2+/EGTA shifts are identical to WT GCAP1 (Sokal et al., 1998).
Figure 6.
A. Ca2+-dependent mobility shift of wild-type GCAP1 (N) and GCAP1(N104K) (M) recombinant proteins. Coomassie blue staining SDS-PAGE gel of GCAP1, GCAP1(N104K) in the presence or absence of Ca2+. B, C. Limited proteolysis of GCAP1 and of GCAP1(N104K) by trypsin in the presence and absence of Ca2+. The digestions were performed at 30°C at ratio of GCAP1/trypsin 100:1, and the digest was analyzed by SDS-PAGE at 0, 5, 10, 20, 40, and 60 min.
We showed previously that, in the absence of Ca2+, GCAP1 is readily susceptible to proteolysis by trypsin (Rudnicka-Nawrot et al., 1998; Sokal et al., 2005), presumably by exposing positively charges residues (sites of trypsin cleavage). Exposure of GCAP1 to trypsin in the absence of Ca2+ leads to complete digestion in less than 10 min (Fig. 6B). Proteolysis is restricted, however, when Ca2+ is bound to the EF hands because cleavage sites become more inaccessible owing to conformational changes (Fig. 6C). In contrast to wild-type GCAP1, GCAP1(N104K) is more susceptible to proteolysis even at 1 mM Ca2+. We conclude that the N104K mutation introduces a structural change that is irreversible even at 1 mM Ca2+.
GCAP1(N104K) stimulates GC1 less efficiently and is incompletely inactivated
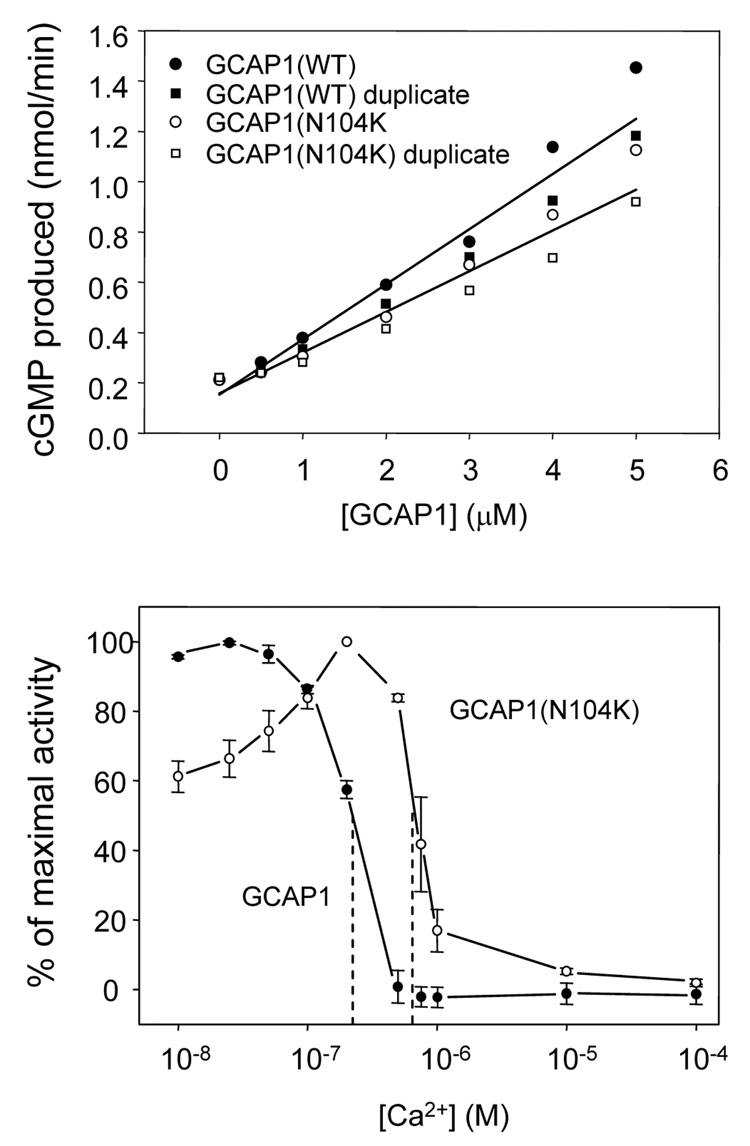
We next compared the ability of GCAP1(N104K) and wild-type GCAP1 to stimulate GC1 in vitro (Fig. 7). Wildtype and mutant GCAPs were assayed for GC1 stimulation in the presence of variable amounts on GCAP1 (Fig. 7A), as well as a function of free Ca2+ (Fig. 7B). The specific activity of the mutant GCAP1 is significantly reduced relative to wildtype (Fig. 7A) suggesting that the mutation affects GCAP1-GC1 interactions (see discussion). Previous analyses of GCAP1 EF-hand loop or flanking mutations (Sokal et al., 1998; Dizhoor et al., 1998; Wilkie et al., 2001; Nishiguchi et al., 2004; Sokal et al., 2005) showed incomplete inactivation of GC1 at physiological dark Ca2+ levels present in dark-adapted photoreceptors. As shown in Fig. 7B, GCAP1(N104K) is active at low [Ca2+] with a peak activity at 200 nM [Ca2+], dropping to about 60% at very low Ca2+ (10 nM). With increasing free [Ca2+], the mutant GCAP1 remains active at Ca2+ levels at which wildtype GCAP1 is inactive. Consistent with other EF-hand mutations, the findings indicate that mutations affecting EF3 will partially impair GC inhibition, leading to persistent stimulation in dark-adapted photoreceptors, and raising [cGMP] to toxic levels.
Figure 7.
GC stimulation by wildtype versus mutant GCAP1. The effect of wildtype GCAP1 and GCAP1(N104K) on GC activity. A. The effect of increasing amounts of wildtype GCAP1 (●,■) and GCAP1(N104K) (○,□) on GC activity in washed membranes of HEK293 cells overexpressing GC1. The free Ca2+ concentration was 50 nM. B. The calcium titration of wildtype GCAP1- or GCAP1(N104K)-mediated GC1 activity. The GC1 activity was assayed in washed cell membranes of HEK293 cells overexpressing GC1 in the presence of 5 µM wildtype GCAP1 (●) or GCAP1(N104K) (○) and varying free Ca2+ concentrations set by Ca2+/EGTA buffering (Materials and Methods). The experiments were performed in triplicate. Error bars are standard deviation.
Discussion
Heterogeneous cone and cone-rod dystrophies (CD and CRD, respectively), inherited in a dominant, recessive, or X-linked fashion, are rare diseases characterized primarily by progressive dysfunction of the cone-mediated visual system. Major hallmarks are photophobia, reduced central visual acuity, achromatopsia, but initially preserved peripheral vision mediated by rod photoreceptors (Simunovic & Moore, 1998; Hamel, 2007). CD and CRD are diagnosed mainly on the basis of changes in the photopic and scotopic electroretinogram, but also by fundoscopy and optical coherence tomography (Jiang et al., 2005; Wolfing et al., 2006; Baraas et al., 2007; Wissinger et al., 2008). The best characterized genes linked to dominant CD/CRD are GUCY2D, encoding photoreceptor guanylate cyclase 1 (retGC-1 or GC1), and GUCA1A, encoding the Ca2+-binding protein GCAP1, which are both expressed exclusively in photoreceptors (Perrault et al., 2000; Palczewski et al., 2004).
The phenotypes of the two patients from the GCAP1(N104K) family are comparable to other patients with CD. In young patients, the rod ERG can be normal. Both patients reported here had developed dense central scotomas, which is typical of the disease and can modestly affect the rod ERG. Nevertheless, rod function in the periphery remained normal in both patients on all visits. Normal peripheral rod function in these patients distinguishes them from patients with CRD, where rod function shows progressive decline throughout the retina (Birch et al., 1999). The cone ERG was more severely affected than the rod ERG in both patients and showed progressive decline over 12 years of follow-up in one patient. Both patients showed substantial increases in Tsat, the period of rod saturation that precedes recovery from an intense flash. This delayed recovery in the rod ERG photoreceptor response has been shown to be analogous to a delay in inactivation in single photoreceptor responses (Pepperberg et al., 2000). This delay presumably reflects a defect in the ability of mutant GCAP to respond to the drop in free [Ca2+] following cGMP-gated channel closure.
Other photoreceptor-specific genes linked to adCD or adCRD are CRX, RIM1, PITPNM3, and UNC119 (Table 1). The transcription factor CRX, a factor essential for the maintenance of mammalian photoreceptors (Freund et al., 1997), regulates the expression of several outer segment proteins such as visual pigments and arrestin. Human RIM1, a putative effector protein for rab3 proteins which are small GTPases involved in synaptic exocytosis (Wang et al., 2002; Johnson et al., 2003). RIM1 is a protein localizing to the synaptic ribbon and possibly involved in regulation of glutamate release (Schoch et al., 2002). CRD genes like those encoding a membrane-associated phosphatidylinositol transfer protein (PITPNM3), a human homolog of the D. melanogaster rdgB gene, and UNC119 encoding the retina specific gene RG4 with a possible function in synaptic transmission (Kobayashi et al., 2000) are less well characterized. Considering the diversity of genes and mutations linked to dominant CD, mutations in the GUCA1A gene are probably the best understood in terms of disease mechanism. In these GCAP1 mutants, residues involved in Ca2+ coordination or in stabilization of the loop structure are replaced by inappropriate amino acids lacking the desired properties. The consequence is inability of the mutant GCAP to assume an inactive conformation at physiological dark [Ca2+] (~500 nM). The crystal structure of GCAP1 with bound Ca2+ (Stephen et al., 2007) shows a molecule consisting of an N-terminal and C-terminal half, each with a pair of EF hands, as expected from molecular modeling (e.g., Fig 8 in (Sokal et al., 1999)). The N-terminal half forms a hydrophobic pocket to accommodate the myristoyl group attached to Gly-2, which is also contacted by the C-terminal helix stabilizing the link between the two halves. The N-terminal region, including EF1 which does not coordinate Ca2+, is thought to be instrumental for interaction with GCs. Under physiological conditions, the C-terminal half containing EF3 and EF4 is likely responsible for monitoring cytoplasmic [Ca2+] and triggering conformational changes that convert the inactive form into an activator. Introduction of a Trp fluorescent reporter residue flanking EF3 (Y99W) into GCAP1 and monitoring Ca2+ binding to EF3 revealed a strong conformational change (Km ~ 200 nM) suggesting that binding of Ca2+ to EF3 may be instrumental in forming the GC-activator. Tyr99 is a hydrophobic amino acid that does not distort the helix N-terminal to EF3. However, replacement of Y99 by a Cys residue (Y99C), the first mutation in GCAP1 linked to dominant cone dystrophy, had adverse effects on the structure of EF3 and Ca2+-binding (Sokal et al., 1998; Dizhoor et al., 1998). N104 occupies a position in the EF3 loop that is critical for Ca2+ coordination by providing oxygen of the amide side chain for coordination. Both Y99C and N104K likely weaken Ca2+ coordination at EF3 under physiological conditions preventing formation of the Ca2+ bound structure that is essential for inhibiting GC catalytic activity. The GCAP1 crystal structure implies that mutations in EF3 may distort contacts of the kinked C-terminal helix with the N-terminal helix of GCAP1.
Table 1.
Genetic loci associated with dominant cone dystrophies. Column 1, chromosomal localization. Column 2, disease nomenclature according to RetNet. Column 3, Online Mendelian Inheritance in Man (OMIM) nomenclature. Column 4, gene symbol. Column 5, function of the gene product. Column 6, references.
| Locus | Symbol | OMIM | Gene | Function | Reference |
|---|---|---|---|---|---|
| 6p21.1 | CORD3 | 602093 | GUCA1A | Guanylate cyclase activator | Review: (Baehr & Palczewski, 2007) |
| 6q13 | CORD7 | 603649 | RIMS1 | Function unknown; ribbon synapse-associated | Johnson et al., 2003) |
| 17p13.1 | CORD6 | 601777 | GUCY2D | Photoreceptor guanylate cyclase | (Perrault et al., 1996); review: (Baehr & Palczewski, 2007) |
| 17p13.2 | CORD5 | 600977 | PITPNM3 | involved in photoreceptor membrane renewal; Drosophila homolog is retinal degeneration B (rdgB) | (Kohn et al., 2007) |
| 17q11.2 | 604011 | UNC119 | Function unknown; localizes to rod and cone cytoplasm and ribbon synapses | (Kobayashi et al., 2000) | |
| 18q21.1-q21.3 | CORD1 | 600624 | QRX | Transcription factor | (Wang et al., 2004) |
| 19q13.3 | CORD2 | 120970 | CRX | Transcription factor | (Swain et al., 1997) |
Acknowledgments
This work was supported by National Institute of Health grants EY008061 (KP), EY09076 (DB), EY08123 (WB), EY014800-039003 (NEI core grant), by center grants of the Foundation Fighting Blindness, Inc., to the University of Utah and to the Retina Foundation of the Southwest, a Foundation Fighting Blindness grant to KP, and an unrestricted grant to the Department of Ophthalmology at the University of Utah from Research to Prevent Blindness (RPB; New York).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
No conflict of interest
References
- Baehr W, Karan S, Maeda T, Luo DG, Li S, Bronson JD, Watt CB, Yau K-W, Frederick JM, Palczewski K. The function of Guanylate Cyclase 1 (GC1) and Guanylate Cyclase 2 (GC2) in rod and cone photoreceptors. J.Biol.Chem. 2007;282:8837–8847. doi: 10.1074/jbc.M610369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehr W, Palczewski K. Guanylate cyclase-activating proteins and retina disease. Subcell.Biochem. 2007;45:71–91. doi: 10.1007/978-1-4020-6191-2_4. [DOI] [PubMed] [Google Scholar]
- Baraas RC, Carroll J, Gunther KL, Chung M, Williams DR, Foster DH, Neitz M. Adaptive optics retinal imaging reveals S-cone dystrophy in tritan color-vision deficiency. J Opt.Soc.Am.A Opt.Image Sci Vis. 2007;24:1438–1447. doi: 10.1364/josaa.24.001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch DG, Anderson JL, Fish GE. Yearly rates of rod and cone functional loss in retinitis pigmentosa and cone-rod dystrophy. Ophthalmology. 1999;106:258–268. doi: 10.1016/S0161-6420(99)90064-7. [DOI] [PubMed] [Google Scholar]
- Birch DG, Fish GE. Rod ERGs in retinitis pigmentosa and cone-rod degeneration. Invest Ophthalmol.Vis.Sci. 1987;28:140–150. [PubMed] [Google Scholar]
- Birch DG, Hood DC, Nusinowitz S, Pepperberg DR. Abnormal activation and inactivation mechanisms of rod transduction in patients with autosomal dominant retinitis pigmentosa and the pro-23- his mutation. Invest Ophthalmol.Vis.Sci. 1995;36:1603–1614. [PubMed] [Google Scholar]
- Burns ME, Arshavsky VY. Beyond counting photons: trials and trends in vertebrate visual transduction. Neuron. 2005;48:387–401. doi: 10.1016/j.neuron.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Dizhoor AM, Boikov SG, Olshevskaya E. Constitutive activation of photoreceptor guanylate cyclase by Y99C mutant of GCAP-1. J.Biol.Chem. 1998;273:17311–17314. doi: 10.1074/jbc.273.28.17311. [DOI] [PubMed] [Google Scholar]
- Ermilov AN, Olshevskaya EV, Dizhoor AM. Instead of binding calcium, one of the EF-hand structures in guanylyl cyclase activating protein-2 is required for targeting photoreceptor guanylyl cyclase. J.Biol.Chem. 2001;276:48143–48148. doi: 10.1074/jbc.M107539200. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Drake SK, Hazard AL, Peerson OB. Molecular tuning of ion binding to calcium signaling proteins. Quantative Review Biophysics. 1994;27:219–290. doi: 10.1017/s0033583500003012. [DOI] [PubMed] [Google Scholar]
- Freund CL, Gregory EC, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis AA, Jacobson SG, Cepko CL, Bhattacharya SS, McInnes RR. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem.J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- Gorczyca WA, Gray-Keller MP, Detwiler PB, Palczewski K. Purification and physiological evaluation of a guanylate cyclase activating protein from retinal rods. Proc.Natl.Acad.Sci.U.S.A. 1994a;91:4014–4018. doi: 10.1073/pnas.91.9.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczyca WA, Van Hooser JP, Palczewski K. Nucleotide inhibitors and activators of retinal guanylyl cyclase. Biochemistry. 1994b;33:3217–3222. doi: 10.1021/bi00177a011. [DOI] [PubMed] [Google Scholar]
- Hamel CP. Cone rod dystrophies. Orphanet.J Rare.Dis. 2007;2:7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Katz BJ, Yang Z, Zhao Y, Faulkner N, Hu J, Baird J, Baehr W, Creel DJ, Zhang K. Autosomal dominant cone dystrophy caused by a novel mutation in the GCAP1 gene (GUCA1A) Mol.Vis. 2005;11:143–151. [PubMed] [Google Scholar]
- Johnson S, Halford S, Morris AG, Patel RJ, Wilkie SE, Hardcastle AJ, Moore AT, Zhang K, Hunt DM. Genomic organisation and alternative splicing of human RIM1, a gene implicated in autosomal dominant cone-rod dystrophy (CORD7) Genomics. 2003;81:304–314. doi: 10.1016/s0888-7543(03)00010-7. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Higashide T, Hamasaki D, Kubota S, Sakuma H, An W, Fujimaki T, McLaren MJ, Weleber RG, Inana G. HRG4 (UNC119) mutation found in cone-rod dystrophy causes retinal degeneration in a transgenic model. Invest Ophthalmol.Vis.Sci. 2000;41:3268–3277. [PubMed] [Google Scholar]
- Kohn L, Kadzhaev K, Burstedt MS, Haraldsson S, Hallberg B, Sandgren O, Golovleva I. Mutation in the PYK2-binding domain of PITPNM3 causes autosomal dominant cone dystrophy (CORD5) in two Swedish families. Eur.J Hum.Genet. 2007:664–671. doi: 10.1038/sj.ejhg.5201817. [DOI] [PubMed] [Google Scholar]
- Kozma P, Hughbanks-Wheaton DK, Locke KG, Fish GE, Gire AI, Spellicy CJ, Sullivan LS, Bowne SJ, Daiger SP, Birch DG. Phenotypic characterization of a large family with RP10 autosomal-dominant retinitis pigmentosa: an Asp226Asn mutation in the IMPDH1 gene. Am.J Ophthalmol. 2005;140:858–867. doi: 10.1016/j.ajo.2005.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov DM, Niemi GA, Dizhoor AM, Hurley JB. Mapping sites in guanylyl cyclase activating protein-1 required for regulation of photoreceptor membrane guanylyl cyclases. J Biol.Chem. 1999;274:10833–10839. doi: 10.1074/jbc.274.16.10833. [DOI] [PubMed] [Google Scholar]
- Li N, Sokal I, Bronson JD, Palczewski K, Baehr W. Identification and functional regions of guanylate cyclase-activating protein 1 (GCAP1) using GCAP1/GCIP chimeras. Biol.Chem. 2001;382:1179–1188. doi: 10.1515/BC.2001.148. [DOI] [PubMed] [Google Scholar]
- Marmor MF, Holder GE, Seeliger MW, Yamamoto S. Standard for clinical electroretinography (2004 update) Doc.Ophthalmol. 2004;108:107–114. doi: 10.1023/b:doop.0000036793.44912.45. [DOI] [PubMed] [Google Scholar]
- Nakatani K, Chen C, Yau KW, Koutalos Y. Calcium and phototransduction. Adv Exp Med Biol. 2002;514:1–20. doi: 10.1007/978-1-4615-0121-3_1. [DOI] [PubMed] [Google Scholar]
- Newbold RJ, Deery EC, Walker CE, Wilkie SE, Srinivasan N, Hunt DM, Bhattacharya SS, Warren MJ. The destabilization of human GCAP1 by a praline to leucine mutation might cause cone-rod dystrophy. Hum.Mol.Genet. 2001;10:47–54. doi: 10.1093/hmg/10.1.47. [DOI] [PubMed] [Google Scholar]
- Nishiguchi KM, Sokal I, Yang L, Roychowdhury N, Palczewski K, Berson EL, Dryja TP, Baehr W. A Novel Mutation (I143NT) in Guanylate Cyclase-Activating Protein 1 (GCAP1) Associated with Autosomal Dominant Cone Degeneration. Invest Ophthalmol.Vis.Sci. 2004;45:3863–3870. doi: 10.1167/iovs.04-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto-Bruc A, Buczylko J, Surgucheva I, Subbaraya I, Rudnicka-Nawrot M, Crabb J, Arendt A, HArgrave PA, Baehr W, Palczewski K. Functional reconstitution of photoreceptor guanylate cyclase with native and mutant forms of guanylate cyclase activating protein 1. Biochemistry. 1997;36:4295–4302. doi: 10.1021/bi963000d. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Sokal I, Baehr W. Guanylate cyclase-activating proteins: structure, function, and diversity. Biochem.Biophys.Res.Commun. 2004;322:1123–1130. doi: 10.1016/j.bbrc.2004.07.122. [DOI] [PubMed] [Google Scholar]
- Patton C, Thompson S, Epel D. Some precautions in using chelators to buffer metals in biological solutions. Cell Calcium. 2004;35:427–431. doi: 10.1016/j.ceca.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Payne AM, Downes SM, Bessant DA, Taylor R, Holder GE, Warren MJ, Bird AC, Bhattacharya SS. A mutation in guanylate cyclase activator 1A (GUCA1A) in an autosomal dominant cone dystrophy pedigree mapping to a new locus on chromosome 6p21.1. Hum.Mol.Genet. 1998;7:273–277. doi: 10.1093/hmg/7.2.273. [DOI] [PubMed] [Google Scholar]
- Pepperberg DR, Birch DG, Hood DC. Electroretinographic determination of human rod flash response in vivo. Methods Enzymol. 2000;316:202–223. doi: 10.1016/s0076-6879(00)16725-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault I, Rozet JM, Gerber S, Ghazi I, Ducroq D, Souied E, Leowski C, Bonnemaison M, Dufier JL, Munnich A, Kaplan J. Spectrum of retGC1 mutations in Leber's congenital amaurosis. Eur.J.Hum.Genet. 2000;8:578–582. doi: 10.1038/sj.ejhg.5200503. [DOI] [PubMed] [Google Scholar]
- Perrault I, Rozet J-M, Calvas P, Gerber S, Camuzat A, Dollfus H, Chatelin S, Souied E, Ghazi I, Leowski C, Bonnermaison M, Le Paslier D, Frezal J, Dufier J-L, Pittler SJ, Munnich A, Kaplan J. Retinal-specific guanylate cyclase gene mutations in Leber's congenital amaurosis. Nature Genet. 1996;14:461–464. doi: 10.1038/ng1296-461. [DOI] [PubMed] [Google Scholar]
- Persechini A, Moncrief ND, Kretsinger RH. The EF-hand family of calcium-modulated proteins. Trends Neurosci. 1989;12:462–467. doi: 10.1016/0166-2236(89)90097-0. [DOI] [PubMed] [Google Scholar]
- Peshenko IV, Dizhoor AM. Activation and inhibition of photoreceptor guanylyl cyclase by guanylyl cyclase activating protein 1 (GCAP-1): the functional role of Mg2+/Ca2+ exchange in EF-hand domains. J Biol.Chem. 2007;282:21645–21652. doi: 10.1074/jbc.M702368200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polans A, Baehr W, Palczewski K. Turned on by Ca2+! The physiology and pathology of Ca2+ binding proteins in the retina. Trends Neurosci. 1996;19:547–554. doi: 10.1016/s0166-2236(96)10059-x. [DOI] [PubMed] [Google Scholar]
- Pugh EN, Jr, Nikonov S, Lamb TD. Molecular mechanisms of vertebrate photoreceptor light adaptation. Curr.Opin.Neurobiol. 1999;9:410–418. doi: 10.1016/S0959-4388(99)80062-2. [DOI] [PubMed] [Google Scholar]
- Ridge KD, Abdulaev NG, Sousa M, Palczewski K. Phototransduction: crystal clear. Trends Biochem.Sci. 2003;28:479–487. doi: 10.1016/S0968-0004(03)00172-5. [DOI] [PubMed] [Google Scholar]
- Rudnicka-Nawrot M, Surgucheva I, Hulmes JD, Haeseleer F, Sokal I, Crabb JW, Baehr W, Palczewski K. Changes in biological activity and folding of guanylate cyclase-activating protein 1 as a function of calcium. Biochemistry. 1998;37:248–257. doi: 10.1021/bi972306x. [DOI] [PubMed] [Google Scholar]
- Schoch S, Castillo PE, Jo T, Mukherjee K, Geppert M, Wang Y, Schmitz F, Malenka RC, Sudhof TC. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415:321–326. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- Simunovic MP, Moore AT. The cone dystrophies. Eye. 1998;12(Pt 3b):553–565. doi: 10.1038/eye.1998.145. [DOI] [PubMed] [Google Scholar]
- Sokal I, Dupps WJ, Grassi MA, Brown J, Jr, Affatigato LM, Roychowdhury N, Yang L, Filipek S, Palczewski K, Stone EM, Baehr W. A GCAP1 missense mutation (L151F) in a large family with autosomal dominant cone-rod dystrophy (adCORD) Invest Ophthalmol.Vis.Sci. 2005;46:1124–1132. doi: 10.1167/iovs.04-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal I, Li N, Surgucheva I, Warren MJ, Payne AM, Bhattacharya SS, Baehr W, Palczewski K. GCAP1(Y99C) mutant is constitutively active in autosomal dominant cone dystrophy. Mol.Cell. 1998;2:129–133. doi: 10.1016/s1097-2765(00)80121-5. [DOI] [PubMed] [Google Scholar]
- Sokal I, Otto-Bruc AE, Surgucheva I, Verlinde CL, Wang CK, Baehr W, Palczewski K. Conformational changes in guanylyl cyclase-activating protein 1 (GCAP1) and its tryptophan mutants as a function of calcium concentration. J.Biol.Chem. 1999;274:19829–19837. doi: 10.1074/jbc.274.28.19829. [DOI] [PubMed] [Google Scholar]
- Stephen R, Bereta G, Golczak M, Palczewski K, Sousa MC. Stabilizing function for myristoyl group revealed by the crystal structure of a neuronal calcium sensor, guanylate cyclase-activating protein 1. Structure. 2007;15:1392–1402. doi: 10.1016/j.str.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen R, Filipek S, Palczewski K, Sousa MC. Ca(2+)-dependent Regulation of Phototransduction. Photochem.Photobiol., Mar 12 [Epub ahead of print] 2008 doi: 10.1111/j.1751-1097.2008.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, Sieving PA, Zack DJ. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Wang QL, Chen S, Esumi N, Swain PK, Haines HS, Peng G, Melia BM, McIntosh I, Heckenlively JR, Jacobson SG, Stone EM, Swaroop A, Zack DJ. QRX, a novel homeobox gene, modulates photoreceptor gene expression. Hum.Mol.Genet. 2004;13:1025–1040. doi: 10.1093/hmg/ddh117. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu X, Biederer T, Sudhof TC. A family of RIM-binding proteins regulated by alternative splicing: Implications for the genesis of synaptic active zones. Proc.Natl.Acad.Sci U.S.A. 2002;99:14464–14469. doi: 10.1073/pnas.182532999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie SE, Li Y, Deery EC, Newbold RJ, Garibaldi D, Bateman JB, Zhang H, Lin W, Zack DJ, Bhattacharya SS, Warren MJ, Hunt DM, Zhang K. Identification and functional consequences of a new mutation (E155G) in the gene for GCAP1 that causes autosomal dominant cone dystrophy. Am.J Hum.Genet. 2001;69:471–480. doi: 10.1086/323265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissinger B, Dangel S, Jagle H, Hansen L, Baumann B, Rudolph G, Wolf C, Bonin M, Koeppen K, Ladewig T, Kohls S, Zrenner E, Rosenberg T. Cone dystrophy with supernormal rod response is strictly associated with mutations in KCNV2. Invest Ophthalmol.Vis.Sci. 2008;49:751–757. doi: 10.1167/iovs.07-0471. [DOI] [PubMed] [Google Scholar]
- Wolfing JI, Chung M, Carroll J, Roorda A, Williams DR. High-resolution retinal imaging of cone-rod dystrophy. Ophthalmology. 2006;113:1019. doi: 10.1016/j.ophtha.2006.01.056. [DOI] [PubMed] [Google Scholar]
- Woodruff ML, Olshevskaya EV, Savchenko AB, Peshenko IV, Barrett R, Bush RA, Sieving PA, Fain GL, Dizhoor AM. Constitutive excitation by Gly90Asp rhodopsin rescues rods from degeneration caused by elevated production of cGMP in the dark. J Neurosci. 2007;27:8805–8815. doi: 10.1523/JNEUROSCI.2751-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Alvarez BV, Chakarova C, Jiang L, Karan G, Frederick JM, Zhao Y, Sauve Y, Li X, Zrenner E, Wissinger B, Hollander AI, Katz B, Baehr W, Cremers FP, Casey JR, Bhattacharya SS, Zhang K. Mutant carbonic anhydrase 4 impairs pH regulation and causes retinal photoreceptor degeneration. Hum.Mol.Genet. 2005;14:255–265. doi: 10.1093/hmg/ddi023. [DOI] [PubMed] [Google Scholar]
- Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, Chien J, Dewan A, Harmon J, Bernstein PS, Shridhar V, Zabriskie NA, Hoh J, Howes K, Zhang K. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]