Abstract
The expression of the cellular form of the prion protein (PrPc) gene is required for prion replication and neuroinvasion in transmissible spongiform encephalopathies. The identification of the cell types expressing PrPc is necessary to understanding how the agent replicates and spreads from peripheral sites to the central nervous system. To determine the nature of the cell types expressing PrPc, a green fluorescent protein reporter gene was expressed in transgenic mice under the control of 6.9 kb of the bovine PrP gene regulatory sequences. It was shown that the bovine PrP gene is expressed as two populations of mRNA differing by alternative splicing of one 115-bp 5′ untranslated exon in 17 different bovine tissues. The analysis of transgenic mice showed reporter gene expression in some cells that have been identified as expressing PrP, such as cerebellar Purkinje cells, lymphocytes, and keratinocytes. In addition, expression of green fluorescent protein was observed in the plexus of the enteric nervous system and in a restricted subset of cells not yet clearly identified as expressing PrP: the epithelial cells of the thymic medullary and the endothelial cells of both the mucosal capillaries of the intestine and the renal capillaries. These data provide valuable information on the distribution of PrPc at the cellular level and argue for roles of the epithelial and endothelial cells in the spread of infection from the periphery to the brain. Moreover, the transgenic mice described in this paper provide a model that will allow for the study of the transcriptional activity of the PrP gene promoter in response to scrapie infection.
The cellular isoform of the prion protein (PrPc) is a cell surface sialoglycoprotein with a physiological role that remains largely unknown (1). PrPc plays a pivotal role in transmissible spongiform encephalopathies in animals and humans. In these neurodegenerative disorders, PrPc is converted to a pathological form, the PrP scrapie form, or PrPsc, by a mechanism that has not been clearly established (2, 3). Unlike PrPc, PrPsc is partially resistant to protease digestion and contains a large β-sheet content (4). The finding that PrPsc is the major constituent of infectious preparations and that the protein seems to be necessary for infectivity (5–9) has led to the hypothesis that PrPsc is the infectious agent of the disease (10).
Prnp0/0 animals are resistant to scrapie infection, indicating the necessity for the infected host cells to express PrPc to support conversion or replication (11, 12). Cell types expressing PrPc are therefore candidates for prion replication and spongiform encephalopathy pathogenesis. The only cells that seem to be functionally compromised lie within the nervous system. However, the infectious agent also is present in different lymphoid organs, depending on the host species, route of infection, and agent strain. In oral infection, which is epidemiologically relevant, it is important to know what happens when the agent gains access to the gastrointestinal tract and which pathways are exploited by the agent to spread throughout the body and attain the central nervous system (CNS). In this regard, it is important to identify in peripheral tissues any cell types expressing PrPc that could be involved in propagating prion infectivity.
The PrPc gene has been described as a housekeeping gene with a preferential expression in neurons (13–15). PrPc has been shown to be present in various regions of the hamster brain, including cortex, hippocampus, striatum, olfactory bulb, hypothalamus, midbrain, cerebellum, and brainstem (14). However, some regional differences in the abundance and localization have been found, according to different authors (16, 17). PrPc has been detected by Northern blot and/or Western blot in a large variety of peripheral tissues, including heart, lung, pancreas, testes, and kidney in the rodent (18, 19) and also skeletal muscle and uterus in sheep (15). PrPc gene expression has also been documented at the cellular level by in situ hybridization (20, 21) and immunohistochemistry. The mRNA encoding PrPc has been shown to be present in the ependymal cells, the epithelial cells of the choroid plexus (13), and the glial cells of the CNS (22). However, in peripheral organs, the identification of PrPc-expressing cells is much less documented than in the brain. In hamster, PrPc protein has been found to be present in pneumocytes, in parietal and enteroendocrine cells of the stomach (14), and in cardiomyocytes (13). PrPc also has been detected on the surface of lymphocytes and of activated T lymphocytes in humans and mice (23, 24) and at higher levels on follicular dendritic cells (25).
To improve our knowledge in the identification of PrP-expressing cells, we created transgenic (tg) mice expressing a green fluorescent protein (gfp) reporter gene under the control of the bovine PrP gene regulatory sequences (6.9 kb). In this paper, we show that a 6.9-kb 5′ regulatory region targets the expression of a linked reporter gene to cell populations that express the endogenous murine PrP gene, Prnp, such as the cerebellar Purkinje cells, the B and T lymphocytes, and the keratinocytes. Interestingly, we also show gfp expression in the enteric nervous system and in a limited number of other cells: epithelial cells of the thymic medulla and endothelial cells of the intestine and kidney. These results suggest a possible role of these cells in the spread of the disease from the peripheral tissues to the CNS.
Materials and Methods
Genomic DNA Clones.
A genomic library constructed in λFIXII replacement vector (CLONTECH) was screened with a PCR-amplified fragment corresponding to a previously sequenced region of 800 bp located upstream from the transcription initiation site of the bovine PrP gene (26). Two overlapping λ clones spanning ≈30 kb were isolated and analyzed. A 10-kb EcoRI restriction fragment of the genomic DNA was subcloned into a pBluescript vector (Stratagene) for the further construction of the transgene.
PCR Analysis.
All tissues were dissected from freshly slaughtered cattle and frozen in liquid nitrogen before processing. Total RNA was extracted and purified with the RNA NOW kit (Biogenetex, Seabrook, TX) as recommended by the supplier. For cDNA synthesis, 2 μg of total RNA was heat-denatured for 3 min at 70°C and reverse-transcribed with 200 units of Superscript II RNaseH− Reverse Transcriptase (Life Technologies, Paisley, Scotland) in 20 μl of 1× incubation buffer (provided by the supplier) supplemented with 10 mM DTT, 1 mM dNTP, 1 unit/μl RNAsin (Promega), and 0.5 μg of (dT)12–18 primers (Amersham Pharmacia). Two microliters of each cDNA reaction was subjected to PCR amplification with 25 pmol each of the sense and antisense primers in 50 μl 1× PCR buffer, 0.2 mM dNTP, and 1 unit recombinant Taq DNA polymerase (Life Technologies). PCRs were performed on a Gene Amp System 9600 thermo-cycler (Perkin–Elmer). PCR products were analyzed by agarose gel electrophoresis, Southern blotting, and hybridization to different probes specific to the transcripts being analyzed. For amplification of the 5′ untranslated region (UTR), sense primer (GCCAGTCGCTGACAGCCGCA) designed from the exon 1 genomic sequence and antisense primer (ACATGGCCACAAAGAGAACC) designed from the ORF genomic sequence were used in the following conditions: 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s.
Construction of PrP-gfp Transgene.
The reporter gene construct consisted of the bovine 6.9-kb PrP gene promoter fragment followed by the gfp reporter gene and simian virus 40 poly(A) signal sequence. Briefly, a 10-kb EcoRI genomic fragment containing 4 kb of exon 1, intron 1, and exon 2 upstream from the transcription initiation site of the bovine PrP gene was subcloned in the pBluescript II vector (Stratagene) and double-digested with NheI and XhoI. A 3.4-kb fragment containing 800 bp upstream from exon 1, followed by the two untranslated exons previously was amplified by PCR on bovine genomic DNA, cloned into TA cloning vector (Invitrogen; data not shown), and double-digested with NheI and PstI. A 4.1-kb XhoI/NheI fragment and a 2.8-kb NheI/PstI fragment generated from the two digested constructs were ligated together into the XhoI/PstI-digested pEGFP-1 vector (CLONTECH). The 6.9-kb fragment was entirely sequenced by using double-stranded templates, the ABI Prism dye terminator cycle sequencing kit (Perkin–Elmer), and either forward or reverse M13 sequencing primers or specific oligonucleotides.
Production of tg Mice.
tg mice were generated as described before (27). Briefly, tg mouse lines were established by injecting the cesium chloride-gradient-purified fragment of PrP-gfp into the pronuclei of outbred C57BL6/SJL fertilized eggs. The integration of the transgene was investigated by dot-blot analysis using 5 μg of tail DNA and screening with a 32P-labeled fragment corresponding to the gfp ORF. In 84 newborn mice, 4 were tg. Subsequent mating of heterozygous mice resulted in the development of a homozygous line of tg mice for further histological examination of the reporter gene expression in various organs.
Histology.
Under phenobarbital anesthesia, 4-week-old animals were perfused intracardially with 4% paraformaldehyde in 0.1 M PBS, pH 7.5. The organs of interest were removed, fixed for 2 h in 4% paraformaldehyde in PBS, and then incubated overnight in 20% sucrose in PBS, frozen (at −70°C), and embedded in Tissue-Tek OCT compound (Sakura Finetek Europe, The Netherlands). Frozen sections (7 μm thick) were cut at −20°C with a Jung Frigocut 2800E (Reichert–Jung, Leika Instrument, Germany) and mounted either unstained in Mowiol 4–88 (Calbiochem) or after counterstaining with Mayer's hematoxylin and eosin solution. The fluorescence of gfp was viewed under a Zeiss Axiophot 2 epifluorescence microscope equipped with an HbO 100-W UV source using filter sets 10 or 24 (Zeiss).
Immunohistology.
A biotin-conjugated monoclonal antibody, 8G8, directed against the human PrP molecule (28) was used to probe for PrP in cryosections prepared as described before. Sections were immersed in acetone at −20°C for 10 min and dried for 2 h at room temperature. Endogenous biotin and endogenous peroxidase were quenched by incubating the sections for 10 min with Biotin Blocking System (Dako) and with 3% hydrogen peroxide, respectively. After washing, the sections were incubated for 1 h at room temperature with biotinylated 8G8 antibody diluted 1:70. Staining was developed by using Dako labeled streptavidin-biotin 2 kit peroxidase as recommended by the supplier.
Flow Cytometry.
Ficoll-isolated lymphocytes from peripheral blood were washed and resuspended in PBS containing 1% BSA (Sigma). Cells then were incubated for 30 min at 4°C with R-phycoerythrin-conjugated anti-mouse CD19, Cy-chrome-conjugated anti-mouse CD3 (Becton Dickinson), or biotinylated anti-PrP antibody (8G8). Cell preparations were washed, treated with FITC-conjugated streptavidin to reveal biotinylated anti-PrP antibody, and analyzed by flow cytometry using a FACScan (Becton Dickinson). The FL1 emission channel normally used for FITC was used for monitoring green fluorescence of the reporter gene.
Results
Isolation and Characterization of Genomic Clones.
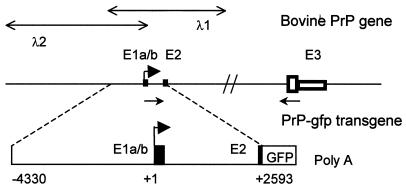
To isolate the bovine PrP gene 5′ flanking region, a λFIXII bovine genomic library (CLONTECH) was screened with a probe generated from the 5′ region of the bovine PrP gene (29). Library screening resulted in the isolation of two λ clones spanning ≈30 kb and including 10 kb upstream from the 5′ initiation start site (Fig. 1). The location of the two individual untranslated exons (1 and 2) and the length of intron within the bovine PrP gene 5′ flanking region were determined by a combination of Southern blot analysis, PCR, and DNA sequencing and confirmed the data of Yoshimoto et al. (29). A 6.9-kb genomic fragment containing 3.4 kb of the promoter region followed by the two untranslated exons was subcloned and sequenced in full. The 3′ region of the genomic fragment contained 3400 bp that were identical to the reported sequence of the bovine PrP gene. The 5′ region of 3500 bp represents the newly sequenced fragment (GenBank accession no. AF163764).
Figure 1.
Schematic representation of the bovine PrP gene and structure of the transgene used in this study. Exons (E) are numbered, and exon 1 and exon 2 are indicated by solid boxes. Within exon 3, the PrP ORF is shown by an open large box, and the mRNA 3′ UTR is represented by an open small box. The lengths of exons 1a, 1b, 2, and 3 are 53, 115, 98, and 4554 bp, respectively. The transcription initiation site is designated with a solid arrow (30). The positions of the two phages (λ1 and λ2) spanning the PrP gene are indicated. The primers used for the analysis of the 5′ UTR are shown by thin arrows (not to scale). The transgene encompasses 6.9 kb of the upstream sequences fused to the gfp reporter gene. GFP, gfp gene ORF; Poly(A), the simian virus 40 polyadenylylation signal. Positions are bp with respect to the transcription initiation site.
5′ UTR Exons and Alternative Splicing.
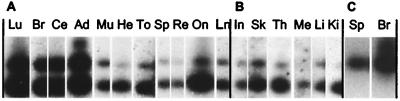
The identification of an 115-nt alternative exon (1b) located immediately downstream of the 53-nt exon 1 has been reported previously in bovids (30). However, the alternative splicing has been studied on a limited number of organs. We decided to further explore the alternative splicing in RNA samples from 17 different organs from cattle. RNA extracts from cerebrum, cerebellum, heart, liver, lung, skeletal muscle, kidney, intestine, skin, lymph node, mesentery, retina, optic nerve, tonsil, spleen, thymus, and adrenal gland were analyzed by reverse transcription–PCR. The amplification using sense exon 1a- and antisense exon 3-specific primers (Fig. 1) generated a 350-bp fragment in addition to a 250-bp fragment. Southern blot and hybridization with four different exon-specific oligonucleotide probes (exons 1a, 1b, 2, and 3) showed that the 250-bp fragment contained exon 1a and exon 2, and the 350-bp fragment contained exons 1a and 1b and exon 2 (Fig. 2). These two transcripts were present in all of the organs tested, and no other 5′ splice variants were detected. The detection of the two transcripts in the spleen (Fig. 2 A and C) contrasts with previous data showing that the exon 1b-containing fragment was not detected (30). This discrepancy can be explained by the use of exon-specific probes that increased the sensitivity of the method.
Figure 2.
Coexpression of the two different bovine PrP 5′ UTR exons in 17 different tissues. RNA fractions were subjected to reverse transcription–PCR using primers specific to exon 1a and exon 3. The PCR products were analyzed by Southern blot hybridization with a 32P-labeled oligonucleotide specific for exon 1a (A and B). The hybridization revealed two fragments (250 and 350 bp). Hybridization with a 32P-labeled oligonucleotide specific for exons 3 or 2 gave identical results. Hybridization with an exon 1b-specific oligonucleotide revealed only the 350-bp fragment, as shown for brain and spleen (C). All tissues tested gave identical results (data not shown). Lu, lung; Br, brain; Ce, cerebellum; Ad, adrenal gland; Mu, skeletal muscle; He, heart; To, tonsil; Sp, spleen; Re, retina; On, optic nerve; Ln, lymph node; In, intestine; Sk, skin; Th, thymus; Me, mesentery; Li, liver; Ki, kidney. A and B differ by the autoradiography exposure conditions, 2 h at room temperature or at −80°C, respectively. Autoradiography in C was exposed overnight at −80°C.
Generation of tg Mice Expressing Bovine PrP Gene Promoter.
The splicing analysis of the 5′ flanking region of the bovine PrP gene suggested that this region might play a role in the control of PrP gene expression. Thus, the 6.9-kb fragment previously described was used for the construction of the tg mice. The 5′ flanking region, spanning positions −4330 to +2593 (numbered in relation to the transcription initiation start site), was fused to the 0.8-kb DNA fragment containing the entire coding sequence of gfp, followed by 0.2 kb of DNA containing the simian virus 40 polyadenylylation signal (Fig. 1). Before microinjection, the resulting PrP-gfp construct was tested in vitro by transient transfection assays using Chinese hamster ovary cells. Specific fluorescence was observed for the PrP-gfp construct, but not with the vector alone or with the same PrP construct placed in the opposite orientation (data not shown).
Four different founder mice (L21, L41, L61, and L64) were generated, each of which transmitted the transgene. The integration pattern of the transgene and the copy number were determined by Southern blot analysis as approximately 5, 15, 45, and 80 integrated copies for lines 61, 64, 21, and 41, respectively (data not shown). Subsequent mating of heterozygous mice resulted in the development of a homozygous line of tg mice.
Tissue-Specific Expression of gfp Under the Control of the Bovine PrP Gene Promoter.
The homozygous tg mice were used subsequently to examine the tissue-specific expression of the bovine PrP gene promoter. The four tg lines were analyzed fully and showed a similar fluorescence pattern with a good correlation between the fluorescence intensity and the level of gfp transcripts detected by reverse transcription–PCR (data not shown). This experiment allowed us to demonstrate the presence of two gfp transcripts differing by the alternative splicing of the exon 1b (data not shown). In addition, we did not detect any aberrant transcript, which could be because of the presence of a cryptic splice site present in the transgene that could cause splicing of the gfp ORF. These data indicate that the splicing of the untranslated exons occur specifically in tg mice.
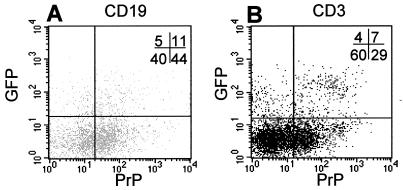
Mouse line 64 was chosen for the following analysis based on preliminary examination of histological sections, which displayed more intense staining than in the other founder mice. To determine whether the transgene expression coincided with the endogenous PrP gene expression, levels of gfp transgene and endogenous PrPc were measured by FACS analysis from B and T lymphocytes in freshly isolated peripheral blood lymphocytes. The results of this experiment can be seen in Fig. 3, where 16% of B lymphocytes and 11% of T lymphocytes displayed gfp fluorescence. Among the gfp-positive cells, 69% and 63% express the endogenous PrPc in B and T lymphocytes, respectively. prnp gene expression is present in lymphocytes shown to express gfp, and these data are in agreement with previous reports showing that B and T lymphocytes express the prnp gene (23, 24). We noted that ≈5% of lymphocytes were gfp-positive and did not express prnp, and ≈35% were PrP-positive and did not express gfp. This result could account for a loss of gfp fluorescence during the experiments and/or a lower activity in these cells of the 6.9-kb PrP gene promoter as compared with the endogenous murine Prnp promoter. Flow cytometric analysis also was performed on lymphoid organs, and we found fluorescent B and T lymphocytes (data not shown). An extensive identification of the subpopulations will need to be performed. These data, taken together with the splicing analysis, suggest that the 6.9-kb 5′ flanking DNA of the bovine PrP gene contains cis-acting elements that bind specific transcription factors and specific splice factors that are conserved between bovids and mice, indicating that the PrP-gfp tg mice could be a relevant model in which to study the cellular expression of the Prnp promoter.
Figure 3.
Analysis of gfp and PrP expression from tg mice (L64) by fluorescence-activated cell sorter on peripheral blood leukocytes, gated for B (A) and T (B) lymphocytes. B cells were stained with R-phycoerythrin-conjugated anti-mouse CD19 antibody, T cells with Cy-chrome-conjugated anti-mouse CD3 molecular complex antibody (Becton Dickinson), and PrP-expressing cells with biotinylated anti-PrP antibody (8G8). The percentage of positive cells is indicated in the upper right corner.
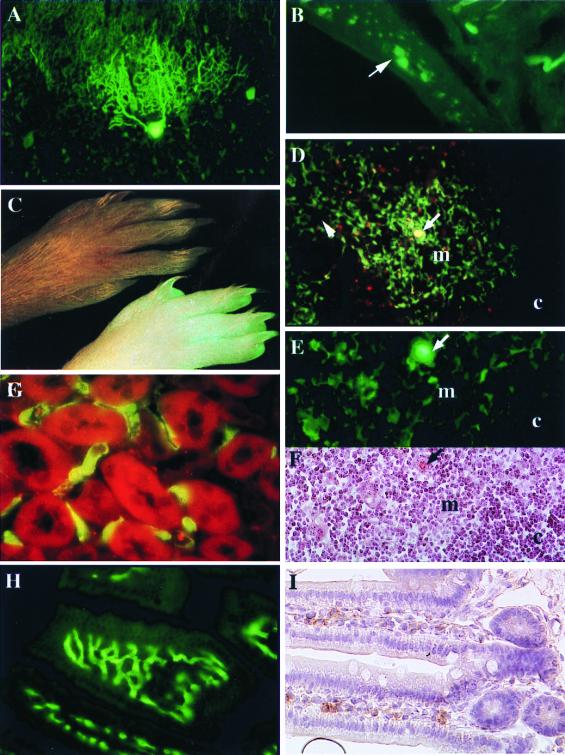
Thus, we investigated the expression of the reporter gene by histologic analysis of frozen sections of various organs. In the brain of tg mice, gfp-expressing neurons were seen in the gray matter of the cortex as well as in subcortical structures. In the cerebellar cortex, intense fluorescence resulting from gfp expression could be detected in Purkinje cells (Fig. 4A). These results will be described extensively by Y.B., C.L.-V., T.S., A. M. Haeberle, G. Bombarde, V.P.-D., J.-P.D., F.B.-G., J.F., J. Grassi, and J.-Y.C. in a distinct report. Concerning the peripheral nervous system, gfp expression was observed in the optic nerve (data not shown), in the sciatic nerve, and at the neuromuscular junction (data not shown). Regarding the autonomic nervous system, large cell bodies belonging to the Auerbach's and Meissner's plexuses of the gut showed a strong green signal (Fig. 4B).
Figure 4.
Analysis of reporter gene expression in PrP-gfp tg mice (L64). (A) Coronal section of cerebellar cortex in which the transgene is expressed in Purkinje neurons. (×200.) (B) Section of the wall of the small intestine showing fluorescence in the ganglion cells in the enteric nervous system. (×200.) (C) Fluorescence picture of the foot from the gfp tg mice (Lower) and their littermate controls (Upper). (D and E) Sections of thymus showing fluorescence in the epithelial cells of the medulla (m) around the Hassal's corpuscles (arrow). c, cortex. [×40 (D) and ×200 (E).] (F) Thymus section stained with eosin and hematoxylin. E and F are serial sections. (G) Sections of renal cortex showing fluorescence in the endothelial cells of the capillaries around the tubules. (×200.) (H and I) Sections of small intestine of gfp-tg mice (H) and wild-type mice stained with anti-PrP 8G8 antibody (1:200) (I). (×200.)
The tg mice exposed under the UV light exhibited very clearly a fluorescent signal in the skin, the nails, and the hair (Fig.4C). Histological analysis of the skin showed a fluorescence signal in basal and spinocellular keratinocytes and in the external root sheet of the hair follicles and their associated sebaceous gland (data not shown). Histological observations of thin sections of the thymus revealed a large number of fluorescent cells with a branching appearance and located mainly throughout the medulla (Fig. 4D). A radiating cluster of fluorescent cells was concentrated around Hassal's corpuscles, which were also highly fluorescent (Fig. 4 D and E). The serial section stained with eosin and hematoxylin in Fig. 4F showed clearly the presence of a large Hassal's corpuscle that was fluorescent (Fig. 4E). Their distribution and morphological properties indicate that they are typical medullar epithelial cells. In the spleen, mesenteric lymph nodes, and Peyer's patches, we observed isolated fluorescent cells, some of them with a dendritic aspect. More extensive identification of these cells with flow cytometric analysis is needed. In the gastrointestinal tract, intensive green fluorescence of endothelial cells was observed in the mucosal capillaries and venules within the lamina propria mucosa of the intestinal villi (Fig. 4H) and in the small vessels of the submucosa. This result was confirmed by immunohistochemistry by using an anti-PrP antibody (Fig. 4I). Nonrelated IgG2a used as a control gave negative results, as did omission of the primary antibody. Fluorescent endothelial cells also were seen in the capillaries along the tubules of the renal cortex (Fig. 4G), as well as along the loop of Henle in the medulla. Microscopic observation of other tissues, including lung, pancreas, liver, and smooth and skeletal muscle, revealed extremely low numbers of positive cells that could not be identified.
Discussion
The mechanism of the PrPc to PrPsc conversion does not occur in scrapie-infected Prnp0/0 animals, indicating the necessity for the infected host cells to express PrP to support conversion or replication (11, 12). The identification of cell types expressing PrPc is necessary to understanding how the agent replicates and spreads from the periphery to the CNS. The mammalian Prnp gene has been described as a ubiquitous gene, with little known regarding gene regulation (26, 31, 32). The cellular specificity of the Prnp gene is based mainly on the data from PrP immunohistochemistry studies (14, 16, 33) and in situ hybridization (13, 20, 21, 34). However, some data are contradictory and may be explained by the techniques used or by the low level of PrP expression in nonneural cells. To better investigate the cellular-specific expression of the Prnp gene, we created tg mouse lines containing 6.9 kb of the bovine PrP gene 5′ regulatory region fused to the gfp reporter gene.
The characterization of the bovine PrP gene allowed us to demonstrate the existence of two transcripts, which were generated by the alternative splicing of one 115-bp 5′ exon to a splice acceptor site within the 5′ UTR of the gene. The two transcripts were detected in all bovine tissues we examined, including splenic tissue. This result contrasts with a report (30) in which only the 250-bp exon 1a transcript was detected in the spleen. A biological explanation for the predominant expression of the shorter transcript over the longer transcript in all tissues tested needs further investigations. At present, it is unclear whether splicing of the bovine PrP mRNA in the 5′ UTR represents an important regulatory mechanism involved in tissue- or cell-specific expression, mRNA stability, or mRNA translatability, as found in other genes (35, 36).
In this context, we choose to express, in tg mice, the gfp reporter gene under the control of the 5′ genomic region of the bovine PrP gene, including the regulatory region upstream from the transcription initiation site followed by the 5′ noncoding exons. We are aware that this model has little chance to reconstitute totally the expression of the endogenous gene because of the lack of large genomic sequences that could putatively regulate the gene, and because of the influence of the insertion site on the tg expression. However, the analysis of the PrP-gfp tg mouse lines shows that the cellular specificity of the reporter gene expression correlates quite well with the endogenous Prnp gene expression. For instance, the cerebellar Purkinje cells (16, 20, 21, 34) in the brain (37), the keratinocytes in the skin (38), and the lymphocytes (23, 24) have been described previously as expressing PrPc and are gfp-fluorescent in the tg mice. This result indicates that the 6.9-kb segment of the 5′ UTR region of the bovine PrP gene is sufficient to direct the expression of gfp in these cells. It is noteworthy that the expression of the transgene, which lacks the large intron, was observed in cerebellar Purkinje cells, because this result is in contrast with the observations of Shmerling et al. (39) and Fischer et al. (40), who propose that there might be a Purkinje cell-specific enhancer in the large intron. Our data suggest that it is not the case.
The expression of the reporter gene in endothelial and epithelial cells strongly suggests that these cell types normally express the Prnp gene. It has been reported in keratinocytes (38), and we observed PrPc on the endothelial cells of the intestinal villi (Fig. 4H). In the thymus, an intense gfp fluorescence of the epithelial cells and Hassal's corpuscles is restricted in the medullary region. In mice, thymus accumulates infectivity very early after the infection by introduction of the scrapie agent into various peripheral sites (41, 42), and Muramoto et al. (43) have shown the presence of PrPsc in the thymus of mice infected with the agent of Creutzfeldt–Jakob disease (CJD). The thymus is devoid of both B lymphocytes and follicular dendritic cells, which have been claimed to support the initial replication of the scrapie agent (25, 44–46). Thus, our data support the hypothesis that epithelial cells could accumulate or replicate prions in the thymus and could provide an interface between the lymphoreticular system and the peripheral nerves that permit neuroinvasion to occur. The fact that only epithelial cells from the skin and the thymus are gfp-positive suggests that these two epithelial cell subtypes contain common transcription factors able to activate the 6.9-kb sequence of the 5′ flanking bovine PrP gene.
The presence of gfp in the endothelial cells of the blood capillaries in the intestinal villi of the digestive tract is of particular interest. These observations correlate well with a recent report demonstrating PrPsc accumulation in the endothelial cells of the blood capillaries in the small intestine of zoo lemurs after an oral infection (47). However, contrary to our results, the same authors also have reported the presence of PrPsc in the epithelial cells lining the lumen of the digestive tract. In our tg mice, we do not observe fluorescence signal in enterocytes, Paneth cells, endocrine cells, or stem cells. The reason for this incongruence is still unclear and could be the low activity of the Prnp promoter in these cells. Importantly, both the report by Bons et al. (47) and our observations suggest that endothelial cells of blood vessels in the intestinal villi could play a role in the propagation of the infectious agent after an oral challenge. In addition, intensive gfp fluorescence was observed in the neurons of the Meissner and Auerbach's plexuses along the digestive tract of tg mice. Thus, this report suggests that PrPc could be expressed in the neurons of the enteric nervous system. This observation is supported by the recent description of the accumulation of PrPsc in the enteric nervous system in scrapie-affected sheep (48). Intramural plexuses are present throughout the entire length of the gastrointestinal tract and have extensive connections with the autonomous nervous system. Thus, our data suggest that the Auerbach's and Meissner's plexuses could also be a way for the agent to reach the nervous system after an oral infection. The endothelial cells of the renal capillaries also express the gfp reporter gene. PrPc has been detected by Western blot in hamster kidney (14), but the authors were unable to determine by immunohistochemical means the cellular localization of PrPc within this tissue. Furthermore, the potential infectivity of this organ has never been reported (43).
In conclusion, we have shown that the 6.9-kb 5′ flanking region of the bovine PrP gene is sufficient to promote gfp expression in the nervous system as well as in peripheral tissues, and that the transgene seems to obey the endogenous murine Prnp regulatory mechanism. These data provide valuable insights on the distribution of PrPc in mammalian tissues and point out the potential importance of epithelial and endothelial cell subtypes in the replication and spread of the agent from the periphery to the CNS. The possible role of these cells in scrapie pathogenesis has not yet been investigated. Our gfp-tg mice provide a model for further analysis of the modulation of transcriptional activity of the Prnp gene in vivo during scrapie infection and in vitro by the use of trans-acting factors, such as cytokines and pharmacological agents. In addition, our tg mice provide an attractive analysis model to those who study a specific subpopulation of cells such as the thymic epithelial cells or the endothelial cells of the intestinal villi.
Acknowledgments
We thank Nathalie Maessian for technical help and Dr. A. Burny for the bovine genomic library. We also thank Prof. J. R. Barta and Dr. S. Saule for critical reading of the manuscript. J.F is supported by a fellowship of the Ministère de la Recherche et de la Technologie Française. This work is supported by the Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, and by a grant from the Ministère de la Recherche et de la Technologie Française “Programme de Recherche sur les Encéphalopathies Spongiformes Subaiguës Transmissibles et les Prions” [Action Concertée Coordonnée no. 1 (1997)], and Economic European Community Food Agro-Industry Research Grant JCT 6022 to E.H. and J.Y.C.
Abbreviations
- PrP
prion protein
- CNS
central nervous system
- gfp
green fluorescent protein
- PrPc
PrP cellular form
- PrPsc
PrP scrapie form
- UTR
untranslated region
- tg
transgenic
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF163764).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080081197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080081197
References
- 1.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner S B, Scott M, Foster D, Pan K M, Groth D, Mirenda C, Torchia M, Yang S L, Serban D, Carlson G A, et al. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 3.Pan K M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, et al. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caughey B W, Dong A, Bhat K S, Ernst D, Hayes S F, Caughey W S. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 5.Bolton D C, McKinley M P, Prusiner S B. Science. 1982;218:1309–1311. doi: 10.1126/science.6815801. [DOI] [PubMed] [Google Scholar]
- 6.Diringer H, Hilmert H, Simon D, Werner E, Ehlers B. Eur J Biochem. 1983;134:555–560. doi: 10.1111/j.1432-1033.1983.tb07602.x. [DOI] [PubMed] [Google Scholar]
- 7.McKinley M P, Bolton D C, Prusiner S B. Cell. 1983;35:57–62. doi: 10.1016/0092-8674(83)90207-6. [DOI] [PubMed] [Google Scholar]
- 8.Prusiner S B, Groth D F, Bolton D C, Kent S B, Hood L E. Cell. 1984;38:127–134. doi: 10.1016/0092-8674(84)90533-6. [DOI] [PubMed] [Google Scholar]
- 9.Gabizon R, McKinley M P, Groth D F, Kenaga L, Prusiner S B. J Biol Chem. 1988;263:4950–4955. [PubMed] [Google Scholar]
- 10.Prusiner S B. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 11.Bueler H, Aguzzi A, Sailer A, Greiner R A, Autenried P, Aguet M, Weissmann C. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 12.Prusiner S B, Groth D, Serban A, Koehler R, Foster D, Torchia M, Burton D, Yang S L, DeArmond S J. Proc Natl Acad Sci USA. 1993;90:10608–10612. doi: 10.1073/pnas.90.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown H R, Goller N L, Rudelli R D, Merz G S, Wolfe G C, Wisniewski H M, Robakis N K. Acta Neuropathol. 1990;80:1–6. doi: 10.1007/BF00294214. [DOI] [PubMed] [Google Scholar]
- 14.Bendheim P E, Brown H R, Rudelli R D, Scala L J, Goller N L, Wen G Y, Kascsak R J, Cashman N R, Bolton D C. Neurology. 1992;42:149–156. doi: 10.1212/wnl.42.1.149. [DOI] [PubMed] [Google Scholar]
- 15.Horiuchi M, Yamazaki N, Ikeda T, Ishiguro N, Shinagawa M. J Gen Virol. 1995;76:2583–2587. doi: 10.1099/0022-1317-76-10-2583. [DOI] [PubMed] [Google Scholar]
- 16.DeArmond S J, Mobley W C, DeMott D L, Barry R A, Beckstead J H, Prusiner S B. Neurology. 1987;37:1271–1280. doi: 10.1212/wnl.37.8.1271. [DOI] [PubMed] [Google Scholar]
- 17.Sales N, Rodolfo K, Hassig R, Faucheux B, Di Giamberardino L, Moya K L. Eur J Neurosci. 1998;10:2464–2471. doi: 10.1046/j.1460-9568.1998.00258.x. [DOI] [PubMed] [Google Scholar]
- 18.Oesch B, Westaway D, Walchli M, McKinley M P, Kent S B, Aebersold R, Barry R A, Tempst P, Teplow D B, Hood L E, et al. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 19.Robakis N K, Sawh P R, Wolfe G C, Rubenstein R, Carp R I, Innis M A. Proc Natl Acad Sci USA. 1986;83:6377–6381. doi: 10.1073/pnas.83.17.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manson J, West J D, Thomson V, McBride P, Kaufman M H, Hope J. Development. 1992;115:117–122. doi: 10.1242/dev.115.1.117. [DOI] [PubMed] [Google Scholar]
- 21.Tanji K, Saeki K, Matsumoto Y, Takeda M, Hirasawa K, Doi K, Matsumoto Y, Onodera T. Intervirology. 1995;38:309–315. doi: 10.1159/000150457. [DOI] [PubMed] [Google Scholar]
- 22.Moser M, Colello R J, Pott U, Oesch B. Neuron. 1995;14:509–517. doi: 10.1016/0896-6273(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 23.Mabbott N A, Brown K L, Manson J, Bruce M E. Immunology. 1997;92:161–165. doi: 10.1046/j.1365-2567.1997.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cashman N R, Loertscher R, Nalbantoglu J, Shaw I, Kascsak R J, Bolton D C, Bendheim P E. Cell. 1990;61:185–192. doi: 10.1016/0092-8674(90)90225-4. [DOI] [PubMed] [Google Scholar]
- 25.McBride P A, Eikelenboom P, Kraal G, Fraser H, Bruce M E. J Pathol. 1992;168:413–418. doi: 10.1002/path.1711680412. [DOI] [PubMed] [Google Scholar]
- 26.Inoue S, Tanaka M, Horiuchi M, Ishiguro N, Shinagawa M. J Vet Med Sci. 1997;59:175–183. doi: 10.1292/jvms.59.175. [DOI] [PubMed] [Google Scholar]
- 27.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 28.Krasemann S, Groschup M H, Harmeyer S, Hunsmann G, Bodemer W. Mol Med. 1996;2:725–734. [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshimoto J, Iinuma T, Ishiguro N, Horiuchi M, Imamura M, Shinagawa M. Virus Genes. 1992;6:343–356. doi: 10.1007/BF01703083. [DOI] [PubMed] [Google Scholar]
- 30.Horiuchi M, Ishiguro N, Nagasawa H, Toyoda Y, Shinagawa M. Biochem Biophys Res Commun. 1997;233:650–654. doi: 10.1006/bbrc.1997.6511. [DOI] [PubMed] [Google Scholar]
- 31.Saeki K, Matsumoto Y, Matsumoto Y, Onodera T. Biochem Biophys Res Commun. 1996;219:47–52. doi: 10.1006/bbrc.1996.0179. [DOI] [PubMed] [Google Scholar]
- 32.Baybutt H, Manson J. Gene. 1997;184:125–131. doi: 10.1016/s0378-1119(96)00600-2. [DOI] [PubMed] [Google Scholar]
- 33.Fournier J, Escaig-Haye F, Billette de Villemeur T, Robain O, Lasmezas C I, Deslys J P, Dormont D, Brown P. Cell Tissue Res. 1998;292:77–84. doi: 10.1007/s004410051036. [DOI] [PubMed] [Google Scholar]
- 34.Harris D A, Lele P, Snider W D. Proc Natl Acad Sci USA. 1993;90:4309–4313. doi: 10.1073/pnas.90.9.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sachs A B. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 36.Brawerman G. Cell. 1987;48:5–6. doi: 10.1016/0092-8674(87)90346-1. [DOI] [PubMed] [Google Scholar]
- 37.Sarkozi E, Askanas V, Engel W K. Am J Pathol. 1994;145:1280–1284. [PMC free article] [PubMed] [Google Scholar]
- 38.Pammer J, Weninger W, Tschachler E. Am J Pathol. 1998;153:1353–1358. doi: 10.1016/S0002-9440(10)65720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shmerling D, Hegyi I, Fischer M, Blattler T, Brandner S, Gotz J, Rulicke T, Flechsig E, Cozzio A, von Mering C, et al. Cell. 1998;93:203–214. doi: 10.1016/s0092-8674(00)81572-x. [DOI] [PubMed] [Google Scholar]
- 40.Fischer M, Rulicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 41.Eklund C M, Kennedy R C, Hadlow W J. J Infect Dis. 1967;117:15–22. doi: 10.1093/infdis/117.1.15. [DOI] [PubMed] [Google Scholar]
- 42.Fraser H, Dickinson A G. J Comp Pathol. 1978;88:563–573. doi: 10.1016/0021-9975(78)90010-5. [DOI] [PubMed] [Google Scholar]
- 43.Muramoto T, Kitamoto T, Tateishi J, Goto I. Am J Pathol. 1992;140:1411–1420. [PMC free article] [PubMed] [Google Scholar]
- 44.Kitamoto T, Muramoto T, Mohri S, Doh-Ura K, Tateishi J. J Virol. 1991;65:6292–6295. doi: 10.1128/jvi.65.11.6292-6295.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein M A, Frigg R, Flechsig E, Raeber A J, Kalinke U, Bluethmann H, Bootz F, Suter M, Zinkernagel R M, Aguzzi A. Nature (London) 1997;390:687–690. doi: 10.1038/37789. [DOI] [PubMed] [Google Scholar]
- 46.Raeber A J, Klein M A, Frigg R, Flechsig E, Aguzzi A, Weissmann C. EMBO J. 1999;18:2702–2706. doi: 10.1093/emboj/18.10.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bons N, Mestre-Frances N, Belli P, Cathala F, Gajdusek D C, Brown P. Proc Natl Acad Sci USA. 1999;96:4046–4051. doi: 10.1073/pnas.96.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Keulen L J, Schreuder B E, Vromans M E, Langeveld J P, Smits M A. J Comp Pathol. 1999;121:55–63. doi: 10.1053/jcpa.1998.0300. [DOI] [PubMed] [Google Scholar]