Abstract
Protein binding to phospholipid surface is commonly mediated by amphipathic α-helices. To understand the role of α-helical structure in protein-lipid interactions, we used discoidal lipoproteins reconstituted from dimyristoyl phosphatidylcholine (DMPC) and human apolipoprotein C-I (apoC-I, 6 kD) or its mutants containing single Pro substitutions along the sequence and differing in their α-helical content in solution (0–48%) and on DMPC (40–75%). Thermal denaturation revealed that lipoprotein stability correlates weakly with the protein helix content: proteins with higher α-helical content on DMPC may form more stable complexes. Lipoprotein reconstitution upon cooling from the heat-denatured state and DMPC clearance studies revealed that protein secondary structure in solution and on DMPC correlates strongly with the maximal temperature of lipoprotein reconstitution: more helical proteins can reconstitute lipoproteins at higher temperatures. Interestingly, at Tc=24 °C of the DMPC gel-to-liquid crystal transition, the clearance rate is independent of the protein helical content. Consequently, if the packing defects at the phospholipid surface are readily available (e.g. at the lipid phase boundary), protein insertion into these defects is independent of the secondary structure in solution. However, if hydrophobic defects are limited, protein binding and insertion is aided by other surface-bound proteins and depends on their helical propensity: the larger the propensity the faster the binding and the broader its temperature range. This positive cooperativity in α-helical binding to phospholipid surface, which may result from direct and/or lipid-mediated protein-protein interactions, may be important for lipoprotein metabolism and for protein-membrane binding.
Keywords: High-density lipoprotein, kinetic stability, lipid fusion, vesicle clearance, atherosclerosis
Protein binding to phospholipid surface is an important step in many biological reactions including apolipoprotein exchange among plasma lipoproteins, activation of lipid-regulated enzymes such as phosphoglycerate kinase (PGK) or CTP:phosphocholine cytidylyltransferase (CCT), synuclein aggregation in Parkinson’s disease, lipid storage in adypocytes, and binding of antimicrobial peptides to cell surfaces. The binding depends on the structural and physicochemical properties of both proteins and lipids and is facilitated by the hydrophobic defects on the lipid surface that form primary protein binding sites (1–4). The common lipid surface-binding motif found in many proteins, including apolipoproteins, perilipin, synucleins, CTT and PGK, is amphipathic α-helix comprised of 11-mer sequence repeats ((5,6) and references therein). The extended apolar face of such a helix is optimized for interactions with apolar lipid moieties while the polar face can interact with phospholipid head groups and solvent molecules.
In solution, amphipathic α-helices may be largely unfolded (in apolipoproteins A-II and Cs, in synucleins, and in short lipid-binding peptides ((7, 8) and references therein), or they may be folded in helix bundles (in apolipoproteins E, A-I, Lp-III, and in perilipin) (9–12). Studies of highly helical apolipoproteins, such as apoE, apoLp-III and their mutant forms showed that the rate of liposome clearance decreases with increasing protein size, thermodynamic stability or degree of self-association in solution, i. e., with increasing tertiary or quaternary helix-helix interactions in solution (1, 13–16). This inverse correlation between the protein stability in solution and its lipid binding ability was interpreted as a requirement for the helix bundle to open and expose apolar helical faces to lipid (15–17). On the basis of lipid binding studies by apoE and apoA-I, a multistep pathway of apolipoprotein binding to phospholipid surface was proposed in which amphipathic α-helices play key roles in initial protein adsorption to the lipid packing defects and in consequent insertion into the phospholipid surface (13,17). At the same time, lipid-induced random coil-to-helix conversion in the lipid binding domains of these and other apolipoproteins has been proposed to provide an energy source for the high-affinity protein-lipid binding (18,19). Thus, in contrast to tertiary or quaternary apolipoprotein structure in solution, the role of the pre-existing secondary structure in the lipid surface binding is not entirely clear.
One way to analyze the effects of helical conformation on protein-lipid interactions is by using large proteins such as apoA-1 (243 a. a.) from which individual helices have been deleted (18). However, in the context of a large protein, such mutations change not only helical content but also protein size, hydrophobicity and tertiary interactions, and the individual contributions of these factors to lipid binding are difficult to resolve. Alternatively, apolipoprotein-mimicking peptides of similar size (20 a. a.) and hydrophobicity but varying helical propensity have been used, revealing a clear correlation between the α-helical propensity and lipid binding ability (20). However, such short peptides are fully unfolded in aqueous solution and thus unsuitable for testing the role of pre-existing helical structure on lipid binding.
Here, we use midway approach by utilizing the smallest human apolipoprotein C-I (57 a. a.) and its point mutants that have similar size and hydrophobicity but differ in their helical content in solution from 0–48% (Fig. 1). ApoC-I, which is a constituent of high-, intermediate- and very low-density lipoproteins, is a secondary activator of lecithin:cholesterol acyltransferase, an inhibitor of cholesterol ester transfer protein, and an important modulator of lipoprotein metabolism ((23–25) and references therein). ApoC-I has high sequence homology and structural and functional similarity to larger apolipoproteins such as apoE or apoA-I but, in contrast to these proteins, apoC-I monomer in solution adopts a fluctuating helix-turn-helix conformation that lacks substantial tertiary structure (22). The helix content in apoC-I in solution can be increased by G15A mutation or reduced by Pro substitutions in the middle of the helices (22). Thus, apoC-I is well-suited for the analysis of the secondary structural effects on lipid binding.
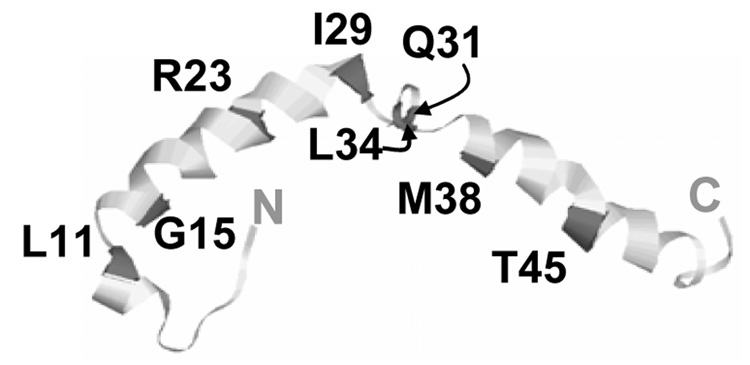
Figure 1.
NMR structure of human apoC-I in “lipid-mimetic” environment (21) (PDB code 1IOJ). Locations of point mutations used in our work are indicated. The effects of these mutations on the secondary structure of lipid-free apoC-I monomer in solution were determined in (22) and are listed in Table 1.
To test the role of secondary structure in protein-induced remodeling of phospholipid surface and in lipoprotein stability, we studied formation and denaturation of macromolecular complexes containing apoC-I and dimyristoyl phosphatidylcholine (DMPC). Such complexes comprise a phospholipid bilayer disk 12–21 nm in diameter surrounded by amphipathic protein α-helices that adopt an extended conformation at the particle perimeter and thereby confer lipoprotein stability and solubility ((26) and references therein). These reconstituted complexes provide useful energetic, structural and functional models for nascent high-density lipoproteins (HDL) whose main constituents are also phosphatidylcholines and exchangeable apolipoproteins such as apoA-I, apoA-II and apoE
Our earlier studies of apoC-I:DMPC disks have revealed a kinetic mechanism of lipoprotein stabilization (27–29). We showed that the energy barriers to lipoprotein denaturation arise from the heat- or denaturant-induced protein unfolding/dissociation and consequent lipoprotein fusion that compensates for the surface depletion of the polar protein moiety (27). Stability studies of DMPC disks containing wild type (WT) apoC-I and its three point mutants, R23P, Q31P and L34P, showed no significant effects of mutations on the lipoprotein stability (28). Interestingly, these studies revealed a correlation between the onset temperature TR of lipoprotein reconstitution upon heating and cooling and the secondary protein structure: the higher the helix content the higher the TR (28). To elucidate the origin of this correlation and relate it to the role of the secondary structure in protein-phospholipid surface interactions, here we analyze reconstitution and denaturation of DMPC complexes containing a wider range of apoC-I point mutants (Fig. 1, Table 1).
Table 1.
Structural and energetic parameters of apoC-I:DMPC complexes. Protein α-helical content was determined from the measured value of [Θ222] according to (29) with 5% accuracy. The temperature TR of lipoprotein reconstitution upon heating and cooling was determined from the light scattering and CD melting curves at 222 nm (Fig. 1). The values of the apparent melting temperature Tm of apoC-I:DMPC disk denaturation were determined from the first derivative d[Θ222]/dT of the CD melting curves measured at 11 K/h heating rate (27). The accuracy in the determination of TR and Tm is about 2 °C. Changes δΔG* in the kinetic lipoprotein stability ΔG* induced by apoC-I mutations were determined from the shifts in the Arrhenius plots (Fig. 3D) with better than 0.5 kcal/mol accuracy. ApoC-I variants reported earlier (28)
| Protein | α-helix content at 24°C, % | TR, °C | Tm, °C | δΔG*, kcal/mol | |
|---|---|---|---|---|---|
| in solution | on DMPC | ||||
| G15A/M38L | 48 | 75 | 40 | 79 | +0.7 |
| WT* | 31 | 65 | 37 | 67 | - |
| Q31P* | 32 | 65 | 38 | 0.0 | |
| G15P | 20 | 44 | 31 | 55 | −1.0 |
| L34P* | 15 | 60 | 31 | 62 | 0.0 |
| L11P | <5 | 45 | 28 | 62 | 0 |
| R23P* | <5 | 40 | 28 | 59 | 0.0 |
| I29P | <5 | 47 | 27 | 58 | −0.9 |
| M38P | <5 | 45 | 27 | 65 | 0 |
| T45P | <5 | 47 | 28 | 58 | −1.0 |
(indicated by asterisks) are shown for comparison.
MATERIALS AND METHODS
Peptide design and lipoprotein preparation
Human apolipoprotein C-1 and its point mutants were obtained commercially by solid state synthesis and purified by HPLC to 95%+ purity at 21st Century Biochemicals as described (22). The peptide termini were not blocked. Structure and stability of WT apoC-I prepared by this method closely resemble those of the plasma protein. The mutants, which were analyzed in their lipid-free state before (22), contain single Pro substitutions 1–2 helical turns apart. Except for charge removal mutation R23P, the sites of Pro substitutions are located in the apolar helical face, which does not significantly alter peptide hydro-phobicity (20) but reduces self-association in solution (30). Depending on the Pro location in the middle of the helix, near the helical kink or terminus, or in the linker region, the mutations induce complete, partial or no helical unfolding in solution, respectively (22). In addition, we used G15A/M38L double mutant that has increased helix content in solution (48% compared to 31% in WT).
The peptides were dissolved in 5 mM sodium phosphate buffer at pH 7.6, which is the standard buffer used throughout this work. Peptide concentration was determined by absorbance at 280 nm and by modified Lowry assay at 750 nm (31). To prepare discoidal particles, apoC-I stock solution of about 0.5 mg/ml protein concentration was mixed with DMPC suspension using1:4 protein:lipid weight ratio and was incubated overnight at the temperature Tc=24 °C of the DMPC gel-to-liquid crystal phase transition at which the protein-lipid association is fastest (1,2).
Circular dichroism (CD) spectroscopy and light scattering
Protein secondary structure as a function of temperature was monitored by far-UV CD using AVIV-215 spectropolarimeter equipped with Peltier temperature control. Temperature-induced changes in the particle size were monitored simultaneously with CD by using fluorescence attachment to record 90° light scattering as described (32). The negative baseline slope in the light scattering curves is an optical artifact of the AVIV-215 model (Jack Aviv, private communication).
ApoC-I:DMPC samples of 20 µg/ml protein concentration placed in 5 mm cells were used to record far-UV CD spectra, melting or kinetic data. The spectra were recorded from 185–250 nm with 15 s/nm data accumulation time. The CD data were normalized to protein concentration and expressed as molar residue ellipticity [Θ]; protein helical content was determined from the value of [Θ222] at 222 nm according to (33). In the melting experiments, CD and light scattering data were recorded simultaneously at 222 nm during sample heating and cooling with 1 °C increment at scan rates of 11 or 80 K/h. In the kinetic temperature-jump (T-jump) experiments, sample temperature was rapidly increased at time t=0 from 25 °C to a higher constant value, and the time course of the protein unfolding was monitored by Θ222 with 30 sec increment for 2–20 h.
Kinetic analysis of the CD data was carried out as described (27–29). Briefly, the T-jump data Θ222(t) recorded at each temperature were approximated by single exponentials to determine the relaxation times τ which is inverse of the unfolding rate k=1/τ. Activation energy (enthalpy) Ea≅H* of the unfolding reaction was determined from the slope of the Arrhenius plot, RT lnτ versus 1/T, where R is universal gas constant and T is temperature in Kelvin. The accuracy in Ea determination is 5–7 kcal/mol, which incorporates fitting errors and deviations among different data sets. Mutation-induced changes δΔG* in the kinetic lipoprotein stability ΔG*=ΔH*-TΔS* = −RT ln(k/K) (where K is the reaction rate in the absence of the barrier) were determined from the shifts in the Arrhenius plots of the DMPC complexes with apoC-I mutants as compared to a similar plot for the WT:DMPC complex, δΔG* = −RT ln(k mutant/ k WT) RT ln(τmutant/τWT) = RT δ lnτ. The accuracy in δΔG* determination is better than 0.5 kcal/mol.
Electron microscopy
ApoC-I:DMPC samples subjected to various thermal treatments were visualized using negative staining technique in a CM12 transmission electron microscope as described (27–29). To trap the intermediates of lipoprotein denaturation and reconstitution and to prevent protein-induced DMPC remodeling during sample cooling to 22 °C and deposition on EM grids, we used 1-naphthol. This is a small fluorescent probe that binds DMPC bilayers at two distinct sites: the interfacial hydrocarbon region and the hydrophobic core region (34). Relative population of these sites depends on bilayer fluidity, making 1-naphtol a useful probe for the analysis of lipid phase transitions and the effects of additives on membrane fluidity (34–36). Importantly, 1-naphtol binding does not alter the phase transition temperature or the polarity of the lipid (34–36) and does not induce morphologic changes in DMPC vesicles or apoC-I:DMPC disks (EM data not shown). However, our EM and clearance studies show that 100 µM concentration of 1-naphthol inhibits apoC-I-induced DMPC clearance and lipoprotein reconstitution at 24 °C. This enables us to use 1-naphthol as an inhibitor of DMPC remodeling by apoC-I. In this work, lipoprotein samples of 20 µg/ml protein and 80 µg/ml lipid concentration were heated and cooled to different final temperatures and a small amount of 1-naphthol stock solution (100 mM in ethanol) was added to the samples at these temperatures to the final concentration of 120 µM.
Analysis of the products of lipoprotein denaturation
HDL denaturation involves lipoprotein separation into two fractions: protein fraction that dissociates from HDL in lipid-poor or lipid-free form, and lipid-rich fraction that comprises fused lipoproteins (27, 37). To establish the presence of protein in the lipid-rich fraction formed upon denaturation of apoC-I:DMPC disks, we isolated and analyzed this fraction as follows. A sample of apoC-I:DMPC disks (0.5 mg/ml protein and 2 mg/ml lipid concentration) was incubated for 15 min at 90 °C and was immediately placed on ice to prevent lipoprotein reconstitution at 22 °C. The lipid-rich fraction was isolated by density gradient centrifugation. The density of the sample and the blank was adjusted to 1.25 mg/ml using solid KBr; the samples were transferred to 4 ml SW-60 tubes, overlaid with 2 ml solution of 1.068 mg/ml KBr, and spun at 4 °C, 50,000 rpm for 22 h. A turbid layer formed at the surface after the spin indicated separation of lipid-rich vesicular fraction. The presence of protein in this fraction was established by Bradford’s assay and by measuring intrinsic Trp fluorescence (280 nm excitation wavelength) as described (29), which was facilitated by the presence of Trp45 in apoC-I.
Clearance studies
ApoC-I-induced clearance of DMPC multilamellar vesicles (MLV) at various temperatures was monitored by turbidity at 325 nm using UV/Vis spectrophotometer Varian Cary-300 equipped with thermoelectroic temperature control. MLV were prepared by solvent evaporation (38). To do so, 5 mg of DMPC was dissolved in one part choloroform, two parts methanol solution that was evaporated slowly under nitrogen to form a uniform lipid film. Excess solvent was removed by overnight incubation under vacuum, and lipid was suspended in 1 ml of standard buffer. In our clearance studies we used 40 µg/ml lipid and 10 µg/ml protein concentration; importantly, at this concentration and buffer conditions, human apoC-I is fully monomeric (39). In our experiments, about 1 ml of DMPC suspension were placed in 1 cm path length quartz cell and equilibrated at a constant temperature for about 5–10 min during which turbidity was recorded with 1 s averaging time and 1 nm bandwidth. No changes in turbidity were detected at this stage, indicating temporal stability of our MLV preparations. Next, the data recording was briefly paused and a small volume of stock protein solution was added to the lipid suspension and mixed gently, followed by resumption of the turbidity measurements for 6–24 h.
All experiments in this study were repeated 3–6 times to ensure reproducibility. ORIGIN software was used for the processing and/or display of the CD, light scattering and turbidity data.
RESULTS
Effects of apoC-I mutations on lipoprotein denaturation upon heating
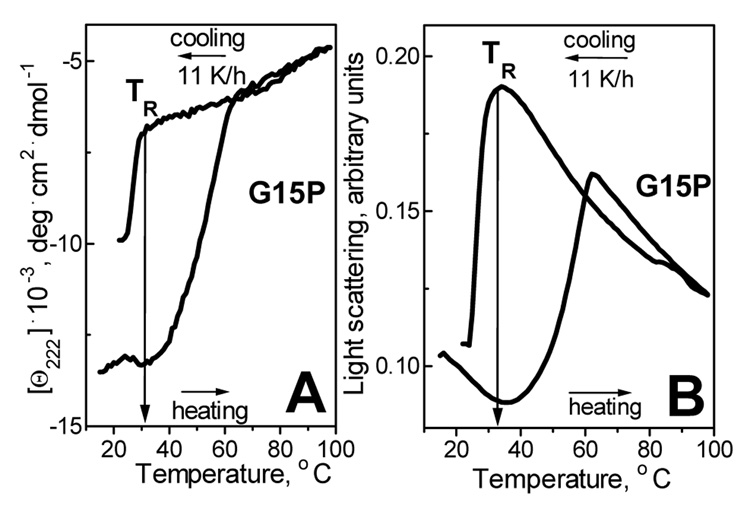
Fig. 2 shows CD and 90° light scattering data that were recorded simultaneously at 222 nm during heating and consecutive cooling of G15P:DMPC complexes. Far-UV CD spectra of such complexes at 25 °C (not shown) indicate that G15P apoC-I, which is largely unfolded in solution (22), acquires 44% helical structure on DMPC disks. Changes in the CD signal Θ222(T) in Fig. 2A, which is proportional to the α-helical content, indicate heat-induced protein unfolding at 35–65 °C, and the concomitant increase in the light scattering (Fig. 2B) indicates disk fusion into vesicles at these temperatures (27). Cooling curves show partial re-folding of the helical structure and concomitant reduction in the particle size which reflects lipoprotein reconstitution; the onset temperature of this transition is TR=32 °C (Fig. 2A, B). Thus, lipoprotein reconstitution upon cooling is observed at significantly lower temperatures than lipoprotein denaturation upon heating, indicating a thermodynamically irreversible reaction (27–29).
Figure 2.
Thermal denaturation and reconstitution of apoC-I:DMPC complexes. Samples of G15P:DMPC disks (20 µg/ml protein, 80 µg/ml lipid in 5 mM Na phosphate buffer, pH 7.6) were heated and cooled from 20–98 °C at a rate 11 K/h. (A) Protein unfolding and refolding upon heating and cooling monitored by CD at 222 nm. (B) Lipoprotein fusion and reconstitution monitored by 90° light scattering at 222 nm simultaneously with the CD signal in panel A. The reconstitution temperature TR is indicated.
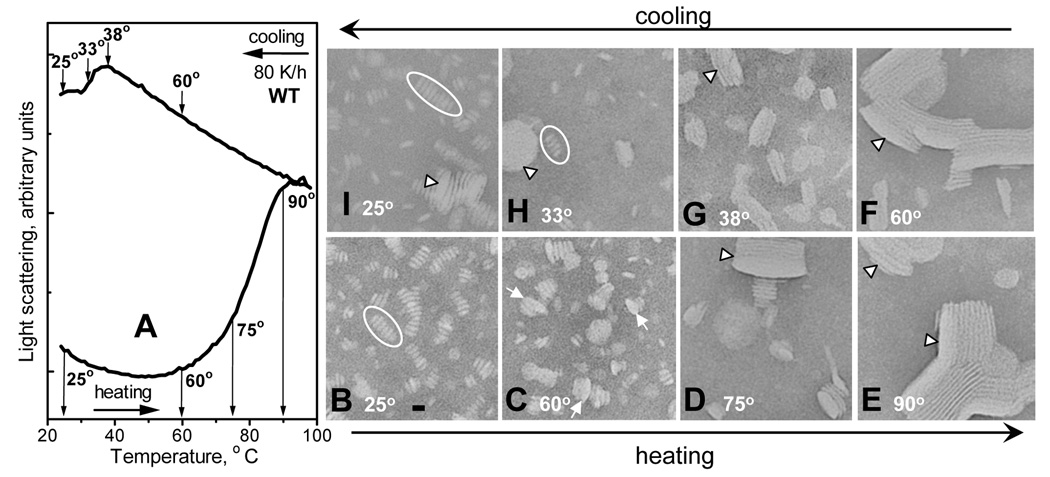
Earlier we showed that the thermodynamically irreversible step in HDL denaturation involves heat-induced protein unfolding/dissociation and particle fusion that compensates for the decrease in the polar surface moiety (27, 37). In this work we used 1-naphthol to trap the products of lipoprotein fusion and reconstitution upon heating and cooling to different temperatures. Samples of WT:DMPC disks were heated and cooled from 20–98 °C at a rate 80 K/h to different final temperatures (depicted in Fig. 3A) at which 1-naphthol was added to stop lipoprotein remodeling. The samples were then cooled to room temperature and visualized by negative staining EM. At 25 °C, addition of naphthol had no detectable effect on the size or morphology of intact disks; in Fig. 3B these disks are seen in edge view stuck in rouleaux, which is typical of negative staining preparation. Heating to 60 °C (early stage of the heat denaturation) leads to formation of enlarged particles including protein-containing small unilamellar vesicles (SUV, Fig. 3C). Heating to 75 °C (middle of the heat denaturation) leads to disappearance of the intact-size disks and formation of large multilamellar vesicles (MLV, Fig. 3D); MLV become the predominant species upon completion of lipoprotein denaturation at 90 °C (Fig. 3E). Thus, the heat-induced increase in the light scattering observed between 60–90 °C (Fig. 3A), which reflects increase in the particle size and/or refractive index, is in excellent agreement with the EM data showing disk fusion into SUV and, eventually, MLV at these temperatures (Fig. 3C–E).
Figure 3.
Complexes of WT apoC-I:DMPC at various stages of thermal denaturation and reconstitution. Bar size is about 15 nm. (A) Light scattering data recorded of WT:DMPC disks during sample heating and cooling at 80 K/h from 20–98 °C; sample conditions are as in Fig. 1. (B–I) Electron micrographs of negatively stained WT:DMPC samples that were heated (lower row) and then cooled (upper row) to different final temperatures (indicated in the panels) that correspond to different stages of lipoprotein denaturation and reconstitution (indicated in panel A). At the final temperature, 1-naphthol was added to each sample (120 µM final concentration) to trap the reaction intermediates. Ovals in B, H indicate disk rouleaux, arrows in C indicate collapsed small unilamellar vesicles (that are thicker and larger in diameter than disks), and arrowheads in D–G indicate collapsed multilamellar vesicles.
Similarly, cooling curves in Fig. 3A are in excellent agreement with the corresponding EM data (Fig. 3F–I). These data show that cooling of the heat-denatured lipoproteins from 90-60 °C leads to no significant changes in the light scattering (Fig. 3A) or particle morphology (Fig. 3F). However, further cooling to 38 °C, i. e. near the onset temperature TR of the lipoprotein reconstitution, leads to a large reduction in the vesicle size (Fig. 3G). Cooling to 33 °C (in the middle of the reconstitution transition) results in a mixture of intact-size disks and vesicles (Fig. 3H). Finally, cooling to 25 °C (near Tc=24 °C of DMPC where lipoprotein reconstitution is complete) results in intact-size disks as the predominant species. Thus, light scattering and EM data in Fig. 3 are in excellent agreement and show disk to SUV to MLV conversion upon heating, followed by MLV fission and disk reconstitution upon cooling to TR and below.
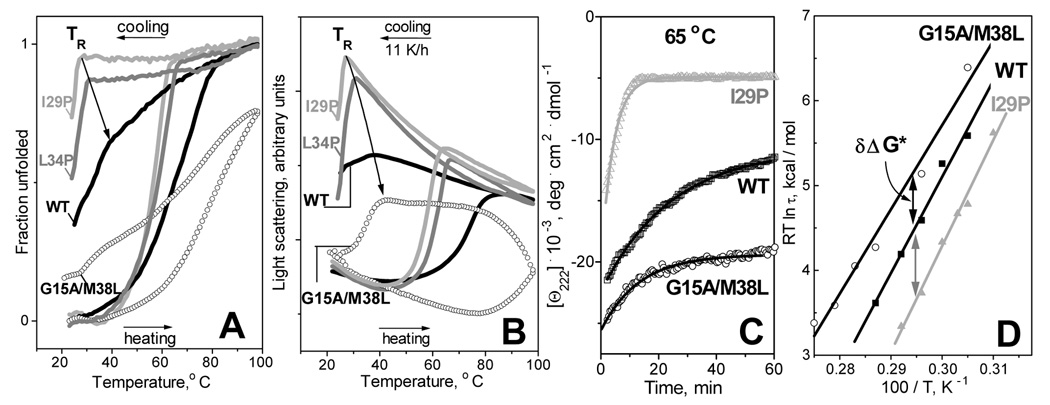
Next, we analyzed the effect of the protein secondary structure on the lipoprotein denaturation and reconstitution upon heating and cooling. To do so, we utilized apoC-I point mutants differing in their helix content in solution and on the disks (Fig. 1, Table 1). Fig. 4A shows normalized CD melting data Θ222(T) and Fig. 4B shows light scattering melting data recorded during heating and cooling of DMPC complexes with these mutants at 11 K/h. These data show that DMPC complexes with I29P apoC-I, which has among the lowest helical contents in solution (<5%) and on the lipid (47%, Table 1), undergo heat-induced thermal unfolding and fusion at relatively low temperatures, yet similar complexes with better-folded proteins, such as L34P, WT or G15A/M38L (which contain 60%, 65% or 75% helix on DMPC, respectively), show progressive increase in their apparent melting temperature. This suggests that apoC-I variants with higher helical content on DMPC form more stable disks.
Figure 4.
Effects of protein mutations on thermal denaturation and reconstitution of apoC-I: DMPC complexes. (A) CD and (B) light scattering heating and cooling data recorded at 222 nm, 11 K/h scan rate of DMPC disks with WT (black line), L34P (grey), I29P (light grey), and G15A/M38L apoC-I (open circles). (C) Time course of lipoprotein denaturation monitored by CD at 222 nm in T-jumps from 25 to 65 °C. Solid lines show monoexponential data fitting that was used to determine the relaxation times τ of protein unfolding. (D) Arrhenius plots, RT ln τ versus 1/T, of DMPC disks containing WT, I29P and G15A/M38L apoC-I. Exponential relaxation times τ(T) at 60–90 °C were determined from the kinetic CD data Θ222(t) recorded in T-jumps from 25 to 60–90 °C. Double arrows indicate changes in the kinetic free energy of the disk stability δΔG* induced by mutations.
To further test the correlation between the disk stability and the protein α-helix content, we analyzed the kinetics of the lipoprotein denaturation in T-jump experiments. The denaturation was triggered at time t=0 by rapid heating from 25 °C to a higher constant temperature; the time course of the protein unfolding at each temperature was monitored by CD at 222 nm. Fig. 4C shows the normalized kinetic CD data Θ222(t) recorded of DMPC complexes with selected apoC-I variants in T-jumps to 65 °C. The unfolding rates for different proteins significantly differ: proteins with lower helix content on DMPC, such as I29P, show faster unfolding, while those with increased helical content, such as WT or G15A/M38L, show progressively slower unfolding. A similar trend is observed in the T-jumps to other temperatures (data not shown). As a result, the Arrhenius plots in Fig. 4D show small but significant shifts that correspond to kinetic stabilization of G15A/M38L:DMPC disks by δΔG*=+0.7 kcal/mol and to destabilization of the I29P:DMPC disks by δΔG*=−0.9 kcal/mol as compared to the WT:DMPC disks. Other apoC-I mutations induce comparable or smaller changes in the disk stability (summarized in Table 1). Taken together, these results suggest weak correlation between the protein helix content on DMPC and the mutation-induced changes in the disk stability δΔG* (r2=0.37): proteins with higher helical content tend to form more stable complexes.
Effects of apoC-I mutations on lipoprotein reconstitution upon cooling
In addition to the differences in the heating curves of DMPC disks containing different mutants, the CD and light scattering data in Fig. 4A, B also show significant differences in the onset temperatures TR of lipoprotein reconstitution upon cooling: DMPC complexes with the less helical I29P mutant show TR=27 °C, while similar complexes with increasingly more helical proteins, such as L34P, WT, and G15A/M38L, reconstitute at increasingly higher TR of 31–41 °C. A similar correlation was observed in our earlier studies of DMPC complexes with L34P and Q31P apoC-I (28): the higher the protein helix content in solution and on DMPC at 25 °C the higher the TR (Table 1). This is confirmed by linear regression analysis of the results in Table 1 showing a good correlation between TR and the helix content in solution (r2=0.96) and on the lipid (r2=0.85). Interestingly, the average helical content observed upon lipoprotein heating and cooling to its respective TR varies from less than 5% for I29P and other mutants that are fully unfolded in solution to about 48% in G15A/M38L. Consequently, no minimal helix content is required for the lipoprotein reconstitution to occur at TR.
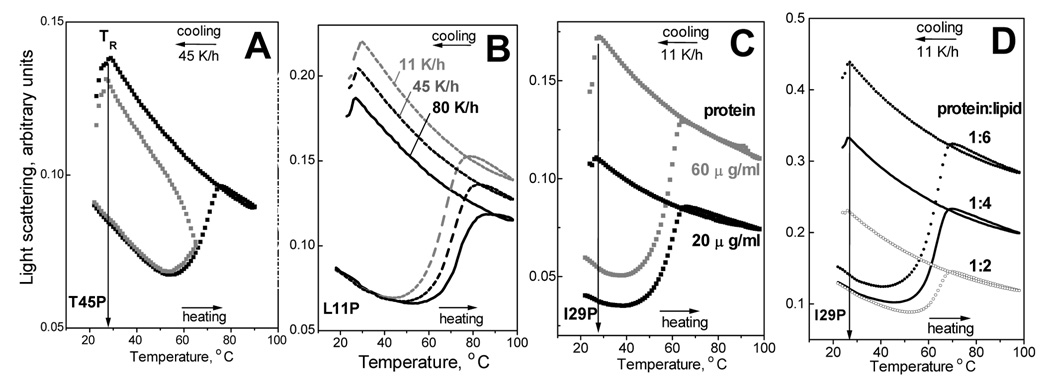
To determine what factors may affect the TR, we first tested whether TR is influenced by the extent of the disk denaturation. CD and light scattering melting data were recorded at 222 nm from several apoC-I:DMPC samples upon heating and cooling at a rate of 45 K/h to the highest temperature of 65–70 °C (in the middle of the heat denaturation) or 90 °C (in the post-transitional range). The cooling curves in Fig. 5A, which were recorded of T45P:DMPC complexes upon heating to 70 °C or 90 °C, show identical TR; similar data for other apoC-I variants also indicated that TR is not affected by the extent of the lipoprotein denaturation.
Figure 5.
Effects of the extent of the disk denaturation, scan rate, sample concentration, and protein-to-lipid ratio on the onset temperature TR of lipoprotein reconstitution. Panels A–D show heating and cooling data recorded by light scattering at 222 nm of DMPC complexes with apoC-I variants (indicated in the figures). Unless otherwise stated, discoidal complexes were prepared as described in Materials and Methods using 1:4 protein-to-lipid weight ratio, and fresh samples under standard conditions (20 µg/ml protein, 80 µg/ml lipid) were used for the data collection at 11 K/h scan rate. (A) T45P:DMPC disks were heated and cooled at a rate 45 K/h from 20 °C to different highest temperatures: 67 °C (grey) or 90 °C (black). (B) L11P:DMPC disks were heated and cooled from 20–98 °C at various scan rates from 11 to 80 K/h (indicated on the lines). (C) I29P:DMPC disks samples of different concentrations (black - 20 µg/ml protein, 80 µg/ml lipid; grey − 60 µg/ml protein, 240 µg/ml lipid) were heated and cooled from 20–98 °C. Protein concentrations are indicated. (D) Heating and cooling data of I29P:DMPC samples containing 20 µg/ml protein and different DMPC concentrations: 40 µg/ml (black), 80 µg/ml (grey), and 120 µg/ml (light grey). Protein-to-lipid weight ratios are indicated.
Next, we tested the scan rate effect on TR. The melting data were recorded during sample heating and cooling from 20–98 °C at various rates from 11–80 K/h. Light scattering heating curves of DMPC complexes with apoC-I variants such as L11P (Fig. 5B) show large shifts (by − 10 °C) upon reduction in the scan rate from 80 to 11 K/h, which is indicative of a kinetically controlled transition with high activation energy Ea (27,40); in contrast, the cooling curves in Fig. 5B show no significant scan rate effects on TR within the accuracy of its experimental determination (about 2 °C). This suggests that the kinetic effects on TR, if any, are too small to be detected experimentally.
Since lipoprotein fusion is a high-order reaction, it may depend on the sample concentration. To test whether disk fusion upon heating and reconstitution upon cooling are affected by the lipoprotein concentration, we recorded heating and cooling data from I29P:DMPC disks (1:4 protein-to-lipid weight ratio) containing 20 or 60 µg/ml protein. The results in Fig. 5C show that the characteristic temperatures of the heating and cooling transitions are independent of the sample concentration. Next, we tested whether the stoichiometry of the protein-lipid complexes affects the temperatures of lipoprotein denaturation and reconstitution. To do so, we prepared I29P:DMPC complexes using three protein-to-lipid weight ratios: 1:2 (which has insufficient amount of lipid to accommodate all protein on the disks, thus part of the protein is probably not associated with discoidal complexes, as suggested by significantly lower α-helical content in these preparations as compared to disks), 1:4 (standard disk preparation in which all protein and lipid are in discoidal complexes) and 1:6 (which contains excess lipid in the form of vesicles). The melting data of these samples show that the protein-to-lipid ratio has no detectable effect on TR (Fig. 5D). Thus, TR is not affected by the sample concentration or by the presence of excess protein or lipid.
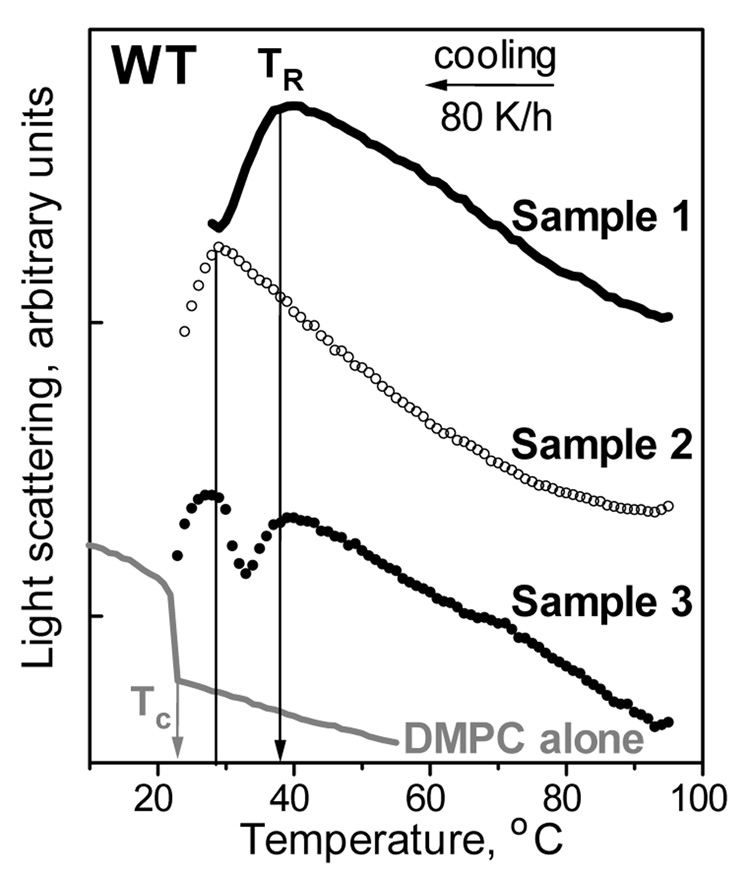
The major determinant for TR is the temperature Tc of the main transition from liquid crystalline (Lα) to rippled gel phase (Pβ’); at Tc, the lipoprotein reconstitution from free protein and lipid is fastest due to the maximal density of the linear hydrophobic defects running along the lipid phase boundary that provide the primary protein binding sites (1–3). Our results show that TR is also affected by the protein, since TR significantly exceeds Tc, particularly for apoC-I variants with higher α-helix content in solution and on the lipid (Fig. 4A; Table 1). To understand the effect of protein on TR, consider two protein fractions that are formed upon HDL denaturation and were observed by non-denaturing gel electrophoresis (37): i) lipid-poor or free protein that dissociates from lipoproteins upon heating, and ii) protein-containing lipid vesicles that are formed upon heat-induced lipoprotein fusion. The presence of the protein in the latter fraction formed upon apoC-I:DMPC disk-to-vesicle fusion was confirmed by isolation and analysis of this fraction by using Bradford assay and intrinsic Trp fluorescence (see Materials and Methods). To identify which protein fraction affects the TR, the cooling data were recorded from WT:DMPC samples that had identical chemical composition but were prepared differently. In sample 1, the disks were formed using our standard protocol (overnight co-incubation of protein and lipid at 24 °C) and were denatured by heating to 98 °C immediately prior to recording the cooling data. In sample 2, free protein and lipid emulsion (that was prepared by solvent evaporation (38) as described in the clearance studies in Materials and Methods) were heated separately to 50 °C or to 98 °C and were mixed at these temperatures which are too high for the spontaneous protein-DMPC complex reconstitution to occur. Thus, at the beginning of the cooling, sample 1 contained lipid-dissociated and vesicle-bound protein, while sample 2 contained lipid-free protein and vesicles that had no significant amount of bound protein (as indicated by non-denaturing gel electrophoresis, data not shown). In addition, sample 3 contained equal parts of samples 1 and 2 that were mixed at 98 °C immediately prior to recording the cooling data. The data in Fig. 6 clearly show that, in contrast to TR=36–38 °C of sample 1, lipoprotein reconstitution in sample 2 does not occur until cooling below 28 °C. Consequently, protein-containing vesicles can reconstitute lipoproteins at significantly higher temperatures than the protein-free vesicles. This implies that the bound protein in the vesicle surface facilitates lipoprotein reconstitution. Furthermore, the cooling data of sample 3 showed two distinct transitions that are coincident with the reconstitution transitions in samples 1 and 2 (Fig. 6). Taken together, these results suggest that protein-containing and protein-free vesicles reconstitute lipoproteins via distinctly different pathways, and that the presence of bound protein in the vesicles facilitates lipoprotein reconstitution at significantly higher temperatures.
Figure 6.
Effect of vesicle-bound protein on the onset temperature TR of lipoprotein reconstitution upon cooling from high temperatures. Light scattering data were recorded during cooling from 98–20 °C at 80 K/h of three samples with identical chemical composition (20 µg/ml WT, 80 µg/ml DMPC in standard buffer) that were prepared differently. Sample 1: protein and lipid were incubated at 24 °C to prepare disks (as described in Materials and Methods), which were heated to 98 °C prior to recording the cooling data. Sample 2: protein solution and lipid emulsion (prepared by solvent evaporation (38)) were heated separately and mixed at 98 °C prior to recording the cooling data. Sample 3: equal amounts of samples 1 and 2 were mixed at 98 °C prior to recording the cooling data. The curves are shifted along the Y-axis to avoid overlap. Cooling data of DMPC emulsion alone (80 µg/ml lipid in standard buffer) is shown for comparison. Protein-containing samples 1–3 show a reduction in the 90° light scattering upon cooling that reflects reduction in the particle size upon lipoprotein reconstitution. In contrast, DMPC alone shows a sharp increase in light scattering upon cooling beyond Tc=24 °C, which is due to higher refractive index (higher density) of the rippled gel phase Pβ’ as compared to the high-temperature liquid crystalline phase Lα of DMPC.
Effects of apoC-I mutations on DMPC clearance at various temperatures
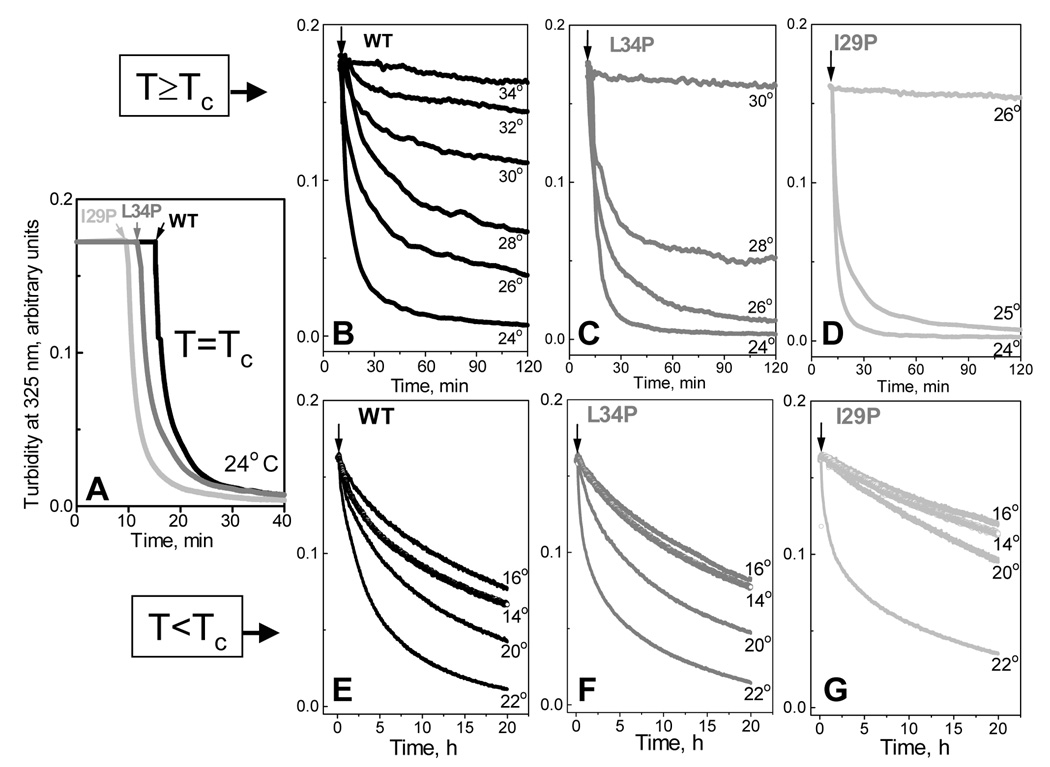
To further test the role of the protein secondary structure in various pathways of lipoprotein reconstitution, we analyzed the time course of DMPC clearance by apoC-I mutants at several constant temperatures near Tc=24 °C of DMPC Lα to Pβ’ phase transition. Liposome clearance, which was triggered by the addition of protein at the final protein-to-lipid weight ratio 1:4, was monitored by turbidity at 325 nm. Importantly, at the solvent conditions and 10 µg/ml protein concentration used in our experiments, human apoC-I is fully monomeric (39). Since Pro substitutions in the apolar face of an amphipathic α-helix produce a kink that reduces peptide self-association (this general phenomenon is described in (30)), Pro-containing apoC-I mutants studied in our work are also monomeric under these conditions. Therefore, the clearance kinetics in our studies is not affected by the peptide self-association. This is consistent with the observation that the DMPC clearance rates by all peptides studied remain invariant upon changing the peptide concentration from 5–20 µg/ml but decelerate at higher concentrations when the apoC-I self-association becomes significant (data not shown).
The data in Fig. 7A show that at 24 °C the clearance kinetics for the I29P, L34P and WT apoC-I are similar despite large differences in the protein α-helical content in solution (5–31%). In contrast, at T>Tc the clearance kinetics for different apoC-I variants is significantly different (Fig. 7B–C). Temperature increase above 24 °C leads to a gradual reduction in the DMPC clearance rate by WT apoC-I, along with a gradual increase in the residual turbidity that suggests the presence of larger particles (Fig. 7B). Interestingly, electron micrographs show that the lipoprotein disks formed at higher temperatures have comparable or smaller diameter than those formed at 24 °C, suggesting that larger residual turbidity observed at higher temperatures reflects the presence of large lipid vesicles in addition to the small disks. Importantly, significant DMPC clearance by WT apoC-I is detected at temperatures up to 32–33 °C, which is close to TR=37 °C observed for this protein in the heating and cooling experiments (Fig. 3A, Table 1). DMPC clearance by less well-folded proteins stops at progressively lower temperatures (30 °C for L34P and for 26 °C I29P (Fig. 7C, D)) that are a few degrees lower than their respective TR (Table 1). Studies of other apoC-I point mutants reveal a similar trend: the clearance rate is similar for different peptides at 24 °C but declines sharply, particularly for less helical proteins, at T>Tc, and completely stops as the temperature approaches TR. At T<Tc, the clearance rates are much slower but show a similar trend (Fig. 7E–G): proteins with higher helical content can clear DMPC in a broader temperature range.
Figure 7.
Effects of apoC-I mutations on the time course of DMPC clearance at various temperatures. After lipid emulsions (prepared by solvent evaporation (38)) were incubated at a constant temperature, the clearance was triggered by addition of protein (indicated by arrows). Final sample concentrations are 10 µg/ml apoC-I and 40 µg/ml DMPC in standard buffer; apoC-I is fully monomeric under these concentrations (22). Clearance by WT (black), L34P (grey), and I29P apoC-I (light grey) at Tc =24 °C of the main DMPC phase transition (A), above (B–D) and below Tc (E–G); the temperatures are indicated. Panels E–G show substantial clearance at 14–16 °C near the pre-transition temperature Tc’=15 °C from the gel (Lβ’) to rippled (Pβ’) phase.
Quantitative Arrhenius analysis of the clearance kinetics is precluded by the inherent caveats in the reaction rate determination (41): turbidity (which is dominated by light scattering of large vesicles) is not proportional to the extent of the reaction, the reaction products at different temperatures are different, and the data are not well-approximated by exponentials. Nevertheless, the data in Fig. 7 clearly show that at T> Tc and T> Tc, proteins with higher helical content clear lipid faster, suggesting lower free energy barrier for lipid penetration by such proteins. Also, proteins with higher helical content show more gradual temperature dependence of the clearance rates above and below Tc, suggesting lower activation energy (enthalpy) of lipid penetration.
DISCUSSION
Lipoprotein stability correlates with protein α-helical content
The results of this work suggest a weak correlation between the protein helix content on DMPC and the lipoprotein stability ΔG* (Table 1). For example, apoC-I mutations such as I29P or G15P that reduce the protein helical content in solution and on the lipid destabilize the apoC-I:DMPC complexes by up to −1 kcal/mol, while G15A/M38L mutation that increases the protein helical content increases the lipoprotein stability by about +0.7 kcal/mol (Fig. 4D). However, not all peptides follow this trend. Thus, L11P, R23P or M38P mutations lead to a large reduction in the α-helix content in solution and on the lipid, yet they cause no large destabilization of the apoC-I:DMPC complexes (Table 1). Therefore, lipoprotein stability depends not only on the protein α-helical content but also on the location and the character of the mutation (Fig. 1). For example, charge removal by R23P substitution may help to optimize the electrostatic interactions that are important for the apoC-I:DMPC disk stability (29); these improved electrostatic interactions may compensate for the destabilizing effect of Pro in the middle of the helix, resulting in similar stability of the R23P and WT-containing DMPC disks.
Earlier studies of 20-residue apolipoprotein peptides showed that Pro substitutions in the middle of the helix, but not near its ends, have large effects on the helical content and peptide-lipid association (20). Our results suggest that some Pro substitutions located in the middle of the helix (T45P), near helical kinks (L11P) or ends (M38), or in the interhelical linker (Q31, L34) follow this general trend, while others (G15P, R23P, I29P) do not (Table 1). For example, despite its terminal location in the helix, I29P mutation causes a large reduction in the helical content in solution and on the lipid and greatly destabilizes apoC-I:DMPC disks (Table 1). Thus, factors affecting protein conformation and lipoprotein stability in the context of a full-size apolipoprotein, even as small as apoC-I, are more complex than in small single-helical peptides.
Weak correlation between the protein helix content on the lipid and lipoprotein stability reported here is consistent with the earlier studies suggesting that increased helix content in apolipoprotein-based 18-mer peptides optimizes protein-lipid interactions (42). This correlation may, at least in part, be an enthalpic effect, since proteins with higher helical content may form more extensive interactions with the lipid. A similar enthalpic effect probably contributes to the correlation between the protein size and the disk stability reported earlier (43,44). However, since apoC-I mutations have no significant effect on the slopes of the Arrhenius plots and hence on the activation energy (enthalpy) Ea of lipoprotein denaturation (Fig. 4D), the enthalpic effects of these mutations may not exceed the accuracy in Ea determination, which is 5–7 kcal/mol. Entropic effect of the protein α-helices on the acyl chain disordering (45) and entropic cost of lipid-induced helical folding may also contribute to the observed correlation between the helix content and lipoprotein stability.
Our results also show that the apparent melting temperature Tm obtained from thermal unfolding at a constant rate does not necessarily provide an accurate measure of the lipoprotein stability. For example, the heating data of L34P:DMPC complexes show significantly lower apparent Tm than similar data of WT:DMPC (Fig. 4A, B), yet these complexes show similar unfolding rates in kinetic T-jump experiments and thus have similar kinetic stability ΔG* (27). This illustrates that the thermodynamic analysis of lipoprotein stability may not always be consistent with the kinetic analysis and should be treated with caution.
Bound α-helices facilitate insertion of additional helices in the lipid surface
The results of this work suggests that the role of the protein secondary structure in lipid surface binding depends on the availability of the hydrophobic defects. If such defects are readily available, as is the case at the phase transition temperature Tc=24 °C of DMPC, the clearance rates by apoC-I variants are identical (Fig. 7A) despite large differences in their solution conformation (0–31% α-helix, Table 1). To our knowledge, this is the first direct observation of the effects of pre-existing secondary structure on lipid clearance; it was facilitated by the identical protein size, by substantial helical content in apoC-I monomer, and by the lack of significant tertiary structure in this small protein. Thus, in contrast to tertiary or quaternary apolipoprotein interactions that correlate inversely with the PC clearance rates (13–16), the secondary protein structure in solution has no detectable effect on the clearance kinetics at Tc (Fig. 7A) and thus on the intrinsic rate constant of protein insertion into the pre-existing lipid packing defects that are maximal at Tc.
As the temperature increases above or decreases below Tc, the perimeter of the lipid phase boundary, and thus the density of the lattice defects running along this boundary that may form protein binding sites, decreases sharply (45, 46); also, the fluctuations in the lipid density that have been proposed to define the size of the dynamic lipid packing defects decrease sharply (47). This greatly decelerates apolipoprotein-lipid binding and clearance at T>Tc and T<Tc (48). Under these conditions, the secondary structure plays an increasingly important role in protein-lipid interactions: proteins with higher helical propensity can clear lipid in a broader temperature range both above and below Tc (Fig. 7B–D and E–G) and also show higher reconstitution temperature TR upon lipoprotein heating and cooling (Table 1). For example, WT apoC-I can clear DMPC at temperatures as high as 32–34 °C (Fig. 7B), which is well beyond the DMPC phase transition range and a few degrees lower than TR=37 °C of WT:DMPC reconstitution upon heating and cooling (Fig. 3A, Table 1). Similarly, the highest temperature of DMPC clearance by other apoC-I variants, such as L34P or I29P (Fig. 7C, D), is a few degrees lower than their respective TR (Table 1). This small difference in the highest temperature of the complex formation observed in the heating/cooling and in the clearance studies may result from different vesicle radii; MLV formed upon disk heating, which are significantly smaller that those obtained by solvent evaporation, may have increased protein reactivity (42,49) and thus form lipoproteins at somewhat higher temperatures.
Importantly, the cooling data in Fig. 6 show that lipoprotein reconstitution at TR≫Tc is facilitated by the vesicle-bound rather than lipid-free protein. This implies that the observed correlation between the TR and the maximal clearance temperature is due to sequential binding and insertion of protein molecules to the lipid surface during clearance.
Taken together, our results suggest that if the hydrophobic defects are abundant (i.e. at the lipid phase boundary), the rate of protein binding and insertion into these pre-existing defects is independent of the secondary structure in solution (α-helix or random coil). However, if the packing defects are scarce, protein insertion in the phospholipid surface is aided by the presence and the helical propensity of other surface-bound proteins: the higher the propensity the faster is the clearance the broader its temperature range (Fig. 7, Table 1) and hence the more readily additional protein molecules insert into lipid surface. Consequently, lipid-bound α-helices can facilitate binding and insertion of additional helices. The positive cooperativity in the binding and insertion of amphipathic α-helices into the lipid surface, which was also suggested in other apolipoprotein studies (19), may result from lipid-mediated interactions (bound α-helices perturb adjacent lipid molecules thereby helping to accommodate additional helices) and, possibly, from direct protein-protein interactions (lipid-bound α-helices may provide a template for folding and insertion of additional helices).
Cooperative lipid surface binding by amphipathic α-helices from the same or from different molecules may be involved in a variety of metabolic lipoprotein reactions, such as apolipoprotein exchange among lipoproteins in plasma or apolipoprotein-mediated lipoprotein binding and activation of specific receptors, transporters or lipolytic enzymes. Also, rapid desorption and re-adsorption of the central helices in apoA-I in response to changes in HDL lipid composition, which leads to formation of HDL subclasses, is facilitated by the lipid-bound helical segments of the molecule (50). This intramolecular cooperativity is a result of apolipoprotein evolution from the apoC-I-related precursor via the duplication and deletion of the 11-mer codon repeats (51). Interestingly, similar 11-mer helical repeats in α-syniclein show cooperative binding to phospholipid surface (52). Thus, the cooperative binding of α-helices to phospholipid surface reported here is not limited to apolipoproteins but may extend to functional reactions in other lipid surface-binding proteins and peptides.
ACKNOWLEDGEMENTS
We thank Dr. David Atkinson for very useful discussions and advice, Donald M. Small for reading the manuscript prior to publication, Donald L. Gantz for invaluable help with electron microscopy, and Cheryl England for general help.
Funding: This work was supported by the National Institutes of Health grants RO1 GM 067260 and HL 026355.
Abbreviations
- HDL
high-density lipoprotein
- apo
apolipoprotein
- DMPC
dimyristoyl phosphatidylcholine
- DPPC
dipalmitoyl phosphatidylcholine
- SUV
small unilamellar vesicle
- MLV
multilamellar vesicle
- WT
wild type
- CD
circular dichroism
- EM
electron microscopy
- T-jump
temperature jump
- PGK
phosphoglycerate kinase
- CCT
CTP:phosphocholine cytidylyltransferase
REFERENCES
- 1.Pownall H, Pao Q, Hickson D, Sparrow JT, Kusserow SK, Massey JB. Kinetics and mechanism of association of human plasma apolipoproteins with dimyristoyl phosphatidylcholine: effect of protein structure and lipid clusters on reaction rates. Biochemistry. 1981;20:6630–6635. doi: 10.1021/bi00526a017. [DOI] [PubMed] [Google Scholar]
- 2.Epand RM. The apparent preferential interaction of human plasma high density apolipoprotein A-I with gel-state phospholipids. Biochim. Biophys. Acta. 1982;712:146–151. doi: 10.1016/0005-2760(82)90096-0. [DOI] [PubMed] [Google Scholar]
- 3.Nuscher B, Kamp F, Mehnert T, Odoy S, Haass C, Kahle PJ, Beyer K. Alpha-synuclein has a high affinity for packing defects in a bilayer membrane: a thermodynamics study. J. Biol. Chem. 2004;279:21966–72195. doi: 10.1074/jbc.M401076200. [DOI] [PubMed] [Google Scholar]
- 4.Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 5.Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 1992;33:141–166. [PubMed] [Google Scholar]
- 6.Bussell R, Jr, Eliezer D. A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J. Mol. Biol. 2003;329:763–778. doi: 10.1016/s0022-2836(03)00520-5. [DOI] [PubMed] [Google Scholar]
- 7.Seelig J. Thermodynamics of lipid-peptide interactions. Biochim. Biophys. Acta. 2004;1666:40–50. doi: 10.1016/j.bbamem.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Anantharamaiah G, Navab M, Reddy ST, Garber DW, Datta G, Gupta H, White CR, Handattu SP, Palgunachari MN, Chaddha M, Mishra VK, Segrest JP, Fogelman AM. Synthetic peptides: managing lipid disorders. Curr. Opin. Lipidol. 2006;17:233–237. doi: 10.1097/01.mol.0000226114.89812.75. [DOI] [PubMed] [Google Scholar]
- 9.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 10.Breiter DR, Kanost MR, Benning MM, Wesenberg G, Law JH, Wells MA, Rayment I, Holden HM. Molecular structure of an apolipoprotein determined at 2.5 Å resolution. Biochemistry. 1991;30:603–608. doi: 10.1021/bi00217a002. [DOI] [PubMed] [Google Scholar]
- 11.Ajees AA, Anantharamaiah GM, Mishra VK, Hussain MM, Murthy HM. Crystal structure of human apolipoprotein A-I: insights into its protective effect against cardiovascular diseases. Proc. Natl. Acad. Sci. USA. 2006;103:2126–2131. doi: 10.1073/pnas.0506877103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Hickenbottom SJ, Kimmel AR, Londos C, Hurley JH. Structure of a lipid droplet protein; the PAT family member TIP47. Structure. 2004;12:1199–11207. doi: 10.1016/j.str.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 13.Segall ML, Phillips MC. Influence of apoE domain structure and polymorphism on the kinetics of phospholipids vesicle solublization. J. Lipid Res. 2002;43:1688–1700. doi: 10.1194/jlr.m200157-jlr200. [DOI] [PubMed] [Google Scholar]
- 14.Garda HA, Arreso EL, Soulages JL. Structure of apolipophorin-III in discoidal lipoproteins. Interhelical distances in the lipid-bound state and conformational change upon binding to lipid. J. Biol. Chem. 2002;277:19773–19782. doi: 10.1074/jbc.M110089200. [DOI] [PubMed] [Google Scholar]
- 15.Weers PM, Narayanaswami V, Choy N, Luty R, Hicks L, Kay CM, Ryan RO. Lipid binding ability of human apolipoprotein E N-terminal domain isoforms: correlation with protein stability? Biophys. Chem. 2003;100:481–492. doi: 10.1016/s0301-4622(02)00300-9. [DOI] [PubMed] [Google Scholar]
- 16.Lu B, Morrow JA, Weisgraber KH. Conformational reorganization of the four-helix bundle of human apolipoprotein E in binding to phospholipid. J. Biol. Chem. 2000;275:20775–20781. doi: 10.1074/jbc.M003508200. [DOI] [PubMed] [Google Scholar]
- 17.Saito H, Dhanasekaran P, Nguyen D, Holvoet P, Lund-Katz S, Phillips MC. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 2003;278(26):23227–23232. doi: 10.1074/jbc.M303365200. [DOI] [PubMed] [Google Scholar]
- 18.Saito H, Dhanasekaran P, Nguyen D, Deridder E, Holvoet P, Lund-Katz S, Phillips MC. Alpha-helix formation is required for high affinity binding of human apolipoprotein A-I to lipids. J. Biol. Chem. 2004;279(20):20974–20981. doi: 10.1074/jbc.M402043200. [DOI] [PubMed] [Google Scholar]
- 19.Arnulphi C, Jin L, Tricerri MA, Jonas A. enthalpy-driven apolipoprotein A-I and lipid bilayer interaction indicating protein penetration upon lipid binding. Biochemistry. 2004;43:12258–12264. doi: 10.1021/bi036118k. [DOI] [PubMed] [Google Scholar]
- 20.Ponsin G, Hester L, Gotto AM, Jr, Pownall HJ, Sparrow JT. Lipid-peptide association and activation of lecithin:cholesterol acyltransferase. Effect of alpha-helicity. J. Biol. Chem. 1986;261(20):9202–9205. [PubMed] [Google Scholar]
- 21.Rozek A, Sparrow JT, Weisgraber KH, Cushley RJ. Conformation of human apolipoprotein C-I in a lipid-mimetic environment determined by CD and NMR spectroscopy. Biochemistry. 1999;38(44):14475–14484. doi: 10.1021/bi982966h. [DOI] [PubMed] [Google Scholar]
- 22.Gursky O. Solution conformation of human apolipoprotein C-1 inferred from praline mutagenesis: far- and near-UV CD study. Biochemistry. 2001;40:12178–12185. doi: 10.1021/bi0111505. [DOI] [PubMed] [Google Scholar]
- 23.Jong MC, Hofker MH, Havekes LM. Role of apoC-I in lipoprotein metabolism: Functional differences between apoC1, apoC2, and apoC3. Arterioscler. Thromb. Vasc. Biol. 1999;19:472–484. doi: 10.1161/01.atv.19.3.472. [DOI] [PubMed] [Google Scholar]
- 24.Shachter NS. Apolipoproteins C-I and C-III as important modulators of lipoprotein metabolism. Curr. Opin. Lipidol. 2001;12:297–304. doi: 10.1097/00041433-200106000-00009. [DOI] [PubMed] [Google Scholar]
- 25.Kolmakova A, Kwiterovich P, Virgil D, Alaupovic P, Knight-Gibson C, Martin SF, Chatterjee S. Apolipoprotein C-I induces apoptosis in human aortic smooth muscle cells via recruiting neutral sphingomyelinase. Arterioscler. Thromb. Vasc. Biol. 2004;24:264–269. doi: 10.1161/01.ATV.0000112036.72200.ac. [DOI] [PubMed] [Google Scholar]
- 26.Lund-Katz S, Liu L, Thuahnai ST, Phillips MC. High density lipoprotein structure. Frontiers Biosci. 2003;8:1044–1054. doi: 10.2741/1077. [DOI] [PubMed] [Google Scholar]
- 27.Gursky O, Ranjana , Gantz DL. Complex of human apolipoprotein C-1 with phospholipid: Thermodynamic or kinetic stability? Biochemistry. 2002;41:7373–7384. doi: 10.1021/bi025588w. [DOI] [PubMed] [Google Scholar]
- 28.Mehta R, Gantz DL, Gursky O. Effects of mutations on the reconstitution and kinetic stability of discoidal lipoproteins. Biochemistry. 2003;42:4751–4758. doi: 10.1021/bi0341253. [DOI] [PubMed] [Google Scholar]
- 29.Benjwal S, Jayaraman S, Gursky O. Electrostatic effects on the kinetic stability of model discoidal high-density lipoproteins. Biochemistry. 2005;44:10218–10226. doi: 10.1021/bi050781m. [DOI] [PubMed] [Google Scholar]
- 30.Ladokhin AS, White SH. Interfacial folding and membrane insertion of a designed helical peptide. Biochemistry. 2004;43:5782–5791. doi: 10.1021/bi0361259. [DOI] [PubMed] [Google Scholar]
- 31.Markwell MAK, Haas SM, Bieber LL, Tolberg NEA. Anal. Biochem. 1985;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 32.Benjwal S, Verma S, Röhm KH, Gursky O. Monitoring protein aggregation during thermal unfolding in circular dichroism experiments. Protein Sci. 2006;15:635–639. doi: 10.1110/ps.051917406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao D, Wallace BA. Differential light scattering and absorption flattening optical effects are minimal in the circular dichroism spectra of small unilamellar vesicles. Biochemistry. 1984;23:2667–2673. doi: 10.1021/bi00307a020. [DOI] [PubMed] [Google Scholar]
- 34.Sujatha J, Mishra AK. Phase transition in phospholipids vesicles. Excited state prototropism of 1-naphthol as a novel probe concept. Langmuir. 1998;14:2256–2262. [Google Scholar]
- 35.Bhattacharyya K. Study of organized media using time-resolved fluorescence spectroscopy. J. Fluoresc. 2001;11(3):167–176. [Google Scholar]
- 36.Sujatha J, Mishra AK. Effect of ionic and neutral surfactants on the properties of phospholipid vesicles: investigation using fluorescent probes. J. Photochem. Photobiol. A. Chem. 1997;104:173–178. [Google Scholar]
- 37.Jayaraman S, Gantz DL, Gursky O. Effects of alt on thermal stability of human plasma high-density lipoproteins. Biochemistry. 2006;45:4620–4628. doi: 10.1021/bi0524565. [DOI] [PubMed] [Google Scholar]
- 38.Jonas A. Reconstitution of high-density lipoproteins. Methods Enzymol. 1986;128:553–582. doi: 10.1016/0076-6879(86)28092-1. [DOI] [PubMed] [Google Scholar]
- 39.Gursky O, Atkinson D. Thermodynamic analysis of human plasma apolipoprotein C-1: high-temperature unfolding and low-temperature oligomer dissociation. Biochemistry. 1998;37:1283–1291. doi: 10.1021/bi971801q. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez-Ruiz JM, Lopez-Lacomba JL, Cortijo M, Mateo PL. Differential scanning calorimetry of the irreversible thermal denaturation of thermolysin. Biochemistry. 1988;27:1648–1652. doi: 10.1021/bi00405a039. [DOI] [PubMed] [Google Scholar]
- 41.Massey JB, Pownall HJ. Role of oxysterol structure on the microdomain-induced microsolubilization of phospholipid membranes by apolipoprotein A-I. Biochemistry. 2005;44(43):14376–14384. doi: 10.1021/bi051169y. [DOI] [PubMed] [Google Scholar]
- 42.Gazzara JA, Phillips MC, Lund-Katz S, Palgunachari MN, Segrest JP, Anantharamaiah GM, Rodrigueza WV, Snow JW. Effect of vesicle size on their interaction with class A amphipathic helical peptides. J. Lipid Res. 1997;38:2147–2154. [PubMed] [Google Scholar]
- 43.Reijngoud DJ, Phillips MC. Mechanism of dissociation of human apolipoproteins A-I, A-II, and C from complexes with dimyristoylphosphatidylcholine as studied by thermal denaturation. Biochemistry. 1984;23:726–734. doi: 10.1021/bi00299a022. [DOI] [PubMed] [Google Scholar]
- 44.Jayaraman S, Gantz DL, Gursky O. Kinetic stabilization and fusion of apolipoprotein A-2: DMPC disks: Comparison with apoA-1 and apoC-1. Biophys. J. 2005;88(4):2907–2918. doi: 10.1529/biophysj.104.055921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koynova R, Caffrey M. Phases and phase transitions of the phosphatidylcholines. Biochim. Biophys. Acta. 1998;1376:91–145. doi: 10.1016/s0304-4157(98)00006-9. [DOI] [PubMed] [Google Scholar]
- 46.Epand RM, Surewicz WK. Effect of phase transitions on the interaction of peptides and proteins with phospholipids. Can. J. Biochem. Cell Biol. 1984;62(11):1167–1173. doi: 10.1139/o84-150. [DOI] [PubMed] [Google Scholar]
- 47.Freire E, Biltonen R. Estimation of molecular averages and equilibrium fluctuations in lipid bilayer systems from the excess heat capacity function. Biochim. Biophys. Acta. 1978;514(1):54–68. doi: 10.1016/0005-2736(78)90076-7. [DOI] [PubMed] [Google Scholar]
- 48.Swaney JB. Mechanisms of protein-lipid interaction. Association of apolipoproteins A-I and A-II with binary phospholipid mixtures. J. Biol. Chem. 1980;255(18):8791–8797. [PubMed] [Google Scholar]
- 49.Wetterau JR, Jonas A. Effect of dipalmitoylphosphatidylcholine vesicle curvature on the reaction with human apolipoprotein A-I. J. Biol. Chem. 1982;257:10961–10966. [PubMed] [Google Scholar]
- 50.Wang L, Atkinson D, Small DM. Interfacial properties of an amphipathic alpha-helix consensus peptide of exchangeable apolipoproteins at air/water and oil/water interfaces. J Biol. Chem. 2003;278:37480–37491. doi: 10.1074/jbc.M303133200. [DOI] [PubMed] [Google Scholar]
- 51.Luo CC, Li WH, Moore MN, Chan L. Structure and evolution of the apolipoprotein multigene family. J. Mol. Biol. 1986;187:325–340. doi: 10.1016/0022-2836(86)90436-5. [DOI] [PubMed] [Google Scholar]
- 52.Bisaglia M, Schievano E, Caporale A, Peggion E, Mammi S. The 11-mer repeats of human alpha-synuclein in vesicle interactions and lipid composition discrimination: a cooperative role. Biopolymers. 2006;84:310–316. doi: 10.1002/bip.20440. [DOI] [PubMed] [Google Scholar]