Abstract
During the last several decades corals have been in decline and at least one-third of all coral species are now threatened with extinction. Coral disease has been a major contributor to this threat, but little is known about the responsible pathogens. To date most research has focused on bacterial and fungal diseases; however, viruses may also be important for coral health. Using a combination of empirical viral metagenomics and real-time PCR, we show that Porites compressa corals contain a suite of eukaryotic viruses, many related to the Herpesviridae. This coral-associated viral consortium was found to shift in response to abiotic stressors. In particular, when exposed to reduced pH, elevated nutrients, and thermal stress, the abundance of herpes-like viral sequences rapidly increased in 2 separate experiments. Herpes-like viral sequences were rarely detected in apparently healthy corals, but were abundant in a majority of stressed samples. In addition, surveys of the Nematostella and Hydra genomic projects demonstrate that even distantly related Cnidarians contain numerous herpes-like viral genes, likely as a result of latent or endogenous viral infection. These data support the hypotheses that corals experience viral infections, which are exacerbated by stress, and that herpes-like viruses are common in Cnidarians.
Keywords: coral reefs, disease, Herpesviridae, viral-like particles, virome
Coral disease is a major cause of the pandemic decline of tropical reefs (1, 2). Increases in coral disease incidence have occurred during the last several decades (3, 4), and, although their devastating effects on coral populations are no longer in question (5), in most cases, it is not yet clear whether specific or opportunistic pathogens are involved (6, 7).
A challenge to identifying causative agents of coral disease is that corals naturally harbor a variety of microbiota including symbiotic algae, fungi, Bacteria and Archaea (8–13). Corals demonstrate species-specific and potentially mutualistic relationships with communities of Bacteria (14), and these associated communities are altered during episodes of coral disease (7, 15). To date, no coral viral diseases have been positively identified, but pathologies, such as yellow blotch and white band disease, have been hypothesized to be the results of viral infection (16).
Virus-like particle abundances shift both across reefs and with proximity to coastal areas and corals themselves (17, 18). On tropical reefs viral numbers range from 0.3 to 1.25 × 107 particles per milliliter in reef water (19) to 0.8–1 × 109 particles per cubic centimeter within the sediment (20). Small-scale variations in viral numbers occur with changing proximity to coral colonies, with viral particles peaking ≈2–4 cm above the coral surface. These abundances are elevated in waters surrounding diseased coral colonies from 1.0 × 107 to 1.5 × 107 particles per milliliter (21). Virus-like particles (VLPs) also occur within corals and their relatives. For example, VLPs have been found within the tissues of anemones and within the symbiotic zoochlorellae and zooxanthellae of various Cnidarians (22–24). While the identity of these VLPs has yet to be established, an increase in viral particles within dinoflagellates has been hypothesized to be responsible for symbiont loss during bleaching (25–27). VLPs also have been identified visually on several species of scleractinian corals, specifically: Acropora muricata, Porites lobata, Porites lutea, and Porites australiensis (28). Based on morphological characteristics, these VLPs belong to several viral families including: tailed phages, large filamentous, and small (30–80 nm) to large (>100 nm) polyhedral viruses (29). Metagenomic data have also shown that both phages and eukaryotic viruses are associated with Porites astreoides (30) and healthy and bleaching Diploria strigosa corals (31).
Environmental stress often results in coral bleaching, disease, and death. Increased temperature, nutrient loading, dissolved organic carbon pollution, and reductions in ambient seawater pH are of particular concern due to their effects on the coral-symbiont relationship, host homeostasis, microbial overgrowth, and skeletal deposition (32–34). To determine whether environmental perturbations shift the eukaryotic viral assemblage present in corals, these 4 parameters were manipulated, and the resulting viral consortia characterized through the generation of 6 metagenomes. We show here that changes in the viral repertoire varied across each treatment, but all treatments contained genomic sequence similarities to specific eukaryotic viral families, most notably the Herpesviridae. Furthermore, a unique herpes-like viral sequence (HLVs) increased up to 6 orders of magnitude in coral specimens exposed to increased temperature, nutrients, and pH stress indicating viral production within the host. From these and other data, we hypothesize that a distantly related herpes-like virus or group of viruses is commonly associated with corals and Cnidarians in general and are produced as a result of stressor events.
Results and Discussion
Experimental Design and Rationale.
To characterize coral-associated viruses, specimens of the finger coral, Porites compressa, were collected in the Hawai'i Marine Laboratory Refuge. This species of coral was chosen because it is a common and endemic species of coral in the Hawaiian archipelago and is the dominant species of coral across reefs in Kane'ohe Bay where Coconut Island and the Hawai'i Institute of Marine Biology are located. Numerous studies have been conducted on this coral and there is a legacy of information on the oceanography and ecology of these reefs, providing pertinent data for insights into the viral ecology of corals across this region.
Coral specimens were exposed to thermal (5° C above ambient), dissolved organic carbon (25 mg·mL−1 glucose), nutrient (10 μM nitrate, nitrite, phosphate, and ammonium), and pH (1.7 units below ambient) stress for 64 hours [supporting information (SI) Fig. S1]. In brief, 3 corals were broken into fingers and placed in aquaria containing a given treatment. Coral samples or “nubbins” were collected just before the experiment (“time zero”) to measure shifts in the viral consortia that resulted from isolation from the reef, and to provide several unadulterated samples for comparison. Subsequent samples were taken after 1, 4, 16, and 64 h of stressor exposure. Six viromes were generated by pooling 1 μg of DNA from each time point: time zero, “aquarium control,” “temperature,” “DOC,” “pH,” and “nutrient” (Fig. S1). Pyrosequencing was conducted at 454 Life Sciences on a GS20. Sequence reads were parsed and analyzed using bioinformatics.
Library Statistics and Bioinformatic Analysis.
The sequenced viromes contained between 30,000 and 51,000 reads with an average length of 105 bases, a dataset of >25 million bases. Several bioinformatic techniques identified changes in the eukaryotic viral consortia across each library. Sequence reads were first compared using BLASTn to the nonredundant database at the National Center for Biotechnology Information (Table S1). A sequence was assigned as “known” if it had a similarity (e ≤ 10−4) to the nonredundant database. These low stringency parameters were chosen because viral DNA often has little similarity to known sequences in the non-redundant database (35). “Knowns” were then taxonomically sorted (viral, bacterial, archaeal, and eukaryotic) based on their highest similarity. In agreement with previous findings, these metagenomes contained few similarities to known viral sequences (Table S1) (36). A majority of sequences were found to be similar to bacterial and eukaryotic annotations. Analyses of 16S and 18S rDNA PCR and similarities to the human genome (see Methods) demonstrated that eukaryotic and microbial DNA contamination did not contribute to these sequences. This high percentage of known sequences similar to Bacteria and Eukaryota is likely the result of (i) protein conservation across viral, microbial, and eukaryotic genomes; (ii) the larger size and larger number of microbial genomes at the National Center for Biotechnology Information database; (iii) prophage and retrotransposon-like sequences within microbial and eukaryotic genomes; and (iv) the genetic mosaicism of viruses. The entire library was also compared with 4 other marine viromes (Table S2) to determine whether the coral viromes resembled seawater viruses. Few similarities were found between the coral viromes and water samples, suggesting that seawater contamination was not an issue.
Coral-Associated Viral Consortia Change with Experimental Manipulation.
One goal of this study was to determine the types of eukaryotic viruses infecting the coral holobiont. Phage data will be presented elsewhere. As few sequences (Table S1) were found to be similar to any viral sequences at the National Center for Biotechnology Information, reanalysis of the metagenomic reads was conducted using tBLASTx (e ≤10−6) to a boutique database of the fully sequenced eukaryotic viral genomes from the National Center for Biotechnology Information database.
Metagenomic sequences with significant similarities (e ≤10−6) were assigned as “knowns” based on the sequence's best similarity (i.e., the top hit) to each reference viral genome. To compare and contrast the overall differences between each library, sequences were first sorted at the family level, and the percentage of known similarities to each viral family was calculated.
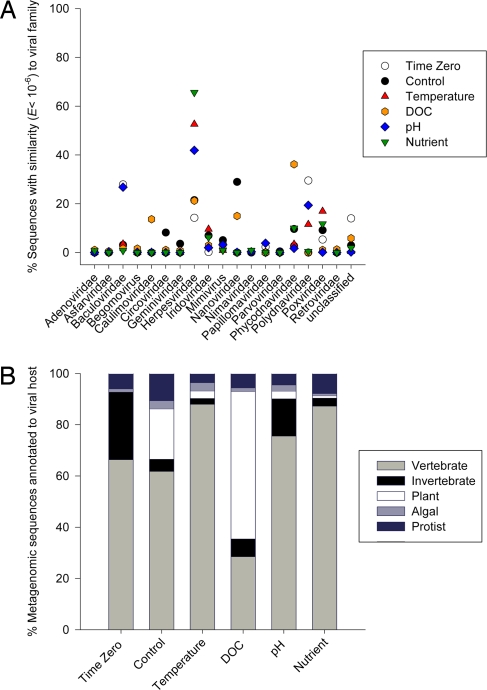
Of the ≈100 viral families described thus far (37), 19 were found in the combined coral-associated viromes (Fig. 1A). For example, the time zero samples contained 23% Baculoviridae-like and 30% Polydnaviridae-like sequences. Baculoviridae-like sequences also accounted for 23% of the similarities in the pH (blue diamonds) sample. The aquarium control (filled circles) contained 30% and the carbon-loaded DOC samples (orange hexagons) 18% sequences similar to the Nanoviridae. A large proportion of the best sequence similarities in the stressed samples were related to the Herpesviridae family, with the nutrient, pH, and temperature-perturbed samples containing 64%, 53%, and 70% herpes-like sequences, respectively.
Fig. 1.
A) Percentages of best similarities (tBLASTx, e ≤10−6) to 19 families of viruses in each individual coral-associated viral metagenome: Time Zero (open circles), Aquarium Control (filled circles), Temperature (red triangles), pH (blue diamonds), Nutrient (green arrows), and DOC (orange hexagons). (B) Predicated host ranges across each metagenome, with libraries containing vertebrate (light gray) and invertebrate (black bars) associated viral sequences and plant (white bars) and algal (dark-gray bars) viruses. Blue bars represent similarity to viral genomes that are associated with Protists.
Coral-Associated Viruses Infect a Range of Hosts.
A discriminating characteristic in viral taxonomy is host range (38). Corals are a collective of cnidarian tissue, microbial symbionts, fungi, and other microeukaryotes. To determine the potential hosts of the coral-associated viruses, sequences were categorized according to a predicted generalized host range defined by the known host of the similar reference genome. These host ranges were modified from the Universal Virus Database from the International Committee on the Taxonomy of Viruses (37). Using these eukaryotic viral assignments, the predicted host range of the coral-associated viruses changed with each stressor treatment (Fig. 1B). In each of the viromes, >38% of the similarities were associated with vertebrate hosts, including representatives of most of the herpesviruses, poxviruses, adenoviruses, papillomaviruses, circoviruses, and iridoviruses (light gray bars). The Aquarium Control and DOC samples contained a large fraction of sequences similar to ssDNA viruses from plants (white bars) such as geminiviruses and nanoviruses (39). Invertebrate infecting viruses (black bars) were also abundant (>60%) within the time zero library and were mostly similar to the baculoviruses and polydnaviruses, 2 viral families that predominantly infect arthropods (40–42).
However, a caveat to this analysis is that the predicted host range is biased by the larger number of completed vertebrate-associated viral genomes. This analysis is also based on the sequences' best similarity, but almost certainly represents distant relatives of the reference viruses. These data suggest that coral-associated viruses possibly have many target hosts, including metazoans, protists, fungi, and plants.
Stressors Increase the Abundance of Herpes-Like Viral Sequences.
Each virome sequence may have multiple similarities due to gene conservation. Therefore, each library was also analyzed for the total number of sequence similarities (as opposed to the best similarity in Fig. 1) to each of the eukaryotic viral genomes at the National Center for Biotechnology Information database, using tBLASTx (e ≤10−6). Individual viromes had between 300 and 11,000 predicted protein level sequence similarities to all eukaryotic viral genomes (Table 1). However, the number of these sequences that had homology to the Herpesviridae was different between the stressors. The percentage of all of the viral similarities to the Herpesviridae was similar in the time zero (≈19%) and the control (≈19%) but was elevated to 30%, 32%, and 32%, in the temperature, nutrient, and pH samples, respectively. The DOC sample had few sequences (< 2%) related to Herpesviridae.
Table 1.
All sequence similarities (e < 10−6) based on tBLASTx analysis to a boutique database containing the fully sequenced eukaryotic viral genomes from the National Center for Biotechnology Information database and the abundance and percentage of those related to herpesviruses (HV)
| Sample | Sequences | Eukaryotic Virus | HV | HV, % |
|---|---|---|---|---|
| Time zero | 39,270 | 11,826 | 2272 | 19.21 |
| Control | 39,340 | 414 | 81 | 19.57 |
| Temperature | 39,036 | 667 | 201 | 30.13 |
| DOC | 35,680 | 373 | 7 | 1.88 |
| pH | 50,368 | 11,831 | 3791 | 32.04 |
| Nutrient | 34,433 | 858 | 277 | 32.28 |
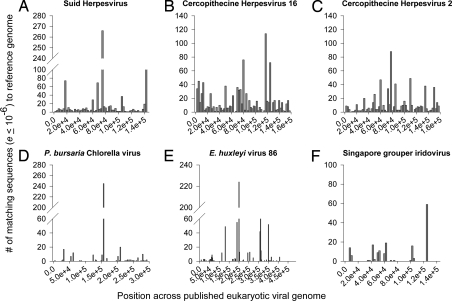
The large abundance of Herpesviridae similarities in the temperature, pH, and nutrient samples indicated that these stressors induced production of viruses with distant relation to the herpesviruses. A caveat to this analysis, however, is that the short read length combined with low sequence homology limits taxonomic resolution (43). To verify if herpes-like viruses were present in the samples, sequences from all of the libraries were combined, compared using tBLASTx (e ≤10−6) to the eukaryotic viral boutique database, and then plotted along the corresponding reference genomes. The combination library contained both broad and deep coverage to some herpesvirus (44–48)genomes (Fig. 2 A–C and Table S3), but low coverage to other marine and cnidarian-associated viral genomes (Fig. 2 E–H and Table S3), such as a panaeid shrimp virus, a zoochlorella infecting phycodnavirus (2D), a phytoplankton virus (2E), and a fish iridovirus (2F) genome (49). From these analyses, we hypothesized that herpes-like viruses (HLVs) are present in corals and become more abundant as a result of stress events.
Fig. 2.
Coverage of selected Herpesviridae genomes and other marine associated viral genomes. Libraries were combined and sequences analyzed for similarity (tBLASTx e ≤10−6) to a boutique database of 2,020 fully sequenced eukaryotic viral genomes collected from the National Center for Biotechnology Information. Genomes were separated into 2,500 base pair bins. (A–C) Broad and high fold coverage was found for many mammalian associated Herpesviridae genomes, such as a suid herpesvirus (A) and 2 cercopithecine/macaque herpesviruses (B and C). (D–F) Low and uneven coverage was found for the symbiont associated chlorella phycodnavirus (D), a coccolithophorid virus (E), and a fish iridovirus (F).
Real-Time PCR on Original Temporal Samples Confirms the Production of HLVs.
Viral production, or the generation of new virions by the host, has been effectively quantified using real-time PCR to genes of various RNA and DNA viruses (50). To determine temporal changes in viral abundance across the manipulation experiments, metagenomic sequences were assembled, annotated, and given a taxonomic classification according to the sequence's best protein similarity using PSI-BLAST to the non-redundant database (see SI Methods for details). Sequences with significant homology (e ≤10−6) to herpesviruses were chosen, and primers were generated for both conventional and real-time PCR. One sequence was 62% identical (e <4 × 10−11) to a single copy thymidylate synthase gene from Saimiriine herpesvirus 2 (Fig. S2). This sequence was amplified, and the resulting PCR product cloned and Sanger sequenced to verify the assembly (GenBank accession no. EU660955). No differences were found between the original assembly and the cloned nucleotide sequence (data not shown).
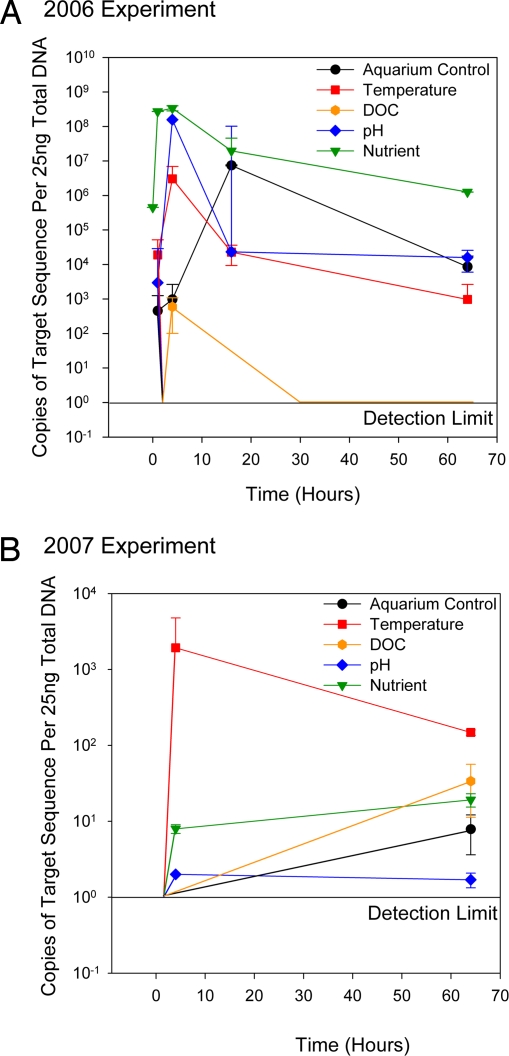
These primers were used for real-time PCR, and it was found that the copy number of this putative herpes-like viral thymidylate synthase sequence increased during the original stressor experiment (Fig. 3A). The target sequence was below the detection level in all of the zero hour time points with the exception of the Nutrient (green triangles). The target sequence was also elevated in all of the 1-, 4-, 16-, and 64-h treated corals. The abundance of this target sequence typically peaked at 4 h after treatment exposure, and the maximum increase was ≈6 orders of magnitude. For example, the pH (blue diamonds) and Temperature (red triangles) stressors increased the abundance of this viral sequence from below the detection limit (≈10 copies) to >105 and 107 copies in only 4 h. Viral abundance was then reduced to 103 for the remainder of those experiments.
Fig. 3.
Herpes-like viral sequence abundance across two 64-h experiments. (A and B) Real-time PCR on a putative herpes-like virus thymidylate synthetase gene was conducted on all temporal samples collected during the original metagenomic experiment (A) and samples from a second experiment conducted 1 year later (B). In the original experiment, coral specimens were exposed to thermal (5° C above ambient), dissolved organic carbon (25 mg·mL−1 glucose), nutrient (10 μM nitrate, nitrite, phosphate, and ammonium), and pH (1.7 units below ambient) stress for 64 hours. (B) In the second 2007 experiment, corals were acclimated to the tanks for 2 weeks before treatment with stressor, thermal stress was an elevation of seawater temperature 3°C, acidity stress was a 0.2 unit reduction of ambient pH, and samples were collected at 0, 4, and 64 h.
In the Aquarium Control sample (Fig. 3, black circles) large numbers (≈107) of this HLV specific target gene were detected after 16 h and demonstrated that even in the absence of applied exogenous abiotic stress, physical manipulation of the corals led to viral production. The viral sequence was detected in only 1 of the 5 DOC (orange hexagons) time points, corroborating the low abundance of sequences in the DOC metagenome. Both this low copy number in the DOC samples and the large copy number in the nutrient zero time point indicate that external herpesvirus contamination was not a confounding factor.
Second Experiment Conducted One Year Later Confirms that HLVs Are Commonly Associated with Stressed Corals.
The possibility that the viruses discovered in the corals were unique to the original metagenomic experiment and/or the result of unnatural stress conditions was tested by conducting a second experiment 1 year later. This second repeat experiment used the same sampling scheme (Fig. S1) with minor modifications to reflect more likely environmental conditions: (i) corals were acclimated to the tanks for 2 weeks before treatment with stressor, (ii) thermal stress was an elevation of seawater temperature 3°C, (iii) acidity stress was a 0.2 unit reduction of ambient pH, and 4) nubbins were only collected at 0, 4, and 64 h. Identical primer sets were used to amplify the HLV thymidylate synthase sequence.
These analyses showed an increase in the herpes-like virus target sequence when stressors were applied to the corals (Fig. 3B). The target sequence was not detected at the zero hour time point in any of the tissue samples, but was found in the temperature (red squares), pH (blue diamonds), and nutrient (green triangles) samples at 4 and 64 h. The DOC exposed and aquarium control corals contained no detectable target sequence until 64 h.
Together, these results show that, although the temporal changes in HLV abundance were analogous in both experiments, the magnitude of this change was different. Compared with the 2006 metagenomic experiment, the total abundance of HLVs was lower in the second 2007 experiment. This may have resulted from the more environmentally relevant levels of stressors applied in the repeat experiment. However, environmental conditions were also milder during the second 2007 experiment with no extended rainy or windy periods before or during the experiment. In contrast, during the original 2006 experiment, precipitation along with significant runoff occurred. As previously mentioned, the abundance of the HLVs was found to be high in the original experiment's nutrient zero hour sample. This indicates that production of virus had already begun on the reef, perhaps as a result of degraded environmental conditions.
A two-way ANOVA conducted on the real-time data indicated significant effects of both treatment (F = 9.80, P < 0.01) and time (F = 4.82, P < 0.01), and a significant interaction effect (F = 4.54, P < 0.01). The combination of the two main effects and the interaction term explains >70% of the variance. These results indicated that the different treatments applied to corals stimulated herpes-like viral production and that the level of production varied significantly over the time course of the experiment. A significant interaction term suggested that the effects of both treatment and time on viral production were amplified multiplicatively. This implies that just studying each treatment individually over time or all treatments simultaneously over 1 time point would not have shown the same differences in production.
Additional Evidence for Herpes-Like Viruses in Corals and All Cnidarians.
In addition to our datasets, transmission electron micrographs (TEM) and sequences from the Nematostella genome and hydra EST project provide further evidence that Cnidarians, not just corals, are hosts for herpes-like viruses. Corals contain large enveloped icosahedral virions morphologically similar to the Herpesviridae (28, 29). Herpesvirus essential genes are also present in Cnidarian genomes (28, 51, 52). Within the Nematostella draft genome, alkaline exonucleases, herpes major outer envelope proteins, herpes DNA primases, herpes ubiquitin-specific proteases as well a herpes surface glycoprotein and transcription activation factor were identified (Table S4). Also, several copies of a latent membrane protein were found suggesting the presence of endogenous or latent herpes-like viruses in Nematostella. Additionally, expressed sequence tags from Hydra magnipapillata contain herpes specific tegument proteins and herpes-associated proteases, suggesting that herpes-like viral transcripts were produced.
Herpes-like viral sequences were also recently identified in metagenomic libraries from P. astreoides and D. strigosa corals. In the P. astreoides metagenome, most of the sequences with similarity to eukaryotic viruses were related to fish iridoviruses, but the second most abundant viral similarities were to Herpesviridae (30). In addition, >50% of the all viral sequence similarities (including phages and Archaeal viruses) from linker amplified shotgun libraries of healthy and bleached D. strigosa were to Herpesviridae, indicating that herpesviruses are commonly associated with corals and not a result of our methodology.
Caveats.
The initial metagenomic experiment was designed both to address questions about the viral consortia present in corals and to determine whether this high-throughput metagenomic approach is amenable for empirical experiments. Therefore, treatments were designed to be harsh and as a result did not reflect realistic stressor levels. However, the second 2007 experiment was redesigned to represent more environmentally relevant levels of stress and used to confirm trends from the metagenomes.
In addition, equal amounts of DNA were combined from each replicate coral at each time point to generate individual stressor libraries that contained between 36 and 45 separately extracted viromes. This pooling approach provided a reduction in the variance, or normalizing shifts in the viral consortia over-time. The metagenomes therefore represent an underestimation of the true changes in the viral consortia. The individual temporal samples were then used for real-time PCR to verify identified trends.
The method used to collect the coral subsamples or nubbins required removal of the corals from the aquaria and repeated tissue damage to the coral finger. Corals from the no treatment aquarium control had elevated numbers of herpes-like viral sequences in both experiments. It was apparent from the real-time PCR that this method was disruptive enough to induce herpes-like viral particle production albeit at a reduced level compared with the other stressors. However, naturally occurring damage to corals results from predation, weathering events like hurricanes and earthquakes, and human activities, such as the aquarium coral and ornamental fish trade, recreational diving, and research (53). Our experimental design may have inadvertently mimicked fragmentation and small scale predation on corals. It is therefore likely that these activities and events are stressful enough to induce production of herpes-like viruses on the reef and may result in short or long term reductions in coral health that have until now gone unnoticed.
Last, the metagenomic DNA was amplified using multiple displacement amplification with Phi29 polymerase before sequencing, which could artificially inflate the occurrence of rare sequences, ssDNA, and small circular genomes, thus biasing the analysis (54, 55). However, multiple displacement amplification generally provides an even representation of genomes except at the ends (56) and, as evidenced by the elevation of ssDNA viruses in only the DOC sample, did not appear to bias these data.
Conclusions
This experiment used 6 empirically derived metagenomes to investigate shifts in previously unexplored viral consortia. The coral holobiont is a symbiotic community of animal host, zooxanthellar algae, and various other microbiota (57). This diversity of organisms is reflected in the viral consortia present within the tissues of corals, which included representatives of viruses that infect many protist and metazoan phyla (Fig. 1B), and viruses that infect Bacteria and Archaea (data not shown). Due to the predominance of herpes-like viral sequences in both our samples and Nematostella, an aposymbiotic anemome, we hypothesize that these distantly related herpesviruses infects the coral host.
Environmental and Physical Stressors Are Sources of Viral Production.
The metagenomic and temporal experiments presented here demonstrate that exposure to stressors results in the production of a herpes-like virus or a consortium of herpes-like viruses in P. compressa corals. Thermal stress, eutrophication, and decreasing seawater pH have each been shown to disrupt coral health. Increases in sea surface temperature causes coral bleaching and increased coral disease incidence (58, 59). Nutrient addition exacerbates coral diseases, and reduced pH results in loss of corallite deposition (60, 61). This study demonstrates that, in addition to symbiont loss and bacterial and fungal disease (62), temperature and nutrient elevation and pH reduction result in increased HLV production.
A hypothesis that was not directly tested is that the duration of each stressor maybe critical to the overall effect on viral production. Treatments were only 64 h in length, but stress events may last significantly longer on the reef. A reduction in the viral abundance after 4 h in both experiments may suggest that corals can withstand these short stressors, but prolonged stress, such as sustained seawater temperature elevation, may overwhelm a coral's ability to fight off infection.
It is currently unclear how these viruses affect coral physiology or ecology. We hypothesize that viral production can (i) directly cause down stream changes in physiology such as a reduced innate immunity response, secondary opportunist infection by microbes, symbiont expulsion, cell detachment, and apoptosis, or (ii) negatively combine with other stressor induced changes.
Herpesviruses, Human Disease, and the Evolution of Metazoans.
Herpesviruses are characterized by several morphological and genomic commonalities, including large enveloped icosahedral virions, long linear double stranded DNA genomes, and similar life cycle pathways (63). Although herpesviruses were once considered exclusively vertebrate diseases, members of the ostreid Herpesviridae are known to infect and kill invertebrates, specifically industrially cultured bivalves (64). The genomes of these shellfish-infecting herpesviruses contain few gene sequences in common with their vertebrate counterparts. Genome examination has demonstrated that herpesviruses can contain upwards of 70% unique genes (65). Across all of the genomes of Herpesviridae there is only a single subunit of a terminase shared in common (66). These data suggest that Herpesviridae is not a phylogentically related group of viruses, but perhaps consists of 2 phenetic Families, one related to vertebrate disease and the other to invertebrate disease.
Our coral metagenomes contain more sequences similar to the α-herpesviruses (including several human associated herpesviruses), which infect predominantly mammals and often exhibit broad host range (37). The data presented here support the hypothesis that herpes-like viruses are common in Cnidarians and could perhaps represent distant relatives of the vertebrate lineage of Herpesviridae. Our studies may also provide insight into the evolution of these viruses. Herpesviruses typically infect nervous tissue, and it is tempting to suggest that the herpes-like viruses in Cnidarians may have ancient origins, because Cnidarians are the first metazoans to develop rudimentary nervous systems (52).
This was the first experiment to use massively parallel pyrosequencing for empirical experiments, demonstrating that this high-throughput approach can be used as a first step to identify novel viruses in virtually any system. Yet caution should be taken when designing empirical metagenomics. Our statistical analysis on the real-time PCR suggests static analysis of metagenomes may fail to identify critical changes between and among viral consortia.
Overall, we have demonstrated that herpes-like viruses are commonly associated with corals and perhaps all Cnidarians and are produced as a result of changes in ambient water conditions and physical disruption. It is not yet clear how these viruses may affect the physiology or resilience of their hosts, but these data provide a critical foundation for future studies on eukaryotic viral disease ecology of corals.
Materials and Methods
Virome Generation.
Virome DNA from each time point and treatment was isolated and extracted to generate twenty-five temporal samples that were subsampled, amplified, and then pooled by treatment for metagenomic library construction (see Fig. S1).
Virome Purity Verifications.
To verify that the viral libraries contained no human contaminating DNA, sequences were compared with the human genome using BLASTn and a minimum 200 score as measured by the formula (0.267 × raw score-ln(0.041))/ln (2)). Raw scores were the sums of the alignment pairwise scores. Across all 6 viromes low numbers of sequences (<0.8%) were assigned “human” indicting that human contamination was not an issue.
Real-Time PCR.
Real-time PCR on the putative herpes-virus target sequence was conducted using SYBR GreenER qPCR SuperMix from Invitrogen according the to the manufactures' recommendations with exactly 25 ng of total viral DNA. Primers used were: forward, 5′-AAAATAAGATTGGGAGATCTAGGC-3′ and reverse, 5′-TGCCATTTTAGGTAAATCAGAAAC-3′. Standards were made from purified target and standard curves were generated using 0.5, 0.05, 0.005, and 0.0005 ng of the target (GenBank accession no. EU660955). A two-way ANOVA (time and treatment) was conducted using Minitab Version 15 (State College, PA). Both parametric and non-parametric tests provided similar results, and the residuals were randomly distributed with a mean of 0, justifying the assumptions.
Supplementary Material
Acknowledgments.
We thank the Rappè laboratory and all those at Hawai'i Institute of Marine Biology who sponsored RV.T. in Hawaiì. E. Brown, A. Thurber, D. Drumm, and M. Hatay were also instrumental in the collections of the corals and viral particles. R. Schmieder also provided bioinformatic assistance. This work was sponsored by National Science Foundation Postdoctoral Fellowship DBI-0511948 (to R.V.T.) and a Marine Microbial Initiative grant from the Gordon and Betty Moore Foundation (to F.L.R.). Corals were sampled under the auspices of an Hawai'i Institute of Marine Biology collection permit and on Department of Land and Natural Resources Special Activity Permit SAP 2007-72 (to R.V.T.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (genome project and accession nos. 28415, 28417, 28419, 28421, 28423, and 28425 and EU660955).
This article contains supporting information online at www.pnas.org/cgi/content/full/0808985105/DCSupplemental.
References
- 1.Harvell CD, et al. Review: Marine ecology—Emerging marine diseases—Climate links and anthropogenic factors. Science. 1999;285:1505–1510. doi: 10.1126/science.285.5433.1505. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter KE, et al. One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science. 2008;321:560–563. doi: 10.1126/science.1159196. [DOI] [PubMed] [Google Scholar]
- 3.Remily ER, Richardson LL. Ecological physiology of a coral pathogen and the coral reef environment. Microbial Ecol. 2006;51:345–352. doi: 10.1007/s00248-006-9029-9. [DOI] [PubMed] [Google Scholar]
- 4.Ward JR, Lafferty KD. The elusive baseline of marine disease: Are diseases in ocean ecosystems increasing? PLoS Biol. 2004;2:542–547. doi: 10.1371/journal.pbio.0020120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg E, Ben-Haim Y. Microbial diseases of corals and global warming. Environ Microbiol. 2002;4:318–326. doi: 10.1046/j.1462-2920.2002.00302.x. [DOI] [PubMed] [Google Scholar]
- 6.Cooney RP, et al. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ Microbiol. 2002;4:401–413. doi: 10.1046/j.1462-2920.2002.00308.x. [DOI] [PubMed] [Google Scholar]
- 7.Pantos O, et al. The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ Microbiol. 2003;5:370–382. doi: 10.1046/j.1462-2920.2003.00427.x. [DOI] [PubMed] [Google Scholar]
- 8.Ritchie KB. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol-Prog Ser. 2006;322:1–14. [Google Scholar]
- 9.Rohwer F, Breitbart M, Jara J, Azam F, Knowlton N. Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs. 2001;20:85–91. [Google Scholar]
- 10.Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser. 2002;243:1–10. [Google Scholar]
- 11.Kramarsky-Winter E, et al. Identification of a protist-coral association and its possible ecological role. Mar Ecol Prog Ser. 2006;317:67–73. [Google Scholar]
- 12.Wegley L, et al. Coral-associated archaea. Mar Ecol Prog Ser. 2004;273:89–96. [Google Scholar]
- 13.Kellogg CA. Tropical Archaea: Diversity associated with the surface microlayer of corals. Mar Ecol-Prog Ser. 2004;273:81–88. [Google Scholar]
- 14.Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science. 2004;305:997–1000. doi: 10.1126/science.1099128. [DOI] [PubMed] [Google Scholar]
- 15.Kushmaro A, Rosenberg E, Fine M, Loya Y. Bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar Ecol-Prog Ser. 147:159–165. 1997. [Google Scholar]
- 16.Cervino JM, Hayes R, Goreau TJ, Smith GW. Zooxanthellae regulation in yellow blotch/band and other coral diseases contrasted with temperature related bleaching: In situ destruction vs expulsion. Symbiosis. 2004;37:63–85. [Google Scholar]
- 17.Wetz JJ, et al. Presence, infectivity, and stability of enteric viruses in seawater: Relationship to marine water quality in the Florida Keys. Mar Pollut Bull. 2004;48:698–704. doi: 10.1016/j.marpolbul.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Griffin DW, et al. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida Keys. Appl Environ Microb. 1999;65:4118–4125. doi: 10.1128/aem.65.9.4118-4125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seymour JR, Patten N, Bourne DG, Mitchell JG. Spatial dynamics of virus-like particles and heterotrophic bacteria within a shallow coral reef system. Mar Ecol Prog Ser. 2005;288:1–8. [Google Scholar]
- 20.Hewson I, Vargo GA, Fuhrman JA. Bacterial diversity in shallow oligotrophic marine benthos and overlying waters: Effects of virus infection, containment, and nutrient enrichment. Microbial Ecol. 2003;46:322–336. doi: 10.1007/s00248-002-1067-3. [DOI] [PubMed] [Google Scholar]
- 21.Patten NL, Seymour JR, Mitchell AG. Flow cytometric analysis of virus-like particles and heterotrophic bacteria within coral-associated reef water. J Mar Biol Assoc Uk. 2006;86:563–566. [Google Scholar]
- 22.Wilson WH, Chapman DM. Observation of virus-like particles in thin sections of the plumose anemone, Metridium senile. J Mar Biol Assoc Uk. 2001;81:879–880. [Google Scholar]
- 23.Meints RH, Vanetten JL, Kuczmarski D, Lee K, Ang B. Viral-infection of the symbiotic chlorella-like alga present in Hydra-viridis. Virology. 1981;113:698–703. doi: 10.1016/0042-6822(81)90198-7. [DOI] [PubMed] [Google Scholar]
- 24.Van Etten JL, Meints RH. Giant viruses infecting algae. Annu Rev Microbiol. 1999;53:447–494. doi: 10.1146/annurev.micro.53.1.447. [DOI] [PubMed] [Google Scholar]
- 25.Davy SK, et al. Viruses: Agents of coral disease? Dis Aquat Organ. 2006;69:101–110. doi: 10.3354/dao069101. [DOI] [PubMed] [Google Scholar]
- 26.Lohr J, Munn CB, Wilson WH. Characterization of a latent virus-like infection of symbiotic zooxanthellae. Appl Environ Microb. 2007;73:2976–2981. doi: 10.1128/AEM.02449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frias-Lopez J, Zerkle AL, Bonheyo GT, Fouke BW. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl Environ Microb. 2002;68:2214–2228. doi: 10.1128/AEM.68.5.2214-2228.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davy JE, Patten NL. Morphological diversity of virus-like particles within the surface microlayer of scleractinian corals. Aquatic Microb Ecol. 2007;47:37–44. [Google Scholar]
- 29.Patten NL, Harrison PL, Mitchell JG. Prevalence of virus-like particles within a staghorn scleractinian coral (Acropora muricata) from the Great Barrier Reef. Coral Reefs. 2008 doi: 10.1007/s00338-008-0356-9. [DOI] [Google Scholar]
- 30.Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol. 2007;9:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- 31.Marhaver KL, Edwards RA, Rohwer F. Viral communities associated with healthy and bleaching corals. Environ Microbiol. 2008;10:2277–2286. doi: 10.1111/j.1462-2920.2008.01652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruno JF, et al. Thermal stress and coral cover as drivers of coral disease outbreaks. PLoS Biol. 2007;5:1220–1227. doi: 10.1371/journal.pbio.0050124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuntz NM, Kline DI, Sandin SA, Rohwer F. Pathologies and mortality rates caused by organic carbon and nutrient stressors in three Caribbean coral species. Mar Ecol-Prog Ser. 2005;294:173–180. [Google Scholar]
- 34.Kleypas JA, et al. Geochemical consequences of increased atmospheric carbon dioxide on coral reefs. Science. 1999;284:118–120. doi: 10.1126/science.284.5411.118. [DOI] [PubMed] [Google Scholar]
- 35.Shackelton LA, Holmes EC. The evolution of large DNA viruses: Combining genomic information of viruses and their hosts. Trends Microbiol. 2004;12:458–465. doi: 10.1016/j.tim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Angly FE, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:2121–2131. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Classification and Nomenclature of Viruses. Vol VIII. Amsterdam: Elservier; 2005. [Google Scholar]
- 38.Lawrence JG, Hatfull GF, Hendrix RW. Imbroglios of viral taxonomy: Genetic exchange and failings of phenetic approaches. J Bacteriol. 2002;184:4891–4905. doi: 10.1128/JB.184.17.4891-4905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gronenborn B. Nanoviruses: Genome organization and protein function. Vet Microbiol. 2004;98:103–109. doi: 10.1016/j.vetmic.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 40.Herniou EA, Olszewski JA, O'Reilly DR, Cory JS. Ancient coevolution of baculoviruses and their insect hosts. J Virol. 2004;78:3244–3251. doi: 10.1128/JVI.78.7.3244-3251.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoltz DB, Krell P, Summers MD, Vinson SB. Polydnaviridae—a proposed Family of insect viruses with segmented, double-stranded, circular DNA genomes. Intervirology. 1984;21:1–4. doi: 10.1159/000149497. [DOI] [PubMed] [Google Scholar]
- 42.Webb BA, et al. Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology. 2006;347:160–174. doi: 10.1016/j.virol.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 43.Wommack KE, Bhavsar J, Ravel J. Metagenomics: Read length matters. Appl Environ Microb. 2008;74:1453–1463. doi: 10.1128/AEM.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dolan A, Jamieson FE, Cunningham C, Barnett BC, McGeoch DJ. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mcgeoch DJ, et al. The complete DNA-sequence of the long unique region in the genome of Herpes-Simplex Virus Type-1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 46.Tyler SD, Severini A. The complete genome sequence of herpesvirus papio 2 (Cercopithecine herpesvirus 16) shows evidence of recombination events among various progenitor herpesviruses. J Virol. 2006;80:1214–1221. doi: 10.1128/JVI.80.3.1214-1221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perelygina L, et al. Complete sequence and comparative analysis of the genome of herpes B virus (Cercopithecine herpesvirus 1) from a rhesus monkey. J Virol. 2003;77:6167–6177. doi: 10.1128/JVI.77.11.6167-6177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bahr U, Darai G. Analysis and characterization of the complete genome of tupaia (tree shrew) herpesvirus. J Virol. 2001;75:4854–4870. doi: 10.1128/JVI.75.10.4854-4870.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song WJ, et al. Functional genomics analysis of Singapore grouper iridovirus: Complete sequence determination and proteomic analysis. J Virol. 2004;78:12576–12590. doi: 10.1128/JVI.78.22.12576-12590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Legoff J, et al. Real-time PCR quantification of genital shedding of herpes simplex virus (HSV) and human immunodeficiency virus (HIV) in women coinfected with HSV and HIV. J of Clin Microbiol. 2006;44:423–432. doi: 10.1128/JCM.44.2.423-432.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 52.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 53.Smith LD, Hughes TP. An experimental assessment of survival, reattachment, and fecundity of coral fragments. J Exp Mar Biol Ecol. 1999;235:147–164. [Google Scholar]
- 54.Spits C, et al. Optimization and evaluation of single-cell whole, genome multiple displacement amplification. Hum Mutat. 2006;27:496–503. doi: 10.1002/humu.20324. [DOI] [PubMed] [Google Scholar]
- 55.Pinard R, et al. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knowlton N, Rohwer F. Multispecies microbial mutualisms on coral reefs: The host as a habitat. Am Nat. 2003;162:S51–S62. doi: 10.1086/378684. [DOI] [PubMed] [Google Scholar]
- 58.Hoegh-Guldberg O. Climate change, coral bleaching and the future of the world's coral reefs. Mar Freshwater Res. 1999;50:839–866. [Google Scholar]
- 59.Fitt WK, Brown BE, Warner ME, Dunne RP. Coral bleaching: Interpretation of thermal tolerance limits and thermal thresholds in tropical corals. Coral Reefs. 2001;20:51–65. [Google Scholar]
- 60.Jones RJ, Bowyer J, Hoegh-Guldberg O, Blackall LL. Dynamics of a temperature-related coral disease outbreak. Mar Ecol-Prog Ser. 2004;281:63–77. [Google Scholar]
- 61.Ben-Haim Y, Zicherman-Keren M, Rosenberg E. Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl Environ Microb. 2003;69:4236–4242. doi: 10.1128/AEM.69.7.4236-4242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alker AP, Smith GW, Kim K. Characterization of Aspergillus sydowii, a fungal pathogen of Caribbean sea fan corals. Hydrobiologia. 2001;460:105–111. [Google Scholar]
- 63.McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117:90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 64.Davison AJ, et al. A novel class of herpesvirus with bivalve hosts. J Gen Virol. 2005;86:41–53. doi: 10.1099/vir.0.80382-0. [DOI] [PubMed] [Google Scholar]
- 65.Alba MM, Das RJ, Orengo CA, Kellam P. Genomewide function conservation and phylogeny in the herpesviridae. Genome Res. 2001;11:43–54. doi: 10.1101/gr.149801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davison AJ. Evolution of the herpesviruses. Vet Microbiol. 86:69–88. doi: 10.1016/s0378-1135(01)00492-8. 2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.