Abstract
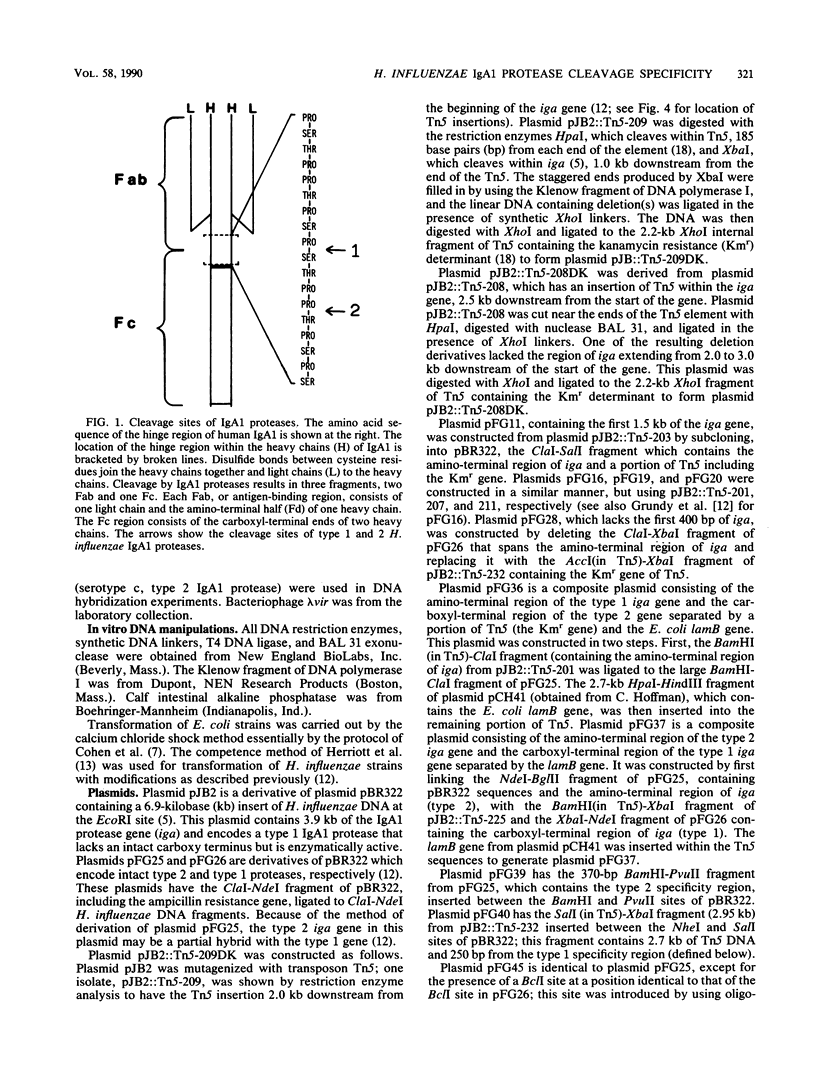
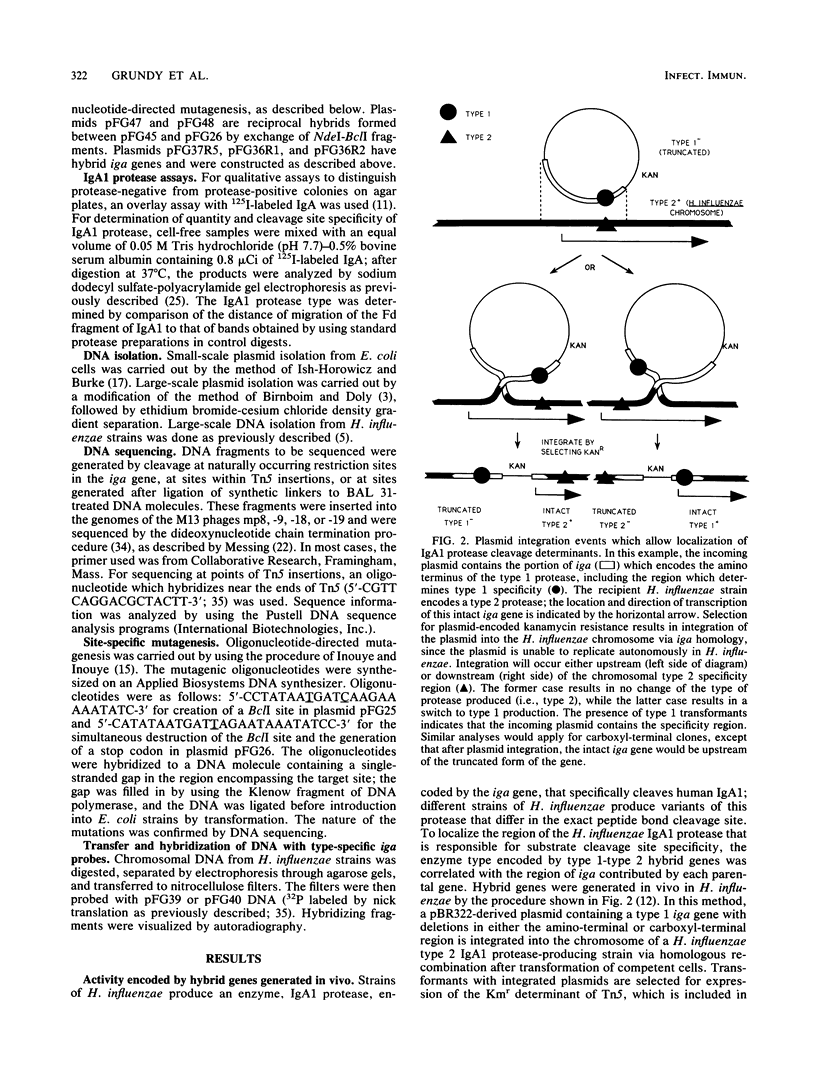
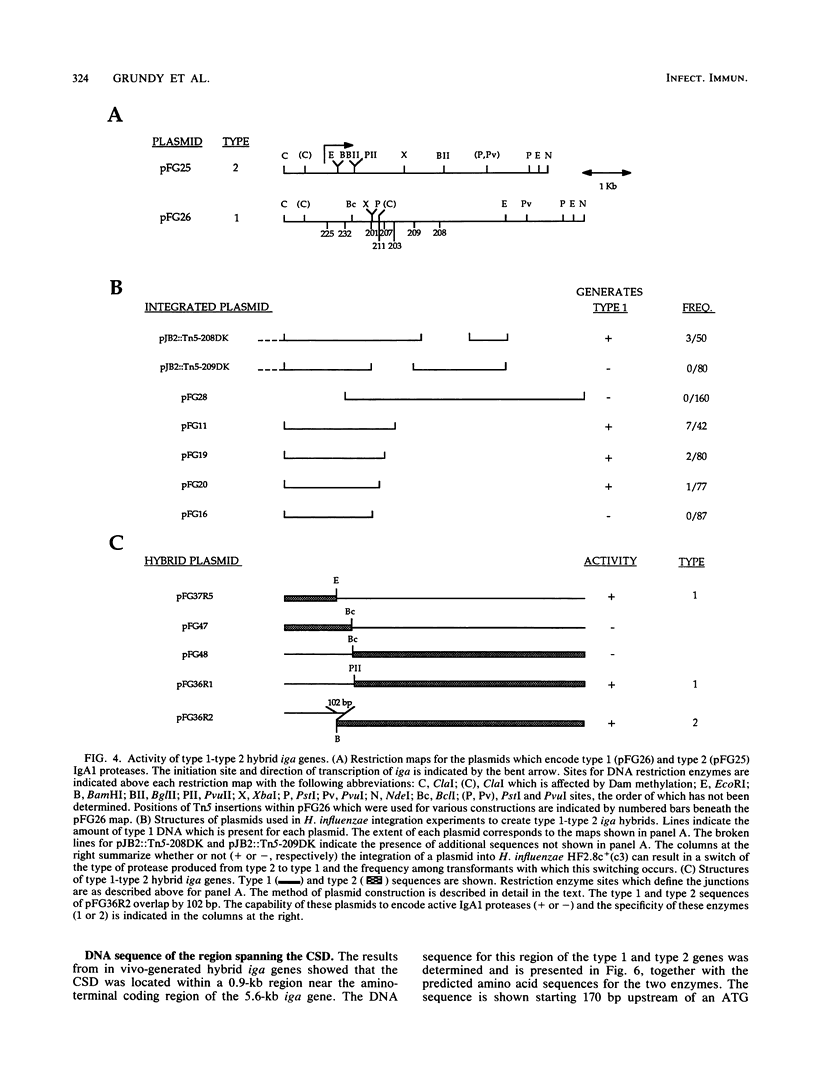
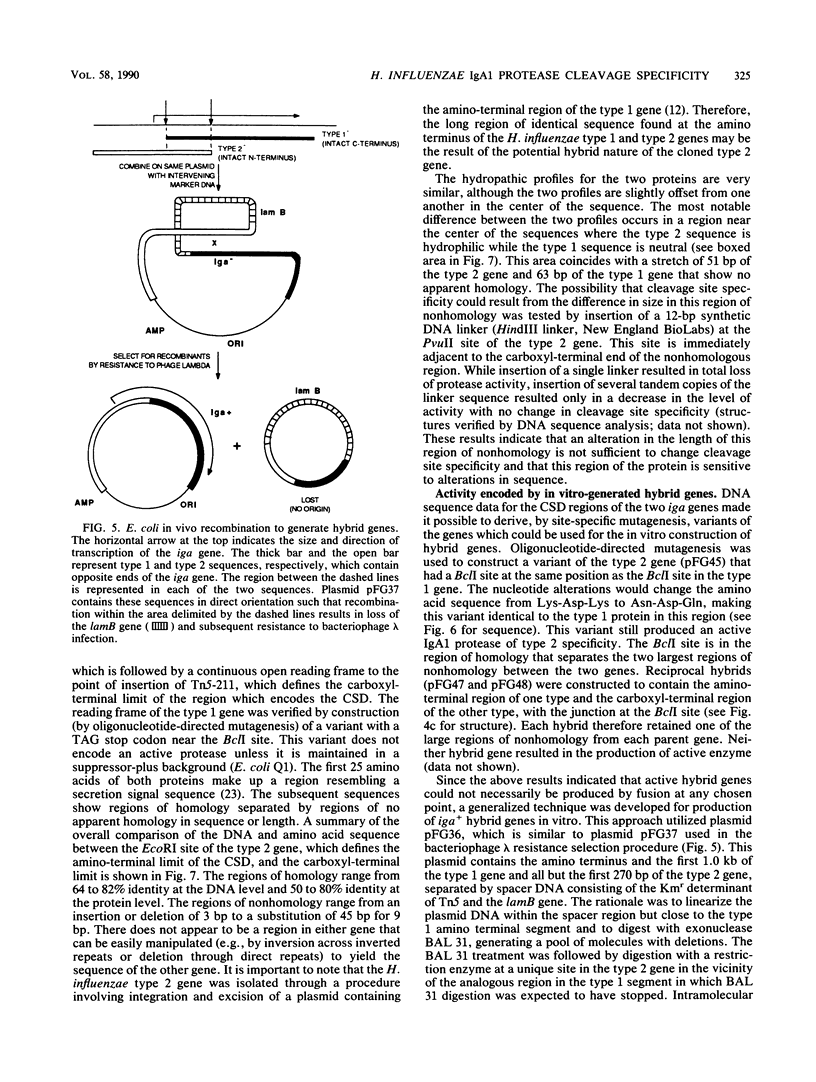
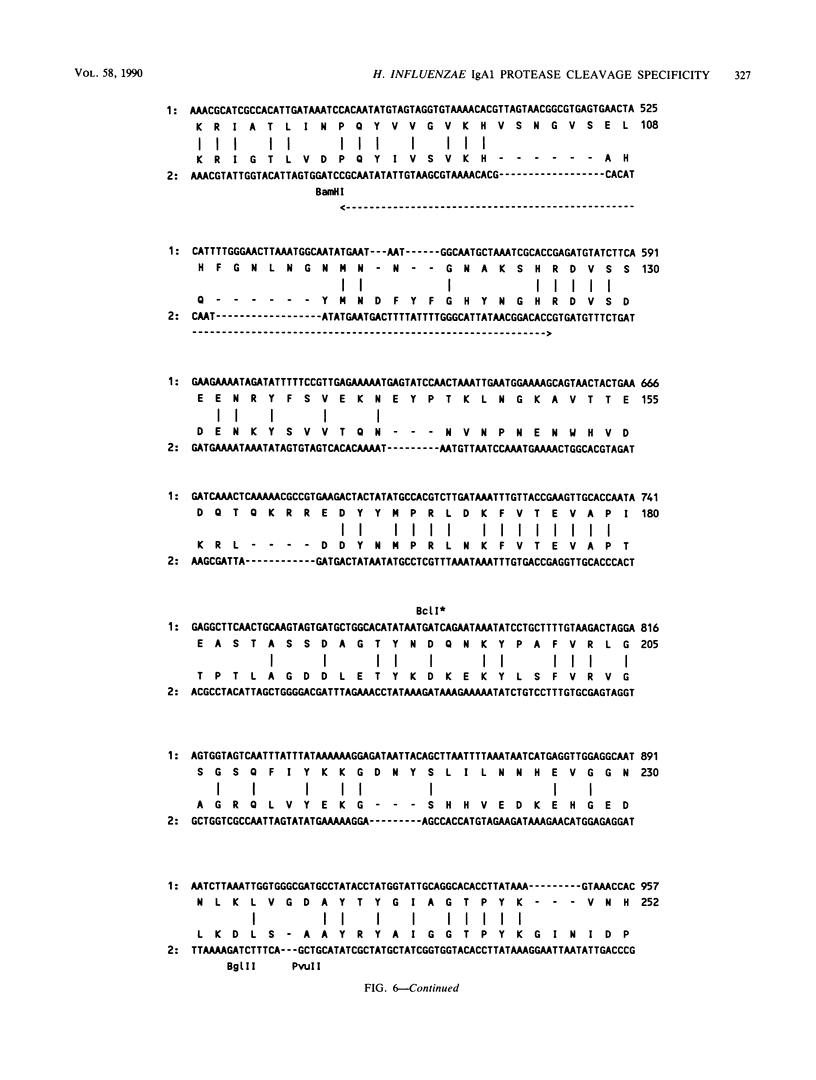
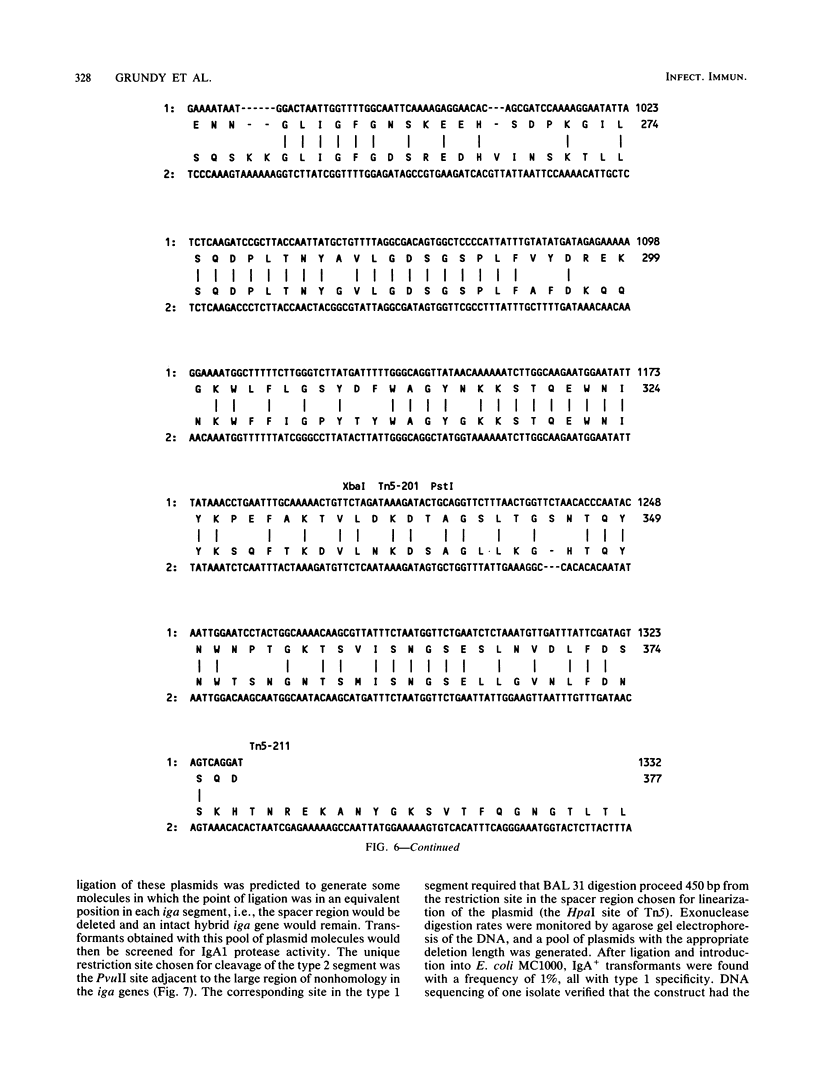
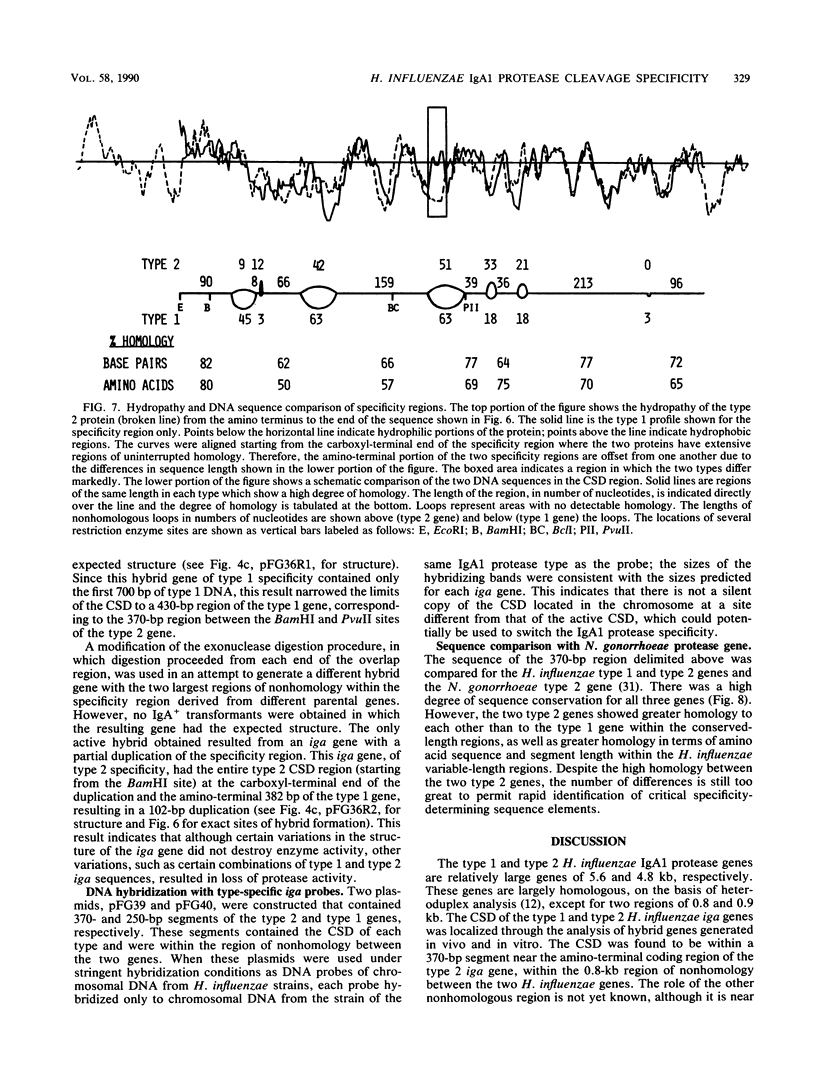
Immunoglobulin A1 (IgA1) proteases are produced by a number of different species of bacteria which cause infection at human mucosal surfaces. The sole substrate of these proteases is human IgA1. Cleavage is within the hinge region of IgA1, although there is variability in the exact peptide bond within the hinge region that is cut by a particular protease. The cleavage site of the Haemophilus influenzae type 1 protease is located four amino acids from the cleavage site of the type 2 enzyme. In this study, the region of the H. influenzae IgA1 protease gene (iga) that determines the cleavage site specificity was localized through the comparison of the type 1 and type 2 genes and the construction and analysis of type 1-type 2 hybrid genes. The hybrid genes were generated by in vivo and in vitro techniques which facilitated the selection and screening of randomly generated hybrids. The cleavage site determinant was found to be within a 370-base-pair region near the amino-terminal coding region, in one of two large areas of nonhomology between the two types of H. influenzae iga genes. DNA sequence analysis of the cleavage site determinant and surrounding regions did not reveal a simple mechanism whereby one enzyme type could be converted to the other type. Comparison of the type 2 gonococcal IgA1 protease gene to the two Haemophilus genes revealed a significant amount of homology around the cleavage site determinant, with the two type 2 genes showing greater homology.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ALEXANDER H. E., LEIDY G. Induction of streptomycin resistance in sensitive Hemophilus influenzae by extracts containing desoxyribonucleic acid from resistant Hemophilus influenzae. J Exp Med. 1953 Jan;97(1):17–31. doi: 10.1084/jem.97.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker J., Mulks M. H., Plaut A. G., Moxon E. R., Wright A. IgA1 proteases of Haemophilus influenzae: cloning and characterization in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1983 May;80(9):2681–2685. doi: 10.1073/pnas.80.9.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker J., Mulks M., Moxon E. R., Plaut A. G., Wright A. Physical and genetic analysis of DNA regions encoding the immunoglobulin A proteases of different specificities produced by Haemophilus influenzae. Infect Immun. 1985 Feb;47(2):370–374. doi: 10.1128/iai.47.2.370-374.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik C. S., Largman C., Fletcher T., Roczniak S., Barr P. J., Fletterick R., Rutter W. J. Redesigning trypsin: alteration of substrate specificity. Science. 1985 Apr 19;228(4697):291–297. doi: 10.1126/science.3838593. [DOI] [PubMed] [Google Scholar]
- Danner D. B., Pifer M. L. Plasmid cloning vectors resistant to ampicillin and tetracycline which can replicate in both E. coli and Haemophilus cells. Gene. 1982 Apr;18(1):101–105. doi: 10.1016/0378-1119(82)90062-2. [DOI] [PubMed] [Google Scholar]
- Frangione B., Wolfenstein-Todel C. Partial duplication in the "hinge" region of IgA 1 myeloma proteins. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3673–3676. doi: 10.1073/pnas.69.12.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. V., Plaut A. G. Detection of IgA protease activity among multiple bacterial colonies. J Immunol Methods. 1983 Feb 25;57(1-3):247–251. doi: 10.1016/0022-1759(83)90084-4. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Plaut A., Wright A. Haemophilus influenzae immunoglobulin A1 protease genes: cloning by plasmid integration-excision, comparative analyses, and localization of secretion determinants. J Bacteriol. 1987 Oct;169(10):4442–4450. doi: 10.1128/jb.169.10.4442-4450.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriott R. M., Meyer E. M., Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970 Feb;101(2):517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M. M. Prophage deletion mapping of bacteriophage Mu-1. Virology. 1973 Jul;54(1):93–101. doi: 10.1016/0042-6822(73)90118-9. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Kett K., Brandtzaeg P., Radl J., Haaijman J. J. Different subclass distribution of IgA-producing cells in human lymphoid organs and various secretory tissues. J Immunol. 1986 May 15;136(10):3631–3635. [PubMed] [Google Scholar]
- Kilian M., Mestecky J., Schrohenloher R. E. Pathogenic species of the genus Haemophilus and Streptococcus pneumoniae produce immunoglobulin A1 protease. Infect Immun. 1979 Oct;26(1):143–149. doi: 10.1128/iai.26.1.143-149.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Male C. J. Immunoglobulin A1 protease production by Haemophilus influenzae and Streptococcus pneumoniae. Infect Immun. 1979 Oct;26(1):254–261. doi: 10.1128/iai.26.1.254-261.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Michaelis S., Beckwith J. Mechanism of incorporation of cell envelope proteins in Escherichia coli. Annu Rev Microbiol. 1982;36:435–465. doi: 10.1146/annurev.mi.36.100182.002251. [DOI] [PubMed] [Google Scholar]
- Mulks M. H., Kornfeld S. J., Frangione B., Plaut A. G. Relationship between the specificity of IgA proteases and serotypes in Haemophilus influenzae. J Infect Dis. 1982 Aug;146(2):266–274. doi: 10.1093/infdis/146.2.266. [DOI] [PubMed] [Google Scholar]
- Mulks M. H., Kornfeld S. J., Plaut A. G. Specific proteolysis of human IgA by Streptococcus pneumoniae and Haemophilus influenzae. J Infect Dis. 1980 Apr;141(4):450–456. doi: 10.1093/infdis/141.4.450. [DOI] [PubMed] [Google Scholar]
- Mulks M. H., Plaut A. G. IgA protease production as a characteristic distinguishing pathogenic from harmless neisseriaceae. N Engl J Med. 1978 Nov 2;299(18):973–976. doi: 10.1056/NEJM197811022991802. [DOI] [PubMed] [Google Scholar]
- Plaut A. G., Gilbert J. V., Artenstein M. S., Capra J. D. Neisseria gonorrhoeae and neisseria meningitidis: extracellular enzyme cleaves human immunoglobulin A. Science. 1975 Dec 12;190(4219):1103–1105. doi: 10.1126/science.810892. [DOI] [PubMed] [Google Scholar]
- Plaut A. G. The IgA1 proteases of pathogenic bacteria. Annu Rev Microbiol. 1983;37:603–622. doi: 10.1146/annurev.mi.37.100183.003131. [DOI] [PubMed] [Google Scholar]
- Plaut A. G., Wistar R., Jr, Capra J. D. Differential susceptibility of human IgA immunoglobulins to streptococcal IgA protease. J Clin Invest. 1974 Dec;54(6):1295–1300. doi: 10.1172/JCI107875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlner J., Halter R., Beyreuther K., Meyer T. F. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. 1987 Jan 29-Feb 4Nature. 325(6103):458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- Randall-Hazelbauer L., Schwartz M. Isolation of the bacteriophage lambda receptor from Escherichia coli. J Bacteriol. 1973 Dec;116(3):1436–1446. doi: 10.1128/jb.116.3.1436-1446.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield P. R., Watson J. M. DNA sequence of Rhizobium trifolii nodulation genes reveals a reiterated and potentially regulatory sequence preceding nodABC and nodFE. Nucleic Acids Res. 1986 Apr 11;14(7):2891–2903. doi: 10.1093/nar/14.7.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi T. B., Grey H. M. Structure and function of immunoglobulin A. Prog Allergy. 1972;16:81–213. [PubMed] [Google Scholar]




