Abstract
IL-6 is an important cytokine that regulates both immune and metabolic functions. Within adipose tissue, preadipocytes produce significant amounts of IL-6, but little is known about the factors or mechanisms that regulate IL-6 production in these cells. Using LS14, a newly developed human adipocyte cell line, our objective was to determine the mechanisms by which insulin stimulates IL-6 production and release in preadipocytes. Insulin increased IL-6 gene expression and secretion in a time- and dose-dependent manner. Insulin decreased cyclic AMP (cAMP) but increased cyclic GMP (cGMP) levels, and IL-6 expression/release was stimulated by a cGMP analog. The stimulatory effect of insulin and cGMP was abrogated by a specific inhibitor of protein kinase G (cyclic GMP-dependent protein kinase). Both insulin and cGMP rapidly induced phosphorylation of cAMP response element binding protein. Insulin also activated the MAPK signaling pathway, and its blockade prevented the insulin-stimulated increases in IL-6 cell content and release, but not IL-6 gene expression. Although inhibition of the proteosome increased IL-6 cell content and release, proteosome activity was unaffected by insulin. These data suggest that the stimulatory effects of insulin on IL-6 release involve several interrelated components: transcription, intracellular releasable pool, and secretion, which are differentially regulated and, thus, determine the size of the releasable pool of IL-6. Insulin-induced IL-6 gene expression is mediated by cGMP/cyclic GMP-dependent protein kinase/cAMP response element binding protein, whereas MAPK is involved in the insulin-stimulated IL-6 synthesis/release.
IL-6 IS A PLEIOTROPIC cytokine that is produced by most cells of the immune system, and is best known for its inflammatory and immune functions, including stimulation of acute phase inflammatory proteins and B cell differentiation (1). In addition, IL-6 is produced by preadipocytes, adipocytes, and macrophages residing within adipose tissue, where it stimulates lipolysis, inhibits lipoprotein lipase activity, and antagonizes insulin-stimulated glucose uptake (2,3,4,5). Among its metabolic functions, IL-6 suppresses the release of adiponectin, an insulin-sensitizing adipokine whose circulating levels are reduced in insulin-resistant and obese patients (6,7). The importance of IL-6 as a metabolic hormone is also supported by the report that IL-6-deficient mice are obese, with impaired glucose tolerance, elevated leptin levels, and leptin resistance (8).
Elevated serum IL-6 levels are associated with increased cardiovascular risk in obese and diabetic patients, and contribute to the low-grade inflammation that accompanies the metabolic syndrome (9,10,11,12). Given the involvement of IL-6 in both immune and metabolic homeostasis, understanding the regulation of its release is of great importance. Insulin, a key regulator of glucose and lipid metabolism in adipose tissue, increases IL-6 release from human adipocytes and 3T3-L1 cells (13,14), but little is known about the underlying mechanism of action.
Our laboratory recently developed a human adipocyte cell line, named LS14, which exhibits many properties of visceral preadipocytes and can be induced to differentiate into functional mature adipocytes (15). The production of large amounts of IL-6 by nondifferentiated LS14 cells presented us with a unique opportunity to study its regulation in a homogeneous population of human cells, as opposed to adipose-derived primary cultures that contain multiple cell types and vary among patients. The goals of this study were to: 1) characterize the time- and dose-dependent effects of insulin on IL-6 gene expression, cell content, and release from LS14 cells; and 2) identify the signaling pathways that mediate these effects.
Materials and Methods
Cell culture and treatment
LS14 cultures were maintained as previously described (15). Briefly, cells were cultured in DMEM-F12 containing 5% fetal bovine serum (Cell Grow, Manassas, VA), 5% FetalClone III (HyClone, Logan, UT), 15 μg/ml bovine pituitary extract (Invitrogen Corp., Carlsbad, CA), 1% ITS+ (insulin, transferrin, selenic acid, and BSA; BD Biosciences, San Jose, CA), 0.5 ng/ml basic fibroblast growth factor (PeproTech, Inc., Rocky Hill, NJ), 1 ng/ml epidermal growth factor (PeproTech), 0.1 ng/ml TGFβ1 (PeproTech), and 50 μg/ml Normocin (Invitrogen). For experimentation, cells were plated at 15,000 cells per cm2 in the aforementioned media on collagen-coated plates. After 8 h, cells were rinsed and maintained overnight in 2% charcoal-stripped serum, 4 mm l-glutamine, 110 mg/ml sodium pyruvate, 750 mg/ml sodium bicarbonate, and 15 mm HEPES (USB Corp., Cleveland, Ohio). Cells were then incubated with vehicle, endotoxin-free recombinant human insulin (Sigma-Aldrich Corp., St. Louis, MO), TNFα (BIOMOL International, L.P., Plymouth Meeting, PA), cyclic GMP (cGMP) (BIOMOL International), forskolin (BIOMOL International), atrial natriuretic peptide (Sigma-Aldrich), a guanylyl cyclase activator, or sodium nitroprusside (NaN) (Sigma-Aldrich), a nitric oxide donor, at equal volumes. For inhibitor studies, cells were pretreated for 30 min with 10 μm U0126 (LC Laboratories, Woburn, MA), a MAPK kinase (MEK)-1 inhibitor, 200 nm wortmannin (LC Laboratories), a phosphatidylinositol 3-kinase (PI3K) inhibitor, 5 μm H89 (BIOMOL International), a cyclic AMP-dependent protein kinase (PKA) inhibitor, 500 nm KT5823 (BIOMOL International), a cyclic GMP-dependent protein kinase (PKG) inhibitor, or 10 μm MG132, (BIOMOL International), a proteasome inhibitor. After treatment, conditioned media (CM) were collected, and the cells were rinsed with cold PBS before being lysed in a buffer (10 mm Tris-HCl, 5 mm EDTA, and 50 mm NaCl).
IL-6 ELISA
IL-6 concentrations in CM or lysates were immediately determined by a fluorescent sandwich ELISA. The assay was optimized in our laboratory using monoclonal antibody pairs against human IL-6 (AHC0562 capture and AHC0469 biotinylated detection; BioSource Europe S.A., Nivelles, Belgium) and verified against commercial plates from the same vendor. Black 96-well plates were coated with the capture antibody and blocked with 0.5% BSA. The plates were coincubated with the detection antibody and recombinant human IL-6 (PeproTech) in triplicate (10–1000 pg/ml) or with CM and lysates in duplicate. After 2 h, bound antibodies were detected using avidin/horseradish peroxidase and a fluorometric horseradish peroxidase substrate (QuantaBlue; Pierce, Rockford, IL). Fluorescence was measured at 325 nm excitation and 420 nm emission using a Gemini fluorescence microplate reader (Molecular Devices, Sunnyvale, CA). The detection limit was 20 pg/ml.
Real-time PCR
Total RNA was isolated using TRI Reagent (Molecular Research Center, Inc., Cincinnati, OH), and cDNA was synthesized as previously described (15). PCR was performed on 200 ng cDNA using intron-spanning primers for human IL-6: forward 5′-ACA AAC AAA TTC GGT AGA TCC TCG-3′, reverse 5′-AGC CAT CTT TGG AAG GTT CAG G-3′. β2-Microglobulin was used as a reference gene: forward 5′-GGC ATT CCT GAA GCT GAC-3′, reverse 5′-GAA TCT TTG GAG TAC GCT GG-3′. Quantitative real-time PCR was performed using Immolase heat-activated Taq DNA polymerase (Bioline USA Inc., Taunton, MA). SYBR Green I (Invitrogen) was used for fluorometric product detection using a SmartCycler I (Cepheid Co., Sunnyvale, CA). Cycle parameters were 96 C for 6 min for polymerase activation, followed by 40 cycles of 95 C for 15 sec, 57 C for 15 sec, and 72 C for 25 sec, and an optical read stage at 83.5 C for 6 sec. Product purity was confirmed by DNA melting curve analysis. After correction for β2-microglobulin, fold changes in gene expression were calculated from the cycle threshold measurements as described (16).
Western blotting
For detection of activated Akt and MAPK, cells were homogenized in lysis buffer containing 50 mm sodium fluoride, 30 mm sodium pyrophosphate, 1% Triton X-100, and protease inhibitors, 200 μm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin, 2 μg/ml leupeptin, and 5 μg/ml aprotinin. For detection of activated cAMP response element binding protein (CREB), cells were homogenized in lysis buffer containing 1% Nonidet P-40, 1% sodium dodecyl sulfate, 5% glycerol, and the protease inhibitors as described previously. Protein concentration was determined by the Pierce bicinchoninic acid assay. Lysates (25 μg for Akt and MAPK, 35 μg for CREB) were electrophoresed on 12% sodium dodecyl sulfate gels. After transfer to a nitrocellulose membrane, protein bands were detected with the following antibodies: total p44/42 Erk1/2, phospho-p44/42 Erk1/2, total Akt, phospho-Akt (Ser473), total CREB, phospho-CREB (Ser133) each at 1:1000 (Cell Signaling Technology, Inc., Danvers, MA). After incubation with horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences Inc., Piscataway, NJ), products were developed on film using SuperSignal chemiluminescence reagents (Pierce) and photographed.
cAMP assay
cAMP was measured using a colorometric competitive ELISA kit from R&D Systems, Inc. (KGE002; Minneapolis, MN). Cells (5 × 105 per 60-mm dish) were treated in serum-free media with vehicle, 125 nm insulin, or 10 μm forskolin for 15 and 30 min. Cell lysates were assayed in duplicate. The detection limit was less than 1 pmol/ml.
cGMP assay
cGMP levels were determined using a colorometric competitive ELISA kit from Cayman Chemical (581021; Ann Arbor, MI). Cells (1 × 106 per 60-mm dish) were treated in serum-free media with vehicle, 125 nm insulin, or 2.5 mm NaN for 15 and 30 min. After lysis in 0.1 m HCl, both samples and standards were acetylated to increase assay sensitivity, and analyzed in duplicate. The detection limit was 0.023 pmol/ml.
Determination of proteosome activity
The chymotrypsin-like activity of the proteosome was measured as previously described (17). Cells (3 × 105 per 24-well plates) were treated with vehicle, 125 nm insulin or 10 μm MG132 for 3 h, and then lysed in a buffer containing 50 mm Tris-HCl, 150 mm NaCl, 0.5% Nonidet P-40, 0.5 mm phenylmethylsulfonyl fluoride, and 0.5 mm dithiothreitol. Cell lysates were incubated with 20 μm of the fluorogenic peptide substrate Suc-Leu-Leu-Val-Tyr-aminomethyl coumarin (LLVY-AMC) (Sigma-Aldrich). After 60 min, free AMC was measured fluorometrically at 380 nm excitation and 460 nm emission. To determine proteosome activity in live cells, LS14 cells were treated with vehicle, insulin, or MG132 for 3 h, followed by a 2-h incubation with 250 μl of 20 μm LLVY-AMC. Free AMC in CM was determined as described previously.
Data analysis
Values are expressed as the mean ± sem. All experiments were repeated two to four times. P values less than 0.05 were considered significant. Statistical differences were determined by one-way ANOVA, followed by Newman-Keuls post hoc analysis.
Results
Time and dose-dependent stimulation of IL-6 expression and release by insulin
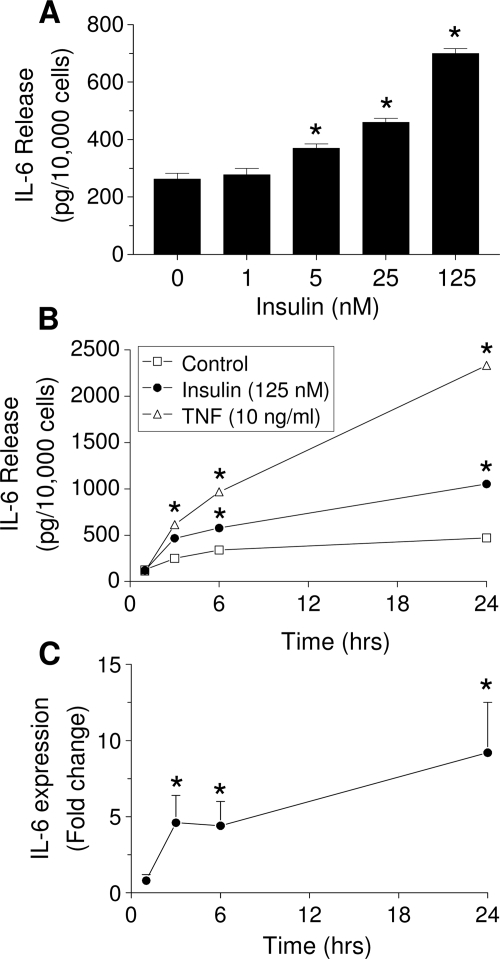
IL-6 concentrations in CM from insulin-treated cells were measured by an ELISA. Figure 1A shows dose-dependent stimulation of IL-6 release after 6 h incubation with insulin. As low as 5 nm insulin significantly (P < 0.05) increased IL-6 release by 40%, whereas 125 nm insulin elevated secretion 240%. Insulin at 125 nm increased IL-6 release over control at 3, 6, and 24 h by 85, 230, and 420%, respectively (Fig. 1B). A total of 125 nm IGF-I had no effect on IL-6 production at any time point (data not shown). TNFα, a potent stimulator of IL-6 release, induced a stronger stimulation than insulin at all time points. A time-dependent induction of IL-6 gene expression by 125 nm insulin is shown in Fig. 1C. Within 3 h, insulin increased IL-6 mRNA levels 4.5-fold. This remained approximately the same after 6 h, increasing to 10-fold over controls after 24 h. A 3-h treatment with insulin was used for the remainder of the studies.
Figure 1.
Time- and dose-dependent effect of insulin on IL-6 release and gene expression in undifferentiated LS14 human adipocytes. A, Cells were treated with insulin for 6 h. B, Cells were incubated with 125 nm insulin (black circles) or 10 ng/ml TNFα (white triangles) for different times. IL-6 in CM was measured by ELISA. C, Cells were incubated with 125 nm insulin, and IL-6 mRNA levels were determined by real-time PCR. Values in A and B are the mean ± sem of four determinations, and are representative of three separate experiments. IL-6 gene expression was calculated as the fold change over controls and represents three separate experiments. *, Significant differences (P < 0.05) from control.
H-89, an inhibitor of both PKA and PKG, blocked insulin-stimulated IL-6 release
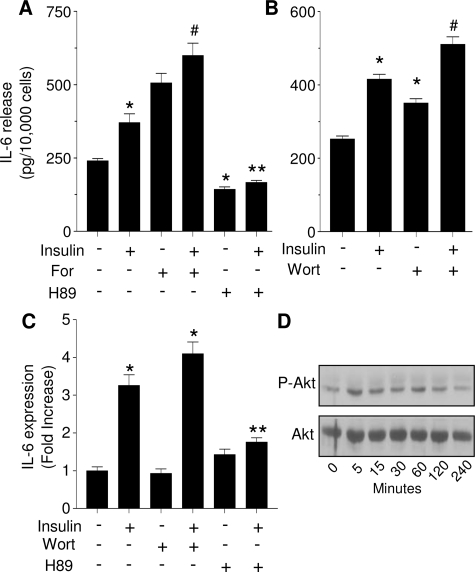
To determine which signaling pathway may mediate the stimulatory effects of insulin, LS14 cells were pretreated for 30 min with 5 μm H89, an inhibitor of PKA/PKG, followed by a 3-h treatment with 125 nm insulin. Forskolin, a potent activator of adenylyl cyclase, served as a control for PKA activation. Figure 2A clearly demonstrates that 10 μm forskolin alone stimulated IL-6 release and showed an additive effect upon incubation with insulin. H89 alone reduced IL-6 secretion below control, and completely blocked the insulin-induced release. As expected, H89 also blocked the stimulatory effect of forskolin on IL-6 secretion (data not shown). Unlike H89, blockade of PI3K with 200 nm wortmannin increased IL-6 release similar to that of insulin (Fig. 2B), with coincubation of wortmannin and insulin resulting in an additive effect. Similar results were obtained with LY294002, another inhibitor of PI3K/Akt (data not shown). The same responsiveness to the various treatments was observed on IL-6 cell content (data not shown). H89 or wortmannin alone had no effect on IL-6 gene expression, whereas insulin-stimulated IL-6 gene expression was only inhibited by H89 (Fig. 2C). Figure 2D shows only weak activation of Akt by insulin, further suggesting that this pathway does not mediate the stimulatory effects of insulin on IL-6 production and release in LS14 cells.
Figure 2.
Effect of PKA and Akt activators or inhibitors on insulin-stimulated IL-6 gene expression and release. A and B, LS14 cells were treated with 125 nm insulin for 3 h in the presence or absence of 10 μm forskolin (For), 0.2 μm wortmannin (Wort), or 5 μm H89. IL-6 was determined by ELISA. Values are expressed as the mean ± sem of four determinations, and are representative of four separate experiments. *, Significant differences (P < 0.01) from control; #, significant differences (P < 0.01) from control, insulin, forskolin, or wortmannin. C, IL-6 gene expression was determined by real-time PCR. Values are expressed as fold change over controls. *, Significant differences (P < 0.01) from control. D, Cells were treated with 125 nm insulin for the designated times, and Akt phosphorylation was determined by Western blotting; total Akt served as a loading control. A, C ** Significant differences (P < 0.05) from insulin.
Insulin increases cGMP but suppresses cAMP levels
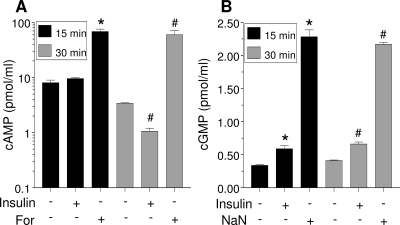
Given the ability of H89 to inhibit both PKA and PKG, we next examined whether insulin affects cAMP or cGMP levels. Short-term incubation with forskolin (15 or 30 min) resulted in a significant (P < 0.05) increase in cAMP levels in LS14 cells (Fig. 3A). On the other hand, insulin did not affect cAMP levels after 15 min and caused a significant (P < 0.05) reduction after 30 min. In contrast, insulin effectively increased cGMP levels at both time points (Fig. 3B). NaN, a potent activator of guanylyl cyclase, induced a 5- to 10-fold increase in cGMP after 15 or 30 min. Note that total cGMP cell content is approximately 20-fold lower than cAMP.
Figure 3.
Insulin decreases intracellular cAMP levels but increases cGMP. Cells were treated with 125 nm insulin for 15 or 30 min, and nucleotides in the lysate were determined by ELISA. Forskolin (For) (10 μm) and NaN (2.5 mm) served as positive controls for cAMP (A) and cGMP (B), respectively. Each value is the mean ± sem of three determinations, and is representative of two separate experiments. *, Significant differences (P < 0.05) from control at 15 min; #, Significant differences (P < 0.05) from control at 30 min.
Inhibition of PKG blocks insulin- and cGMP-stimulated IL-6 expression and release
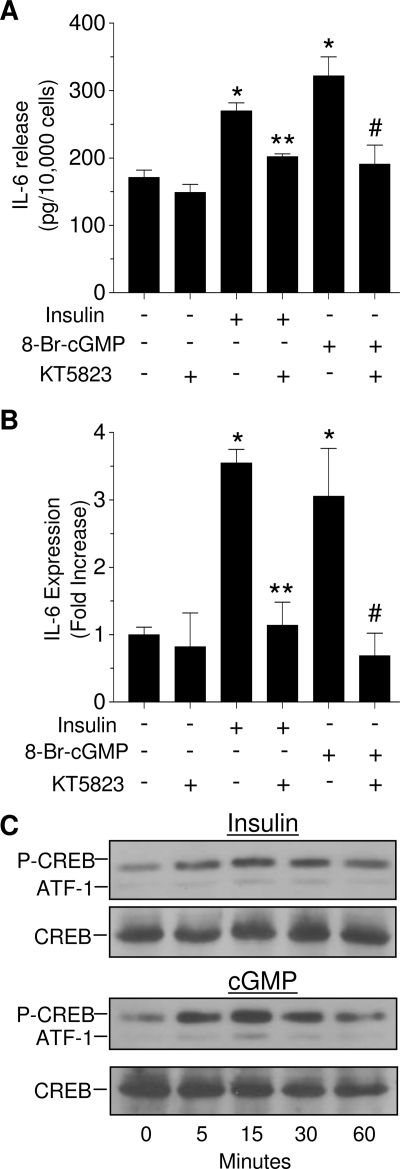
Based on the ability of insulin to increase cGMP levels, we next examined whether cGMP/PKG mediates the effects of insulin on IL-6 expression and release. As shown in Fig. 4, A and B, both insulin and 8-bromo-cGMP, a cGMP analog, increased IL-6 release and gene expression by 50% and 3-fold, respectively, within 3 h. In the presence of 500 nm KT5823, which inhibits PKG (IC50 = 60 nm) but not PKA (IC50 = 10 μm), the stimulatory effects of both insulin and 8-bromo-cGMP on IL-6 release and gene expression were abrogated. Atrial natriuretic peptide, an activator of guanylyl cyclase, as well as NaN, also stimulated IL-6 expression and release (data not shown).
Figure 4.
IL-6 release (A) and gene expression (B) are stimulated by a cGMP and inhibited by a PKG inhibitor. Cells were treated with 125 nm insulin or 10 nm 8-bromo-cGMP (8-Br-cGMP) for 3 h in the presence or absence of 500 nm KT5823. IL-6 release was determined by ELISA, and gene expression was determined by real-time PCR. Each value is the mean ± sem of three determinations, and is representative of two separate experiments. *, Significant differences (P < 0.05) from control; **, significant differences (P < 0.05) from insulin; #, significant differences (P < 0.05) from 8-bromo-cGMP. C, Both insulin (125 nm) and cGMP (10 nm) induced CREB phosphorylation, as determined by Western blotting; total CREB served as a loading control.
Both insulin and cGMP activate CREB
Next, we tested the ability of insulin and cGMP to induce phosphorylation of CREB, which binds to a cAMP response element sequence within the IL-6 promoter. As seen in Fig. 4C, insulin increased CREB phosphorylation within 5 min, and this activation lasted for at least 60 min. A stronger activation of CREB was observed after treatment with 10 nm cGMP. A slight increase in the phosphorylation of activating transcription factor 1 (ATF-1), a CREB-related protein, was seen in both treatments. Total CREB served as a loading control.
MAPK mediates the effects of insulin on IL-6 cell content and release but not gene expression
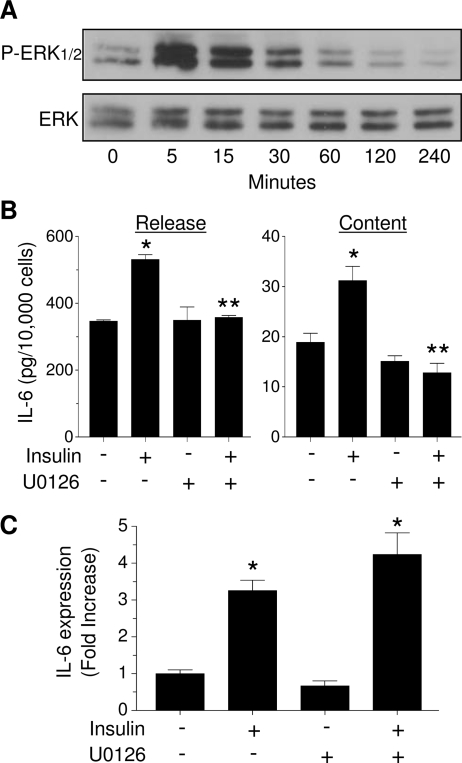
We then explored whether other signaling pathways are also involved in insulin-stimulated IL-6 production. Figure 5A shows that insulin induced a strong activation of MAPK within 5 min, an effect that lasted for 30 min and then returned to basal levels. Next, LS14 cells were pretreated for 30 min with 10 μm U0126, a specific Mek-1 inhibitor, followed by treatment with insulin for 3 h. As depicted in Fig. 5B, insulin increased both IL-6 release and cell content by 50%, an effect that was completely blocked by U0126. Unexpectedly, U0126 failed to block the stimulatory effect of insulin on IL-6 gene expression (Fig. 5C).
Figure 5.
MAPK mediates insulin-stimulated IL-6 cell content/release, but not gene expression. A, LS14 cells were treated with 125 nm insulin, and phosphorylation of ERK1/2 (P-ERK1/2) was determined by Western blotting; total ERK served as a loading control. B and C, Cells were treated with 125 nm insulin for 3 h in the presence or absence of 10 μm U0126. IL-6 cell content and release were determined by ELISA, and IL-6 gene expression by real-time PCR. Values are the mean ± sem of four determinations, and are representative of four separate experiments. Values in C are expressed as fold change over controls. *, Significant differences (P < 0.01) from control; **, Significant differences (P < 0.01) from insulin.
Intracellular IL-6 content is regulated by the proteosome
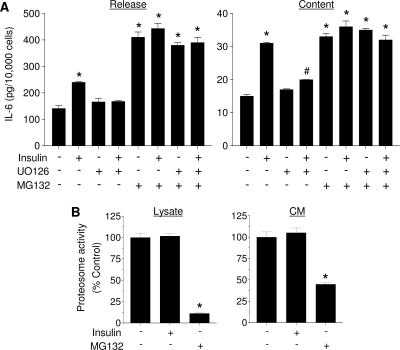
The dissimilar effects of MAPK inhibition on the insulin-stimulated IL-6 gene transcription, cell content, and release suggested that they are differentially regulated. Consequently, we explored whether the releasable IL-6 pool is subjected to additional regulatory processes such as degradation. Cells were pretreated for 30 min with 10 μm MG-132, a proteasome inhibitor, and then incubated with 125 nm insulin. Figure 6A shows a typical increase in IL-6 release and cell content in response to insulin. MG132 alone increased IL-6 release and cell content 3- and 2-fold, respectively, indicating that proteosomal degradation affects IL-6 cell content and, thus, its release. Coincubation of insulin and MG132 did not have an additive effect on either IL-6 cell content or release. In addition, preincubation with U0126 had no effect on IL-6 release/content in the presence or absence of MG132.
Figure 6.
Inhibition of IL-6 degradation increases IL-6 cell content and release. A, LS14 cells were pretreated for 30 min with 10 μm MG132 or U0126 and then treated with 125 nm insulin for 3 h. IL-6 was measured by ELISA. B, Measurement of fluorescent aminomethyl coumarin in the cell lysate or CM. All values are the mean ± sem of four determinations, and are representative of three separate experiments. *, Significant differences (P < 0.01) from control.
To determine whether insulin inhibits IL-6 degradation, proteosome activity in both lysate and intact cells was measured. Cells were treated with insulin or MG132 for 3 h, and then lysates as well as cells were incubated with the fluorogenic proteosome substrate LLVY-AMC. As evident in Fig. 6B, insulin had no effect on proteosome activity, whereas MG132, as expected, significantly inhibited substrate degradation.
Discussion
We are reporting that insulin stimulates IL-6 production and release from nondifferentiated human LS14 adipocytes in a time- and dose-dependent manner. These data suggest that the stimulatory effects of insulin on IL-6 release can be separated into several interrelated components: transcription, intracellular releasable pool, and secretion, which are differentially regulated. Insulin-induced IL-6 expression is mediated by cGMP/PKG/CREB, whereas MAPK is involved in the insulin-stimulated IL-6 synthesis/release. The size/dynamics of the IL-6 releasable pool is determined by a balance between the rates of transcription/translation and proteosomal degradation.
Adipose tissue is comprised of several cell types, with preadipocytes, adipocytes, and macrophages all capable of producing IL-6 (18). Several investigators have reported that IL-6 is primarily released from the stromal vascular fraction, with less than 10% released by mature adipocytes (19,20). Because the stromal vascular fraction includes both preadipocytes and macrophages, it is difficult to determine the fractional release of IL-6 from either cell type. IL-6 production is much higher in preadipocytes than adipocytes because of its down-regulation during adipogenesis (21,22). Yet, a number of studies have used differentiated 3T3-L1 adipocytes to examine IL-6 release (14,23). One study, using differentiated human adipocytes, reported stimulation of IL-6 release by insulin after 48 h treatment, without examining the underlying mechanism (13). Here, we used LS14 cells, whose properties resemble visceral preadipocytes. Because IL-6 expression and release are also down-regulated during the differentiation of LS14 cells (15), only undifferentiated cells were used. A sensitive ELISA enabled us to determine the effects of relatively short exposures to insulin on both IL-6 cell content and release, two endpoints that were not examined in earlier studies using 3T3-L1 adipocytes (14).
A well-characterized effect of insulin on the adipocytes is stimulation of phosphodiesterase 3B, which degrades cAMP, leading to reduced PKA activity (24). The observed blockage of insulin-stimulated IL-6 gene expression and release by H89, a widely used inhibitor of PKA, initially suggested that this effect requires activation, rather than suppression of PKA. Although other secretagogues increase IL-6 production in 3T3-L1 adipocytes by activating PKA (23), thus far, insulin has not been reported to stimulate PKA in any cell type. This led us to measure cAMP levels, which were indeed suppressed by insulin. Therefore, we questioned whether H89 blocks pathways other than cAMP/PKA. A review of the literature revealed that the IC50 values of H89 for PKA and PKG are 0.05 and 0.5 μm, respectively. Because H89 at the commonly used dose of 5 μm likely inhibits both enzymes, we postulated that PKG, rather than PKA, mediates the effects of insulin on IL-6 expression and release. We further explored this possibility by using KT5823, a highly specific inhibitor of PKG that does not block PKA. Because KT5823 abrogated insulin-stimulated IL-6 expression and release, we conclude that PKG mediates the effect of insulin on IL-6 production.
PKG is activated by cGMP, which is generated by two types of guanylyl cyclase, a soluble form stimulated by nitric oxide, and a receptor-mediated form activated by natriuretic peptides (25). Insulin activates PKG by stimulating cGMP production in several cell types (26,27). The rapid, significant increase of cGMP in response to insulin in LS14 cells was similar to the aforementioned reports. The IL-6 promoter contains several regulatory domains, including cAMP response element, which binds the transcription factor CREB (28). Both the cAMP/PKA and cGMP/PKG pathways activate CREB by phosphorylation (29,30). Insulin, as well as cGMP, rapidly phosphorylated CREB in LS14 cells. This, together with the ability of KT5823 to inhibit 8-bromo-cGMP-stimulated IL-6 production, support the notion that insulin acts via cGMP/PKG/CREB to modulate IL-6 expression. Our observations agree well with the report that cGMP/PKG-mediated phosphorylation of CREB up-regulates IL-6 expression in osteoblasts (31).
Binding of insulin to its receptor also activates the MAPK pathway (32). Our data reveal not only a robust phosphorylation of MAPK by insulin, but also the blockade of its stimulatory effect on IL-6 release by the specific MEK inhibitor U0126. Unexpectedly, U0126 failed to inhibit insulin-induced IL-6 gene expression, suggesting that IL-6 gene expression and release are differentially regulated, with MAPK affecting either IL-6 synthesis or release.
The releasable pool of IL-6 represents a balance between the rates of its synthesis, degradation, and secretion. To examine IL-6 degradation, we used MG132, a proteosome inhibitor (33). The marked increase in both IL-6 cell content and release in response to MG132 indicates that degradation of IL-6 by the proteosome is a potential mechanism by which IL-6 secretion is regulated in LS14 cells. The role of protein degradation in adipokine release is supported by the reports that newly synthesized leptin in rat adipose tissue (34) and a truncated form of human leptin (35) are degraded by the proteosome. Nonetheless, insulin and MG132 did not show an additive effect on either IL-6 release or content. This raises the possibility of a shared mechanism, which was explored by measuring proteosome activity. Because insulin did not alter degradation of a proteosome substrate (LLVY-AMC) in the presence or absence of U0126, we conclude that, unlike the situation in other cell types (36), insulin does not appear to alter proteosomal activity in LS14 cells. Nonetheless, it would be interesting to explore whether MAPK mediates insulin-stimulated IL-6 release by altering its translation.
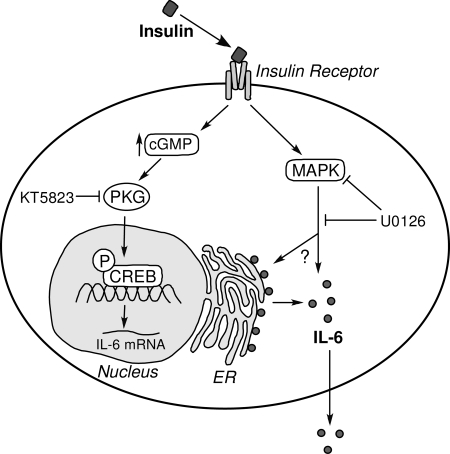
The model shown in Fig. 7 summarizes our current understanding of the mechanisms by which insulin stimulates IL-6 expression and release in LS14 cells. Although the model is incomplete, it highlights the fact that multiple processes regulate secretion in adipocytes, which unlike most neuroendocrine cells, do not contain high-storage capacity secretory granules and have no apparent calcium-dependent exocytosis (37). Although adipokine secretion is commonly described as “constitutive,” the question remains whether adipocytes can regulate the release of proteins destined for secretion once they are synthesized. It has recently been reported that adiponectin is stored within the endoplasmic reticulum of 3T3-L1 adipocytes by chaperone proteins via thiol-mediated retention (38). The authors proposed that regulation of chaperone expression or disulfide bond formation interposes additional steps for the control of adipokine release beyond gene expression. It remains to be determined whether the release of additional adipokines, including IL-6, whose structure is stabilized by two disulfide bridges (39), is also regulated by such novel mechanisms.
Figure 7.
A model illustrating putative mechanisms by which insulin stimulates IL-6 gene expression and release. Insulin binds and activates the insulin receptor, and increases the production of cGMP. This activates PKG, leading to phosphorylation (P) of CREB that increased IL-6 gene transcription. On the other hand, MAPK mediates the effects of insulin at a posttranscriptional step through an unknown mechanism.
Footnotes
This work was supported by National Institutes of Health Grants ES012212 and CA096613, DOD BC05725, and Susan G. Komen Breast Cancer Foundation Grant BCRT87406.
Disclosure Statement: The authors have nothing to disclose.
First Published Online July 10, 2008
Abbreviations: ATF-1, Activating transcription factor 1; cGMP, Cyclic GMP; CM, conditioned media; CREB, cAMP response element binding protein; LLVY-AMC, Suc-Leu-Leu-Val-Tyr-aminomethyl coumarin; MEK, MAPK kinase; NaN, sodium nitroprusside; PI3K, phosphatidylinositol 3-kinase; PKA, cyclic AMP-dependent protein kinase; PKG, cyclic GMP-dependent protein kinase.
References
- Kamimura D, Ishihara K, Hirano T 2003 IL-6 signal transduction and its physiological roles: the signal orchestration model. Rev Physiol Biochem Pharmacol 149:1–38 [DOI] [PubMed] [Google Scholar]
- Path G, Bornstein SR, Gurniak M, Chrousos GP, Scherbaum WA, Hauner H 2001 Human breast adipocytes express interleukin-6 (IL-6) and its receptor system: increased IL-6 production by β-adrenergic activation and effects of IL-6 on adipocyte function. J Clin Endocrinol Metab 86:2281–2288 [DOI] [PubMed] [Google Scholar]
- van Hall G, Steensberg A, Sacchetti M, Fischer C, Keller C, Schjerling P, Hiscock N, Moller K, Saltin B, Febbraio MA, Pedersen BK 2003 Interleukin-6 stimulates lipolysis and fat oxidation in humans. J Clin Endocrinol Metab 88:3005–3010 [DOI] [PubMed] [Google Scholar]
- Trujillo ME, Sullivan S, Harten I, Schneider SH, Greenberg AS, Fried SK 2004 Interleukin-6 regulates human adipose tissue lipid metabolism and leptin production in vitro. J Clin Endocrinol Metab 89:5577–5582 [DOI] [PubMed] [Google Scholar]
- Rotter V, Nagaev I, Smith U 2003 Interleukin-6 (IL-6) induces insulin resistance in 3T3-L1 adipocytes and is, like IL-8 and tumor necrosis factor-α, overexpressed in human fat cells from insulin-resistant subjects. J Biol Chem 278:45777–45784 [DOI] [PubMed] [Google Scholar]
- Fasshauer M, Kralisch S, Klier M, Lossner U, Bluher M, Klein J, Paschke R 2003 Adiponectin gene expression and secretion is inhibited by interleukin-6 in 3T3-L1 adipocytes. Biochem Biophys Res Commun 301:1045–1050 [DOI] [PubMed] [Google Scholar]
- Lagathu C, Bastard JP, Auclair M, Maachi M, Capeau J, Caron M 2003 Chronic interleukin-6 (IL-6) treatment increased IL-6 secretion and induced insulin resistance in adipocyte: prevention by rosiglitazone. Biochem Biophys Res Commun 311:372–379 [DOI] [PubMed] [Google Scholar]
- Wallenius V, Wallenius K, Ahren B, Rudling M, Carlsten H, Dickson SL, Ohlsson C, Jansson JO 2002 Interleukin-6-deficient mice develop mature-onset obesity. Nat Med 8:75–79 [DOI] [PubMed] [Google Scholar]
- Spranger J, Kroke A, Mohlig M, Hoffmann K, Bergmann MM, Ristow M, Boeing H, Pfeiffer AF 2003 Inflammatory cytokines and the risk to develop type 2 diabetes: results of the prospective population-based European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam Study. Diabetes 52:812–817 [DOI] [PubMed] [Google Scholar]
- Bastard JP, Jardel C, Bruckert E, Blondy P, Capeau J, Laville M, Vidal H, Hainque B 2000 Elevated levels of interleukin 6 are reduced in serum and subcutaneous adipose tissue of obese women after weight loss. J Clin Endocrinol Metab 85:3338–3342 [DOI] [PubMed] [Google Scholar]
- Fontana L, Eagon JC, Trujillo ME, Scherer PE, Klein S 2007 Visceral fat adipokine secretion is associated with systemic inflammation in obese humans. Diabetes 56:1010–1013 [DOI] [PubMed] [Google Scholar]
- Yudkin JS 2003 Adipose tissue, insulin action and vascular disease: inflammatory signals. Int J Obes Relat Metab Disord 27(Suppl 3):S25–S28 [DOI] [PubMed] [Google Scholar]
- Vicennati V, Vottero A, Friedman C, Papanicolaou DA 2002 Hormonal regulation of interleukin-6 production in human adipocytes. Int J Obes Relat Metab Disord 26:905–911 [DOI] [PubMed] [Google Scholar]
- Fasshauer M, Klein J, Lossner U, Paschke R 2003 Interleukin (IL)-6 mRNA expression is stimulated by insulin, isoproterenol, tumour necrosis factor α, growth hormone, and IL-6 in 3T3-L1 adipocytes. Horm Metab Res 35:147–152 [DOI] [PubMed] [Google Scholar]
- Hugo ER, Brandebourg TD, Comstock CE, Gersin KS, Sussman JJ, Ben Jonathan N 2006 LS14: a novel human adipocyte cell line that produces prolactin. Endocrinology 147:306–313 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L 2002 Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S, Smith DM, Dou QP 2001 Ester bond-containing tea polyphenols potently inhibit proteasome activity in vitro and in vivo. J Biol Chem 276:13322–13330 [DOI] [PubMed] [Google Scholar]
- Ahima RS 2006 Adipose tissue as an endocrine organ. Obesity (Silver Spring) 14(Suppl 5):242S–249S [DOI] [PubMed] [Google Scholar]
- Fried SK, Bunkin DA, Greenberg AS 1998 Omental and subcutaneous adipose tissues of obese subjects release interleukin-6: depot difference and regulation by glucocorticoid. J Clin Endocrinol Metab 83:847–850 [DOI] [PubMed] [Google Scholar]
- Fain JN 2006 Release of interleukins and other inflammatory cytokines by human adipose tissue is enhanced in obesity and primarily due to the nonfat cells. Vitam Horm 74:443–477 [DOI] [PubMed] [Google Scholar]
- Antunes TT, Gagnon A, Chen B, Pacini F, Smith TJ, Sorisky A 2006 Interleukin-6 release from human abdominal adipose cells is regulated by thyroid-stimulating hormone: effect of adipocyte differentiation and anatomic depot. Am J Physiol Endocrinol Metab 290:E1140–E1144 [DOI] [PubMed] [Google Scholar]
- Harkins JM, Moustaid-Moussa N, Chung YJ, Penner KM, Pestka JJ, North CM, Claycombe KJ 2004 Expression of interleukin-6 is greater in preadipocytes than in adipocytes of 3T3-L1 cells and C57BL/6J and ob/ob mice. J Nutr 134:2673–2677 [DOI] [PubMed] [Google Scholar]
- Antunes TT, Gagnon A, Bell A, Sorisky A 2005 Thyroid-stimulating hormone stimulates interleukin-6 release from 3T3-L1 adipocytes through a cAMP-protein kinase A pathway. Obes Res 13:2066–2071 [DOI] [PubMed] [Google Scholar]
- Elks ML, Manganiello VC 1985 Antilipolytic action of insulin: role of cAMP phosphodiesterase activation. Endocrinology 116:2119–2121 [DOI] [PubMed] [Google Scholar]
- Vaandrager AB, de Jonge HR 1996 Signalling by cGMP-dependent protein kinases. Mol Cell Biochem 157:23–30 [DOI] [PubMed] [Google Scholar]
- Jacob A, Molkentin JD, Smolenski A, Lohmann SM, Begum N 2002 Insulin inhibits PDGF-directed VSMC migration via NO/ cGMP increase of MKP-1 and its inactivation of MAPKs. Am J Physiol Cell Physiol 283:C704–C713 [DOI] [PubMed] [Google Scholar]
- Bergandi L, Silvagno F, Russo I, Riganti C, Anfossi G, Aldieri E, Ghigo D, Trovati M, Bosia A 2003 Insulin stimulates glucose transport via nitric oxide/cyclic GMP pathway in human vascular smooth muscle cells. Arterioscler Thromb Vasc Biol 23:2215–2221 [DOI] [PubMed] [Google Scholar]
- Dendorfer U, Oettgen P, Libermann TA 1995 Interleukin-6 gene expression by prostaglandins and cyclic AMP mediated by multiple regulatory elements. Am J Ther 2:660–665 [DOI] [PubMed] [Google Scholar]
- Gudi T, Casteel DE, Vinson C, Boss GR, Pilz RB 2000 NO activation of fos promoter elements requires nuclear translocation of G-kinase I and CREB phosphorylation but is independent of MAP kinase activation. Oncogene 19:6324–6333 [DOI] [PubMed] [Google Scholar]
- Ciani E, Guidi S, Bartesaghi R, Contestabile A 2002 Nitric oxide regulates cGMP-dependent cAMP-responsive element binding protein phosphorylation and Bcl-2 expression in cerebellar neurons: implication for a survival role of nitric oxide. J Neurochem 82:1282–1289 [DOI] [PubMed] [Google Scholar]
- Broderick KE, Zhang T, Rangaswami H, Zeng Y, Zhao X, Boss GR, Pilz RB 2007 Guanosine 3′,5′-cyclic monophosphate (cGMP)/cGMP-dependent protein kinase induce interleukin-6 transcription in osteoblasts. Mol Endocrinol 21:1148–1162 [DOI] [PubMed] [Google Scholar]
- Martinez-deMena R, Obregon MJ 2005 Insulin increases the adrenergic stimulation of 5′ deiodinase activity and mRNA expression in rat brown adipocytes; role of MAPK and PI3K. J Mol Endocrinol 34:139–151 [DOI] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL 1998 Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol 8:397–403 [DOI] [PubMed] [Google Scholar]
- Lee MJ, Fried SK 2006 Multilevel regulation of leptin storage, turnover, and secretion by feeding and insulin in rat adipose tissue. J Lipid Res 47:1984–1993 [DOI] [PubMed] [Google Scholar]
- Rau H, Reaves BJ, O'Rahilly S, Whitehead JP 1999 Truncated human leptin (δ133) associated with extreme obesity undergoes proteasomal degradation after defective intracellular transport. Endocrinology 140:1718–1723 [DOI] [PubMed] [Google Scholar]
- Bennett RG, Fawcett J, Kruer MC, Duckworth WC, Hamel FG 2003 Insulin inhibition of the proteasome is dependent on degradation of insulin by insulin-degrading enzyme. J Endocrinol 177:399–405 [DOI] [PubMed] [Google Scholar]
- Bradley RL, Cheatham B 1999 Regulation of ob gene expression and leptin secretion by insulin and dexamethasone in rat adipocytes. Diabetes 48:272–278 [DOI] [PubMed] [Google Scholar]
- Wang ZV, Schraw TD, Kim JY, Khan T, Rajala MW, Follenzi A, Scherer PE 2007 Secretion of the adipocyte-specific secretory protein adiponectin critically depends on thiol-mediated protein retention. Mol Cell Biol 27:3716–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huising MO, Kruiswijk CP, Flik G 2006 Phylogeny and evolution of class-I helical cytokines. J Endocrinol 189:1–25 [DOI] [PubMed] [Google Scholar]