Abstract
The solution structure of the N1-(1-hydroxy-3-buten-2(R)-yl)-2′-deoxyinosine adduct arising from the alkylation of adenine N1 by butadiene epoxide (BDO), followed by deamination to deoxyinosine, was determined, in the oligodeoxynucleotide d(CGGACXAGAAG)•d(CTTCTCGTCCG). This oligodeoxynucleotide contained the BDO adduct at the second position of codon 61 of the human N-ras protooncogene, and was named the ras61 R-N1-BDO-(61,2) adduct. 1H NMR revealed a weak C5 H1′ to X6 H8 NOE, followed by an intense X6 H8 to X6 H1′ NOE. Simultaneously the X6 H8 to X6 H3′ NOE was weak. The resonances arising from the T16 and T17 imino protons were not observed. 1H NOEs between the butadiene moiety and the DNA positioned the adduct in the major groove. Structural refinement based upon a total of 394 NOE-derived distance restraints and 151 torsion angle restraints yielded a structure in which the modified deoxyinosine was in the syn conformation about the glycosyl bond, with a glycosyl bond angle of 83°, and T17, the complementary nucleotide, was stacked into the helix, but not hydrogen bonded with the adducted inosine. The refined structure provides a plausible hypothesis as to why these N1 deoxyinosine adducts strongly code for the incorporation of dCTP during trans lesion DNA replication, irrespective of stereochemistry, both in Escherichia coli [Rodriguez, D.A., Kowalczyk, A., Ward, J.B.J., Harris, C.M., Harris, T.M., and Lloyd, R.S. (2001) Environ. Mol. Mutagen. 38, 292–296], and in mammalian cells [Kanuri, M., Nechev, L.N., Tamura, P.J., Harris, C.M., Harris, T.M., and Lloyd, R.S. (2002) Chem. Res. Toxicol. 15, 1572–1580]. Rotation of the N1 deoxyinosine adduct into the syn conformation may facilitate incorporation of dCTP via Hoogsteen-type templating with deoxyinosine, generating A to G mutations. However, conformational differences between the R- and S-N1-BDO-(61,2) adducts, involving the positioning of the butenyl moiety in the major groove of DNA, suggest that adduct stereochemistry plays a secondary role in modulating the biological response to these adducts.
Introduction
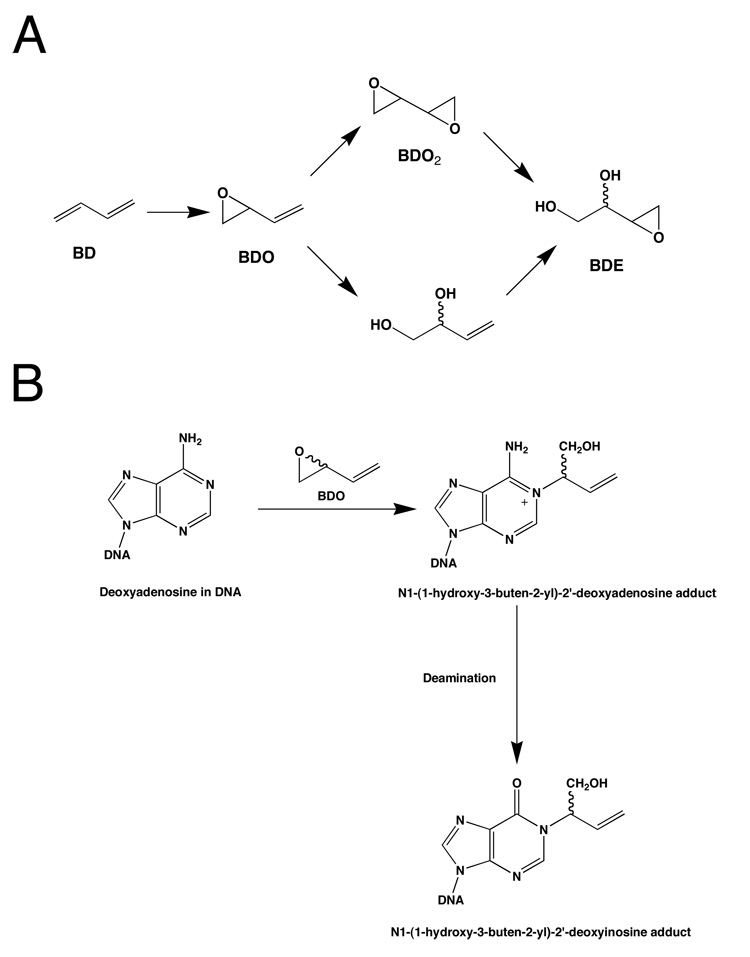
1,3-Butadiene (CAS RN 106-99-0) (BD)1 is used in the manufacture of styrene-butadiene rubber (SBR) (1, 2); several billion lbs/yr are produced in the United States. It is a combustion product from automobile emissions (3) and cigarette smoke (4). BD is genotoxic and is a carcinogen in rodents, particularly in mice (5–7) and also in rats (8). Recently, BD was classified by the United States Environmental Protection Agency as "carcinogenic to humans by inhalation" (9). The International Agency for Cancer Research (IARC) lists BD as a "probable human carcinogen" (Group 2A) (10). Chronic human exposure in the SBR industry may induce genotoxic effects (11–13) and is correlated with increased risk for leukemia (1, 14–22).
BD is epoxidized primarily by cytochrome P450 2E1, but also by cytochrome P450 2A6, to form 1,2-epoxy-3-butenes (BDO) (Scheme 1) (23, 24). BDO is a reactive electrophile that can potentially alkylate nucleophilic sites in DNA, including the imine nitrogen at N1 of deoxyadenosine. Alkylation could occur from either carbon atom of the oxirane; attack by the interior carbon atom, designated Cβ, yields two possible stereoisomers of the N1-(1-hydroxy-3-buten-2-yl)-2′-deoxyadenosine adduct. These initially formed N1-dA adducts are prone to deamination (25), which yields two possible stereoisomeric N1-(1-hydroxy-3-buten-2-yl)-2′-deoxyinosine adducts (Scheme 2).
Scheme 1.
A. Cytochrome P450-Mediated Epoxidation of Butadiene to Mono- and Di-Epoxides. B. Alkylation of Deoxyadenosine N1 by Cβ of Butadiene Mono-Epoxide (BDO) and Subsequent Deamination Yields the N1-(1-hydroxy-3-buten-2-yl)-2′-Deoxyinosine Adduct.
Scheme 2.
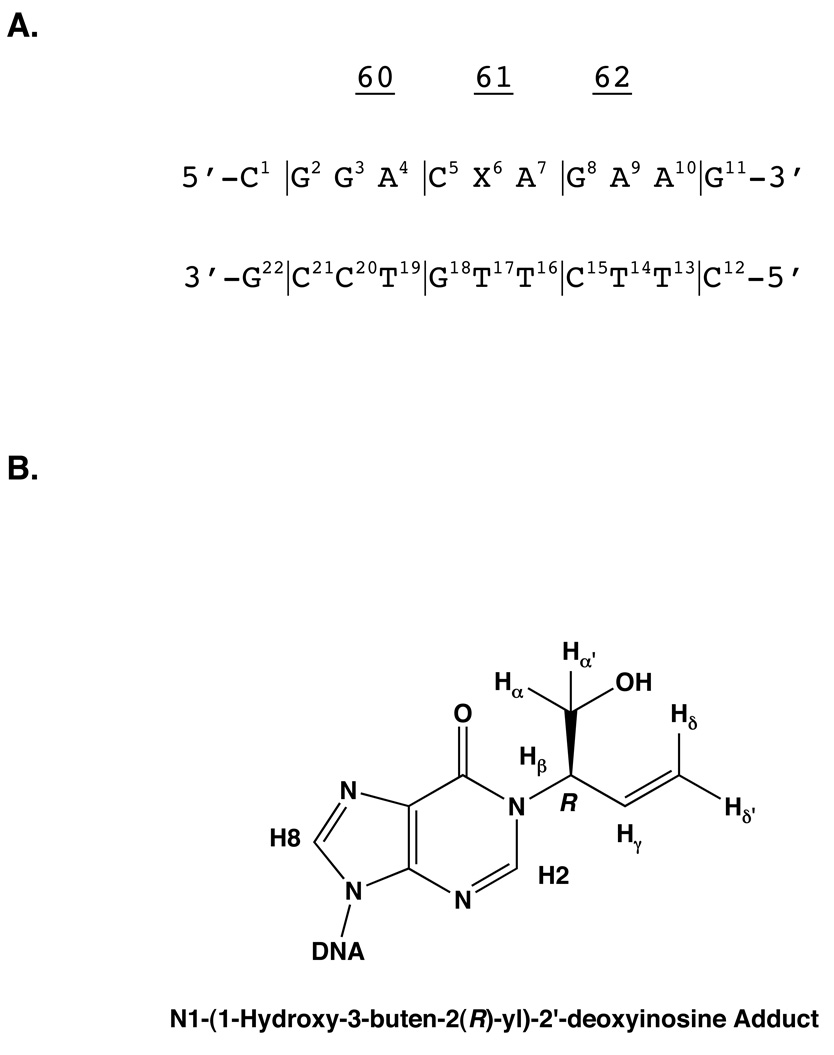
A. The ras61 oligodeoxynucleotide. B. The chemical structures of the N1-(1-hydroxy-3-butene-2(R)-yl)-2′-deoxyinosine adduct.
Deamination of dA represents a pro-mutagenic event because during DNA replication, the resulting dI nucleotide is recognized as dG and preferentially pairs with incoming dCTP during DNA replication. However, in the N1-(1-hydroxy-3-buten-2-yl)-2′-deoxyinosine adducts, dI is alkylated at the N1 position, thus blocking base pairing with dCTP. Nevertheless, when ligated into the single-stranded vector M13mp7L2 that was used to transfect repair-deficient AB2480 (uvrA-, recA-) and SOS-proficient AB1157 Escherichia coli, the N1-(1-hydroxy-3-buten-2(S)-yl)-2′-deoxyinosine adduct strongly coded for incorporation of dCTP (26). Studies of this adduct in COS-7 cells yielded similar results (27).
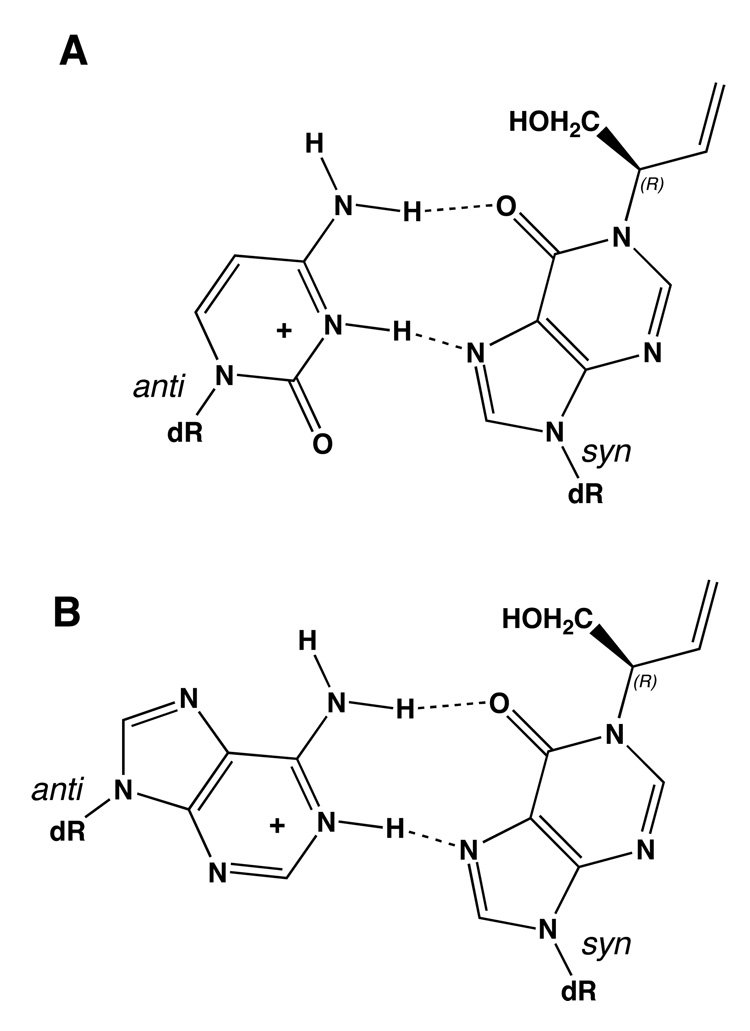
It was proposed that incorporation of dCTP opposite the N1-(1-hydroxy-3-buten-2(S)-yl)-2′-deoxyinosine adduct might occur as a result of rotation of the N1-adducted dI into the syn conformation about the glycosyl bond, thus enabling formation of a protonated dI•dC Hoogsteen pair during trans-lesion synthesis (27) (Scheme 3). Structural studies of the N1-(1-hydroxy-3-buten-2(S)-yl)-2′-deoxyinosine adduct site-specifically incorporated into the ras61 oligodeoxynucleotide 5′-d(CGGACXAGAAG)- 3′•5′-d(CTTCTTGTCCG)-3′, containing the adduct at the second position of codon 61, named the ras61 S-N1-BDO-(61,2) adduct, revealed the rotation of the glycosyl bond of the S-N1-BDO-(61,2) adduct into the high syn conformation, thus placing the butadiene moiety into the major groove (28).
Scheme 3.
A. Formation of a Protonated Hoogsteen Pair Between the N1-(1-Hydroxy-3-buten-2(R)-yl)-2′-deoxyinosine Adduct and Cytosine. B. Formation of a Protonated Hoogsteen-like Pair Between the N1-(1-Hydroxy-3-buten-2(R)-yl)-2′-deoxyinosine Adduct and Adenine.
The present work characterizes the structural perturbation to the ras61 oligodeoxynucleotide caused by the ras61 R-N1-BDO-(61,2) adduct. Its structure is compared to the S-N1-BDO-(61,2) adduct (28). Both the R- and S-N1-BDO-(61,2) adducts induced A→G mutations. However, in the COS-7 system, the mutation frequencies differed with respect to adduct stereochemistry. The R-N1-BDO-(61,2) adduct yielded a mutation frequency of 60%, whereas the S-N1-BDO-(61,2) adduct yielded mutations at a frequency of 90% (27). Molecular dynamics calculations restrained by interproton distances obtained from nuclear Overhauser effects (29) and torsion angle restraints obtained from NMR data indicate that the R-N1-BDO-(61,2) adduct adduct assumes the syn conformation about the glycosyl bond in duplex DNA, whereas the S-N1-BDO-(61,2) adduct glycosyl angle was in the "high-syn" conformation. In both instances the complementary dT remains intrahelical at the adduct site. The results suggest that that the predominance of A→G mutations observed for these adducts may be explained by the syn conformation observed at the glycosyl bond. On the other hand, stereospecific differences in adduct structure may correlate with differences in mutagenesis rate (26, 27).
Materials and Methods
Sample Preparation
The oligodeoxynucleotides 5′-d(CGGACAAGAAG)-3′ and its complementary strand 5′-d(CTTCTTGTCCG)-3′, were synthesized by the Midland Certified Reagent Co. (Midland, TX) and purified by anion-exchange chromatography. The concentrations of the single-stranded oligonucleotides were determined from their calculated extinction coefficients at 260 nm (30). The modified oligodeoxynucleotide (31) and its complement were annealed in 10 mM NaH2PO4, 10 mM Na2HPO4, 0.1 M NaCl, and 50 µM Na2EDTA (pH 7.0). The modified duplex was eluted from DNA Grade Biogel hydroxylapatite (Bio-Rad Laboratories, Hercules, CA) with a gradient from 10 to 200 mM NaH2PO4, pH 7.0. It was desalted using Sephadex G-25. For NMR experiments, the modified duplex was prepared at a concentration of 2 mM. For observation of nonexchangeable protons, the sample was dissolved in 0.5 mL of 99.96% D2O containing 10 mM NaH2PO4, 10 mM Na2HPO4, 0.1 M NaCl, and 50 µM Na2EDTA at pH 7.0. For observation of exchangeable protons, the sample was dissolved in 0.5 mL of 9:1 H2O:D2O in the same buffer.
Capillary gel electrophoresis
The purity of the modified duplex was analyzed using a PACE 5500 (Beckman Instruments, Inc., Fullerton, CA) instrument. Electrophoresis was conducted using an eCAP ssDNA 100-R kit applying 12,000 V for 30 min. The electropherogram was monitored at 254 nm.
Mass Spectrometry
MALDI-TOF mass spectra were measured on a Voyager-DE (PerSeptive Biosystems, Inc., Foster City, CA) instrument in negative reflector mode. The matrix contained 0.5 M 3-hydroxypicolinic acid and 0.1 M ammonium citrate.
Nuclear Magnetic Resonance
1H NMR spectra were recorded at 600.13 MHz and 800.23 MHz. The non-exchangeable protons were monitored at 25 °C; the exchangeable protons were monitored at 17 °C. Chemical shifts were referenced to the water resonance. NMR data were processed using FELIX2000 (Accelyris, Inc., San Diego, CA).
NOESY spectra of the non-exchangeable protons were recorded using TPPI phase cycling with mixing times of 150, 200, and 250 ms. These were acquired sequentially without removing the sample from the magnet. Spectra for the exchangeable protons were recorded using a 150 ms mixing time. These experiments were recorded with 1024 real data points in the d1 dimension and 2048 real data points in the d2 dimension. A relaxation delay of 2.0 s was used. Water suppression was performed using the WATERGATE sequence (32). TOCSY experiments were performed with mixing times of 90 and 150 ms, utilizing the homonuclear Hartman-Hahn transfer with the MLEV17 sequence for mixing. DQF-COSY spectra were zero-filled to give a matrix of 1024 × 2048 real points. A skewed sine-bell square apodization function with a 90° phase shift and a skew factor of 1.0 was used in both dimensions.
Experimental Restraints
(a) Distance Restraints
Footprints were drawn around cross peaks obtained at a mixing time of 200 ms using FELIX2000. Identical footprints were transferred and fit to the crosspeaks obtained at the other two mixing times. The intensities of these cross peaks were determined by volume integration. These were combined as necessary with intensities generated from complete relaxation matrix analysis of a starting DNA structure to generate a hybrid intensity matrix (33, 34). MARDIGRAS (35–37) was used to iteratively refine the hybrid intensity matrix and to optimize the agreement between the calculated and experimental NOE intensities. Calculations were initated using isotropic correlation times of 2, 3, and 4 ns, and with both IniA and IniB starting structures and the three mixing times, yielding eighteen sets of distances. Analysis of this data yielded the experimental distance restraints used in subsequent restrained molecular dynamics calculations, and the corresponding standard deviations for the distance restraints. The distance restraints were divided into five classes, reflecting the confidence level in the experimental data.
(b) Torsion Angle Restraints
Deoxyribose pseudorotation (57) was determined graphically using the sums of 3J 1H coupling constants (58), measured from DQF-COSY spectra. Discrete J1′2″ and J1′2′ couplings were measured from active and passive couplings, respectively, of the H2″ (d2) to H1′ (d1) spectral region. The data were fit to curves relating the coupling constants to the deoxyribose sugar pseudo rotation angle (P), sugar pucker amplitude (φ), and the percentage S-type conformation. The sugar pseudo rotation angle and amplitude ranges were converted to the five dihedral angles ν0 to ν4. Coupling constants measured from 1H-31P HMBC spectra were applied (38, 39) to the Karplus relationship (40) to determine the backbone dihedral angle ε (C4′-C3′-O3′-P), related to the H3′-C3′-O3′-P angle by a 120° shift. The ζ (C3′-O3′-P-O5′) backbone angles were calculated from the correlation between ε and ζ in B-DNA (60).
Restrained Molecular Dynamics Calculations
Classical A-DNA and B-DNA were used as reference structures to create starting structures for the refinement (41). The butadiene adduct was constructed at X6 using the BUILDER module of INSIGHT II (Accelrys, Inc., San Diego, CA). A-form and B-form structures of the appropriate sequence were energy-minimized by the conjugate gradients method for 200 iterations using the AMBER 7.0 force field (42) without experimental restraints to give starting IniA and IniB used for the subsequent relaxation matrix analysis and molecular dynamics calculations. The restraint energy function was comprised of terms describing distances and dihedral restraints, both of which were in the form of a standard square-well potential (43). Bond lengths involving hydrogens were fixed with the SHAKE algorithm (44). The generalized Born approach was used to model solvation (45, 46). The calculations utilized a salt concentration of 0.2 mM. A series of randomly seeded rMD calculations were performed over a time course of 40 ps. These used the SANDER module of AMBER 7.0, and the Cornell et al. force field (47), including the Parm94.dat parameter set. The simulated annealing protocol utilized a starting temperature of 25 K. In the first ps the temperature was increased to 600 K. This was maintained for the next 4 ps, followed by cooling to 298 K over 15 ps. During the final 20 ps the temperature was reduced to 0 K. Temperature was controlled by coupling the molecules to a temperature bath. During the first 1 ps of heating a coupling of 0.4 ps was used. During the next 4 ps of constant temperature dynamics a coupling of 1.0 ps was used. In the first 15 ps of cooling, a value of 1.0 ps was used, followed by a value of 0.5 ps for the second 15 ps. During the final 5 ps of cooling, the coupling was ramped down to 0.01 ps. In the first 1 ps of heating, the experimental force constants were amplified by factors that ranged from 0.5 to 1.00. During the 4 ps of constant temperature dynamics and the first 15 ps of cooling, the amplification factor was increased to 1.75. In the final 20 ps of cooling, the amplification factor was reduced to a value of 1.00. Structure coordinates were archived every 0.1 ps over the final 10 ps of the rMD simulation. Structure coordinates extracted from the final 4 ps of each rMD calculation were averaged and energy-minimized for 200 iterations using the conjugate gradients algorithm. An average structure was obtained from eight randomly seeded rMD calculations.
Back-calculation of 1H NOE data was performed using CORMA (v. 4.0) (48). Helicodial parameters were examined using 3DNA (49).
Results
Sample Properties
The duplex R-N1-BDO-(61,2) oligodeoxynucleotide yielded two sharp bands in a 1:1 ratio, corresponding to the modified and complementary strands, when assessed using capillary gel electrophoresis. MALDI-TOF mass spectrometry showed two signals that corresponded to the anticipated mass units of 3468 (adducted strand) and 3272 (complementary strand). The melting temperature of the R-N1-BDO-(61,2) duplex as determined by UV spectroscopy was 33° C (50), less than the observed 57° C melting temperature of the unmodified ras61 duplex. The R-N1-BDO-(61,2) sample yielded excellent NMR data in the temperature range of 15° C--25°C.
DNA 1H Resonance Assignments
(a) Nonexchangeable Protons
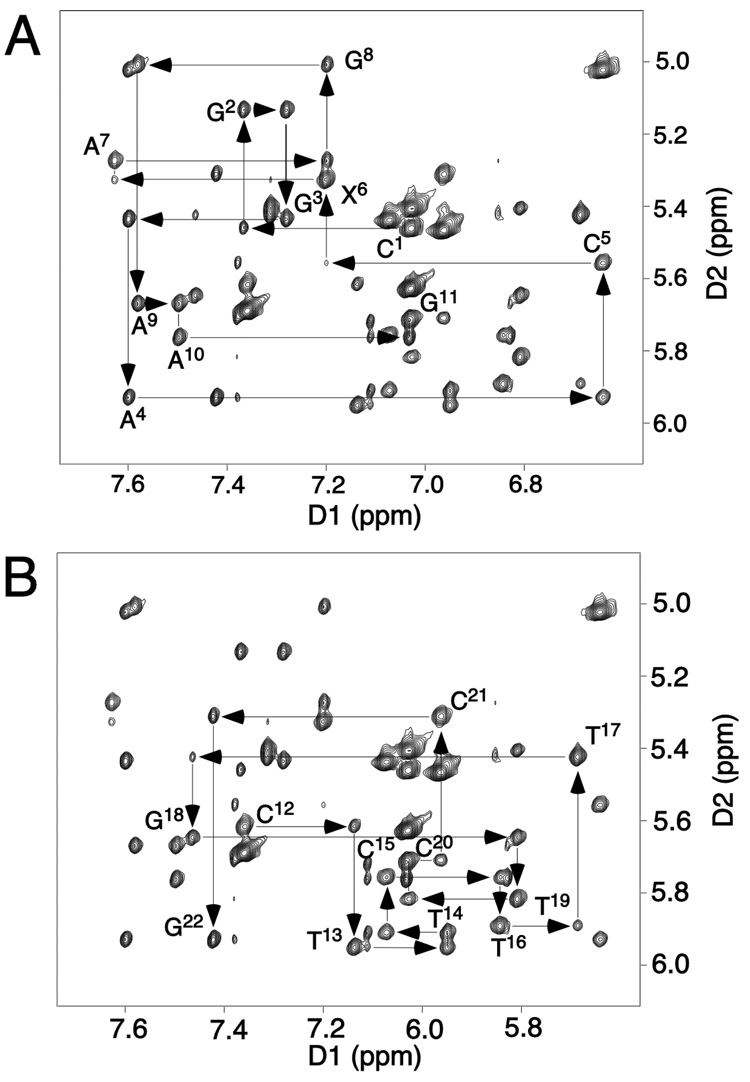
The sequential 1H NOEs between the aromatic and anomeric protons of the R-N1-BDO-(61,2) oligodeoxynucleotide duplex are displayed in Figure 1. The NMR spectrum was well-resolved and yielded resonances with narrow linewidths, indicative of a stable and ordered conformation. These resonances were assigned using standard methods (51, 52). The remarkable features of this region of the 1H NOE spectrum were the weak C5 H1′→A6 H8 and X6 H1′→A7 H8 crosspeaks. With respect to the complementary strand, all sequential NOEs were observed, although the T17 H1′→G18 H8 NOE was weak. The deoxyribose sugar proton resonances were assigned from DQF-COSY spectra. A complete 1H assignment was achieved, with the exception of several of the H5′ and H5″ protons. The 1H NMR assignments are provided in Table S1 of the Supporting Information.
Figure 1.
Expanded plots of a NOESY spectrum at a mixing time of 200 ms showing sequential NOE connectivities from aromatic to anomeric protons. A. Nucleotides C1→G11 of the modified strand of the R-N1-BDO-(61,2) adduct. B. Nucleotides C12→G22 of the R-N1-BDO-(61,2) adduct. The experiment was at 800.23 MHz and 25 °C.
(b) Exchangeable Protons
An expanded region showing the far downfield region of the 1H NOE spectrum, exhibiting cross-peaks between the hydrogen-bonded imino protons is shown in Figure 2. As compared to the unmodified ras61 duplex (31), the primary changes in the spectrum were the lack of signals for base pairs X6•T17 and A7•T16. In the amino region of the 1H NMR spectrum, the NOE between A7 H2→T16 N3H was also not observed, eliminating the possibility that the T14 N3H and T16 N3H resonances were overlapped, as had been observed for the S-N1-BDO-(61,2) duplex (28). Consequently, the sequential NOE connectivity in the imino region of the 1H spectrum was interrupted at base pair C5•G18 and resumed at base pair G8•C15. The NOE between T13 N3H and T14 N3H was weak. It was not observed at the contour level plotted in Figure 2. It was also weak in the spectrum of the unmodified ras61 oligodeoxynucleotide (31), and presumably reflects a sequence-specific rapid exchange of this proton with solvent. The imino protons of the terminal C1•G22 pair and G11•C12 pairs were not detected, presumably due to fraying of the ends of the helices.
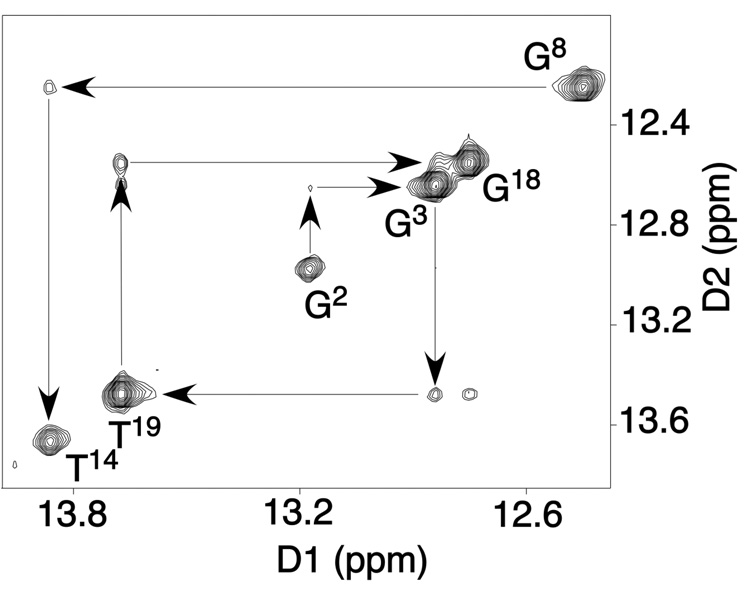
Figure 2.
Expanded plot of a NOESY spectrum at a mixing time of 200 ms showing NOE connectivities for the imino protons for the base pairs from G2•C21 to A10•T13. There were no imino resonances observed for base pairs X6•T17 and A7•T16. The experiment was at 800.23 MHz and 25 °C.
(c) Butadiene Protons
The butadiene proton assignments were achieved using a combination of NOESY and COSY experiments2 (Figure 3). The protons Hγ and Hδ′ overlapped at 5.40 ppm, and the proton Hδ″ resonated at 5.43 ppm. The proton Hβ was assigned at 4.76 ppm. It exhibited NOEs to two protons that resonated at 3.22 and 3.40 ppm, which were assigned as the Hα′,α″ protons. The Hα′,α″ assignments were based upon potential energy minimization calculations, which predicted that the lowest energy conformation of the R-N1-BD moiety placed the hydroxyl at Cα such that the hydroxyl proton was within hydrogen bonding distance of the inosine keto oxygen, X6O6. In this conformation Hα′ was expected to exhibit a stronger NOE to Hδ′ and a weaker NOE to Hδ″, whereas Hα″ was expected to exhibit only weak NOEs to Hδ′ and Hδ″. Accordingly, the resonance at 3.42 ppm was assigned as Hα′ and the resonance at 3.45 ppm was assigned as Hα″. The BD protons exhibited spectral linewidths comparable to those of the oligodeoxynucleotide protons.
Figure 3.
Expanded NOESY spectrum at 200 ms mixing time exhibiting the assignment of adduct protons in the R-N1-BDO-(61,2) duplex. The experiment was at 800.23 MHz and 25 °C.
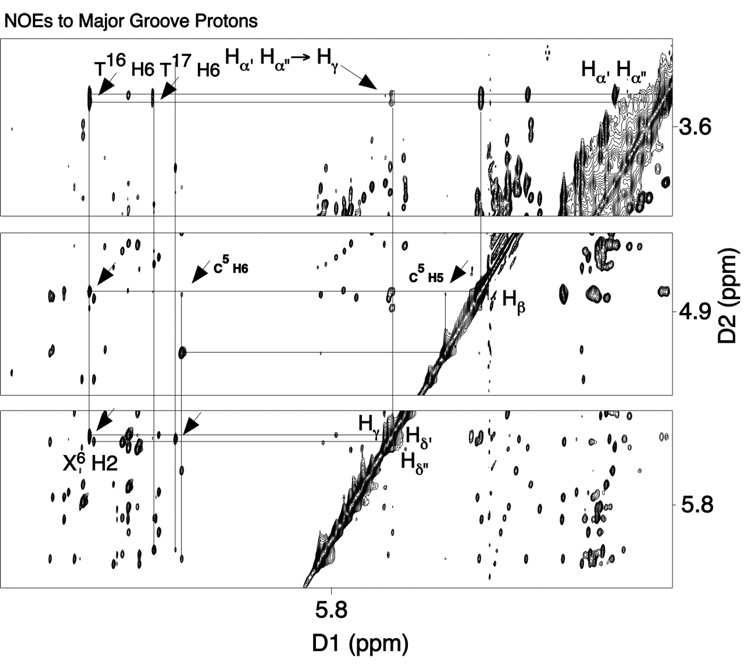
(d) Butadiene-DNA NOEs
These were observed between the BD moiety and major groove protons associated with the modified base pair X6•T17 and its 3′-neighbor base pair, A7•T16 (Figure 3). The proton Hγ exhibited a strong NOE to X6 H2, which may also be observed in Figure 1. The Hγ resonance overlapped with that arising from Hδ′, but the primary NOE was anticipated to arise from the X6 H2→Hγ dipolar interaction. Crosspeaks in the NOESY spectrum were also observed between X6 H2, and BD Hβ and Hα′,α″ protons. The latter NOEs probably resulted from spin diffusion at the 200 ms mixing time shown in Figure 3.
Torsion Angle Measurements
The glycosyl torsion angle conformations were evaluated by inspection of 1H NOESY data. Figure 4 details the relative intensities of intranucleotide NOEs between purine H8 and pyrimidine H6 protons, and anomeric H1′ protons for the adducted strand of the R-N1-BDO-(61,2) adduct. The strong NOE between X6 H8 and X6 H1′ was evident, indicating that the X6 inosine ring was in the syn conformation. Weaker NOEs between the other purine H8 or pyrimidine H6 protons and the anomeric H1′ protons of the attached deoxyribose sugars were consistent with glycosyl torsion angles in the anti conformational range. Analysis of DQF-COSY data suggested that all deoxyribose pseudorotation angles remained in the C2′-endo range anticipated for B-family DNA. There were no unusually shifted 31P resonances.
Figure 4.
Resonance intensities of intranucleotide H1′ and H6/H8 1H NOE crosspeaks at 800.13 MHz and 35°C.
Chemical Shift Perturbations
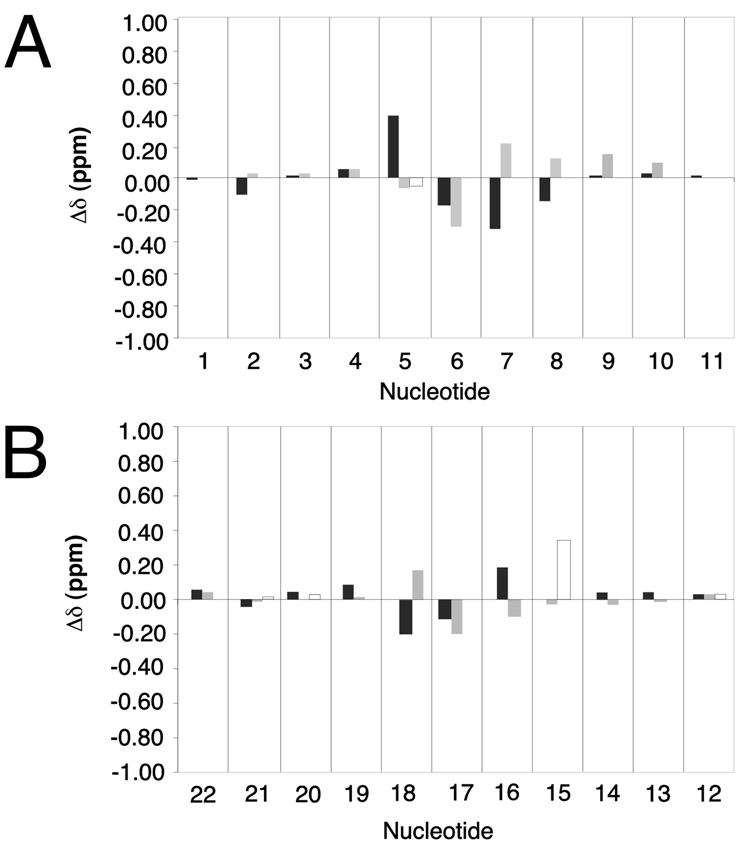
Figure 5 shows chemical shift differences between the R-N1-BDO-(61,2) adduct and the unmodified ras61 duplex oligodeoxynucleotide (31). The changes were observed at the four base pairs C5•G18, X6•T17, A7•T16, and G8•C15. Of the DNA base aromatic protons, A7 H8 shifted 0.2 ppm upfield, C15 H5 shifted 0.4 ppm upfield, and G18 H8 shifted 0.2 ppm upfield. A downfield shift of 0.3 ppm was observed for X6 H8. T16 H6 shifted downfield 0.15 ppm, and T17 H6 shifted downfield 0.2 ppm. Of the deoxyribose H1′ protons C5 H1′ shifted 0.45 ppm upfield, G8 H8 shifted 0.15 ppm upfield, and T16 H1′ shifted 0.2 ppm upfield. In contrast, X6 H1′ shifted 0.2 ppm downfield, A7 H1′ shifted 0.4 ppm downfield, G8 H1′ shifted 0.15 ppm downfield, T17 H1′ shifted 0.15 ppm downfield, and G18 H1′ shifted 0.2 ppm downfield. At X6 there were noticeable changes in chemical shift for the H2′ and H2″ protons.
Figure 5.
Chemical shift differences of protons of the R-N1-BDO-(61,2) adduct duplex relative to the unmodified ras61 oligodeoxynucleotide. A. The modified strand of the R-N1-BDO-(61,2) adduct. B. The complementary strand of the R-N1-BDO-(61,2) adduct. Grey bars represent the deoxyribose H1′ protons; black bars represent the purine H8 or pyrimidine H6 protons, respectively.
Structural Refinement
The experimental distance restraints generated by MARDIGRAS were evenly distributed over the length of the R-N1-BDO-(61,2) oligodeoxynucleotide. The presence of the R-N1-BDO-(61,2) adduct at nucleotide X6 resulted in a greater number of NOE restraints at nucleotides C5 and X6; these served to orient the BD moiety in the duplex DNA. In addition to the distance restraints, eighty deoxyribose pseudorotation restraints were included. Fifty empirical Watson-Crick hydrogen bonding restraints (53) were included in the calculations. Hydrogen-bonding restraints were not used for X6•T17 and its 3′-neighbor A7•T16, as 1H NOESY data did not indicate Watson-Crick base-pairing at these base pairs. There were 71 empirical phosphodiester backbone torsion angle restraints used in the rMD calculations. These restrained the torsion angles ε and ζ at angles of 165° ± 35° and 245° ± 35° respectively (54).
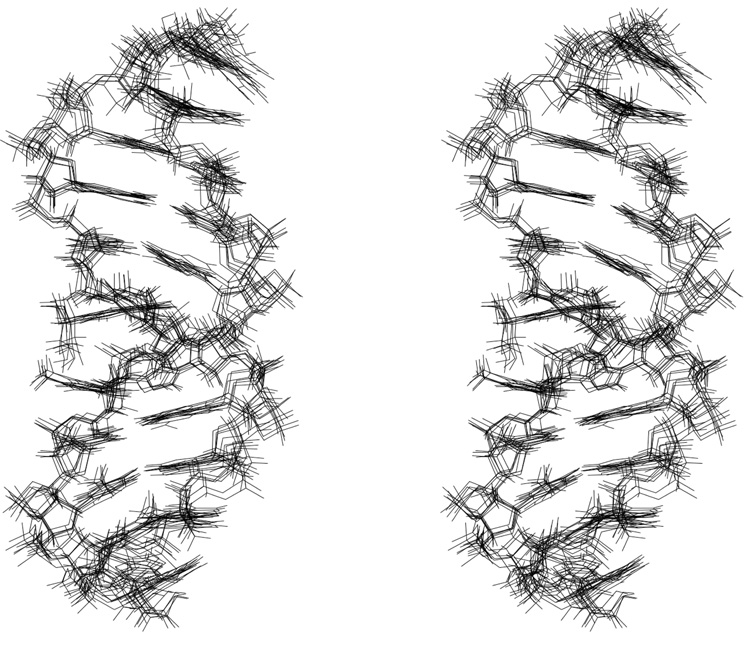
A stereoview of structures originating from a series of randomly seeded rMD calculations is shown in Figure 6. These rMD calculations utilized both A-form and B-form DNA starting structures, which exhibited a difference rmsd of 5.92 Å. Utilizing either starting structure, the rMD calculations converged to similar structures. The maximum pairwise rmsd between structures emergent from the rMD calculations was 1.35 Å. The average refined structure showed a rmsd of 2.17 Å compared to the B-DNA starting structure, whereas it showed a 7.34 Å rmsd compared to the A-DNA starting structure. Thus, the structures emergent from the rMD calculations were different than either A-form or B-form DNA, consistent with the observation that the modified nucleotide X6 was in the syn conformation about the glycosyl bond. The structural refinement statistics are presented in Table 1.
Figure 6.
A stereoview of superimposed structures emergent from the simulated annealing rMD protocol; the structures resulted from randomly seeded calculations.
Table 1.
Analysis of the rMD-Generated Structures of the ras61 R-N1-BDO-(61,2) adduct.
| NMR restraints | |
| total no. of distance restraints | 394 |
| interresidue distance restraints | 125 |
| intraresidue distance restraints | 253 |
| DNA-BDE distance restraints | 16 |
| Total torsion angle restraints | |
| sugar pucker restraints | 80 |
| backbone torsion angle restraints | 71 |
| empirical restraints | |
| H-bonding restraints | 46 |
| dihedral planarity restraints (in XPLOR) | 20 |
| structural statistics | |
| NMR R-factor (R1x)a,b,c | 9.25 ×10−2 |
| rmsd of NOE violations (Å) | 2.32×10−2 |
| no. of DNA NOE violations > 0.15 Å | 6 |
| pairwise rmsd (Å) over all atoms | |
| IniA vs. IniB | 5.92 |
| IniA vs. rMDA | 7.34 |
| IniB vs. rMDB | 2.17 |
| rMDA vs rMDB | 0.99 |
| rMDA vs rMDA | 1.35 |
| rMDB vs rMDB | 0.96 |
| <rMDA> vs. rMDAavg | 0.89 ± 0.28 |
| <rMDB> vs. rMDBavg | 0.62 ± 0.21 |
The mixing time was 200 ms.
R1x = ∑|(ao)i1/6 − (ac)i1/6|/∑|(ao)i1/6|, where ao and ac are the intensities of observed (non-zero) and calculated NOE cross-preaks.
<rMDA>, represents a group of 8 converged structures starting from IniA; <rMDB>, represents a group of 8 converged structures starting from IniB. rMDAavg represents the potential energy minimized average structure of all 8 rMD calculations starting with A-form DNA rMDBavg represents the potential energy minimized average structure of all 8 rMD calculations starting with B-form DNA. The comparisons: rMDA vs rMDB, rMDA vs rMDA, rMDB vs rMDB, represent the maximum observed pairwise rmsd over all atoms between these groups
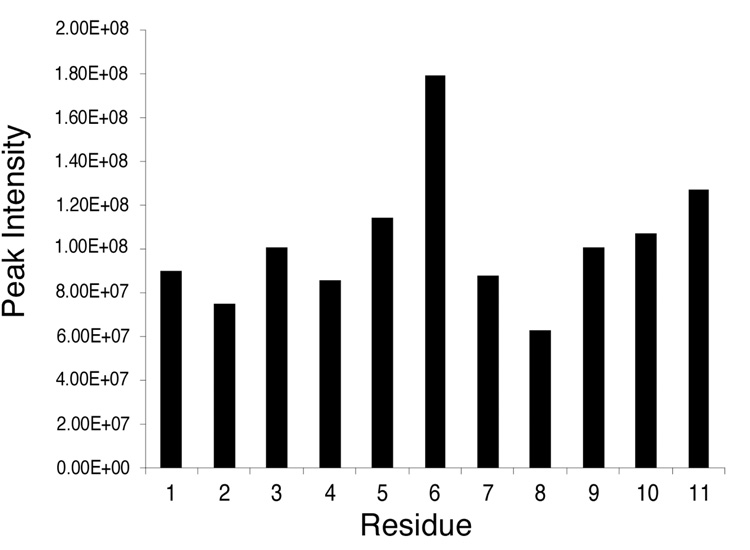
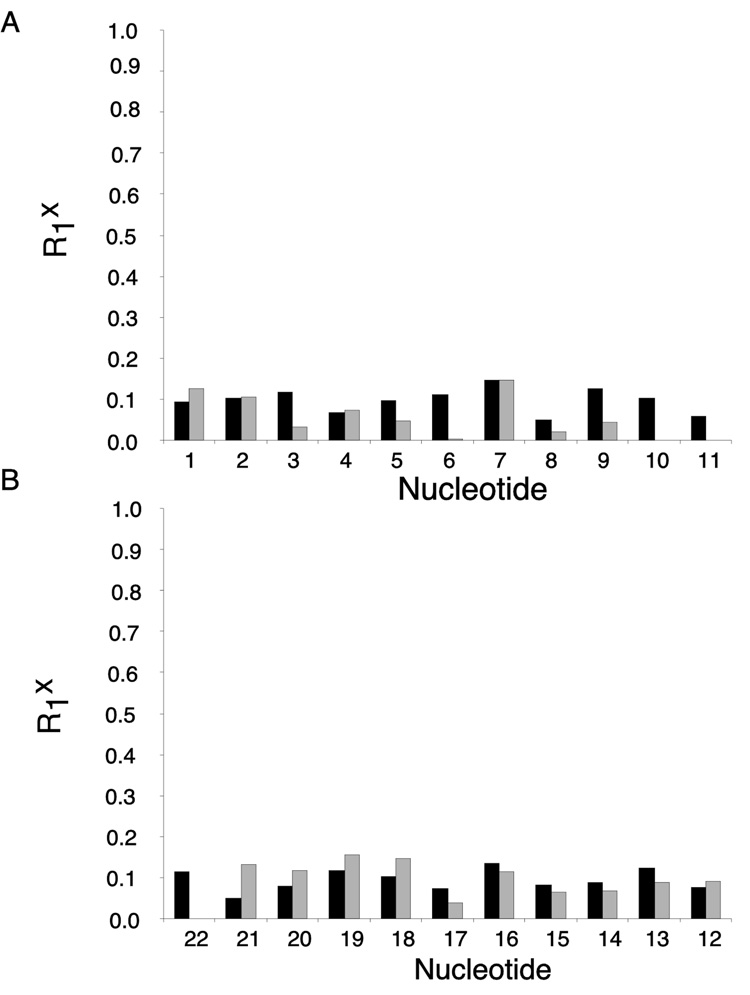
The accuracies of the refined structures were evaluated using complete relaxation matrix calculations, and comparison of the resulting theoretical crosspeak intensities with the 1H NOE data (Figure 7). These yielded sixth root residual (R1x) values of 9.2 × 10−2 for intranucleotide and 9.3 × 10−2 for internucleotide NOEs. The residual values were reasonably consistent over the length of the oligodeoxynucleotide.
Figure 7.
Distribution of sixth root residual (R1x) values calculated using CORMA. A. the modified strand of the R-N1-BDO-(61,2) duplex. B. the complementary strand of the R-N1-BDO-(61,2) duplex. The dark bars represent intranucleotide R1x values. The light bars represent internucleotide R1x values.
Structure of the R-N1-BDO-(61,2) Adduct
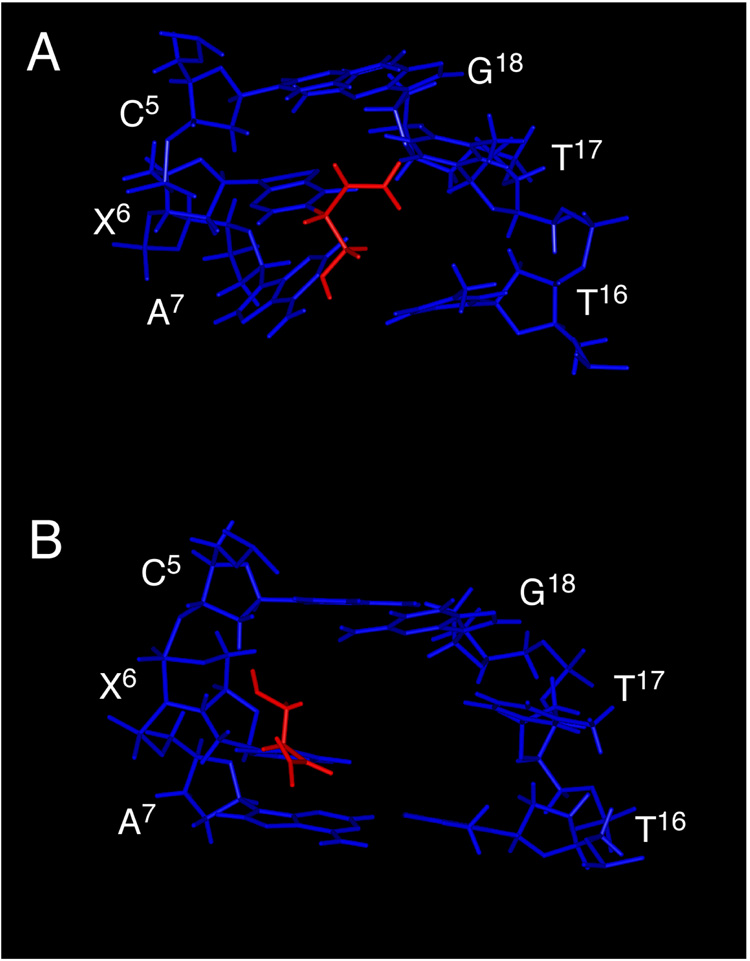
The N1 BD adduct induced a significant structural perturbation to the modified oligodeoxynucleotide (Figure 8). The predominant feature was the syn conformation at the N1-adducted inosine X6. This placed the BD moiety into the major groove of the DNA duplex. The vinyl group was positioned in the 5′ direction of the duplex, while the hydroxymethyl group was positioned in the 3′ direction. The nucleotide complementary to X6, T17, tilted into the minor groove and did not hydrogen bond with X6. Base pair A7•T16 was also disrupted. The purine ring of A7 tilted toward the major groove and was out-of-plane with its complement T16, thus precluding Watson-Crick hydrogen bonding at this base pair. Thus, Watson-Crick hydrogen bonding was disrupted both at the modified base pair X6•T17, and at its 3′-neighbor, A7•T16.
Figure 8.
A. R-N1-BDO-(61,2) Adduct: View from the major groove of X6•T17 and the surrounding base pairs C5•G18, A7•T16 and G8•C15. B. S-N1-BDO-(61,2) Adduct: View from the major groove of X6•T17 and the surrounding base pairs C5•G18, A7•T16 and G8•C15. In both instances the duplex DNA is shown in blue, the BD moiety is in red.
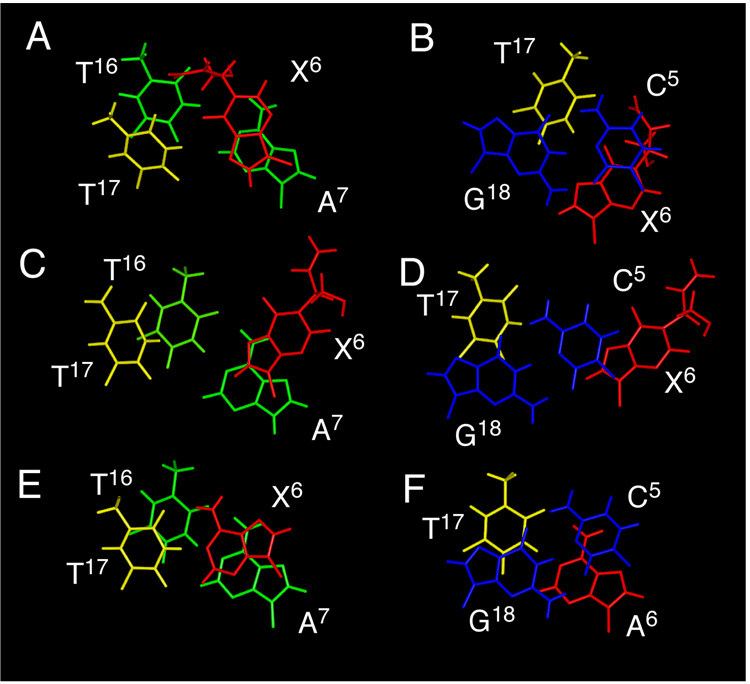
Figure 9 shows the base stacking orientations, as predicted by rMD calculations, at base pairs C5•G18, X6•T17, and A7•T16 of the R-N1-BDO-(61,2) duplex. The rotation of X6 into the syn conformation about the glycosyl bond, placing the N1 BD adduct into the major groove, can be observed. T17, the nucleotide complementary to X6, remained stacked within the double helix. In the modified strand, the imidazole ring of X6 stacked above the pyrimidine ring of A7. The stacking patterns in the complementary strand of the duplex were relatively unperturbed. Figure 9 also compares stacking interactions of the R-N1-BDO-(61,2) duplex with the S-N1-BDO-(61,2) duplex (28) and the unmodified ras61 duplex (31). In both the R- and S-N1-BDO-(61,2) duplexes, the modified nucleotide X6 rotated about the glycosyl bond, in both instances placing the BD moiety into the major groove. The major difference in stacking between the two diastereomeric N1 adducts was an increased base pair opening at X6•T17 for the S-N1-BDO-(61,2) adduct, which was in the high syn conformation (28). The S-N1-BDO-(61,2) adduct exhibited essentially no intrastrand stacking interactions between nucleotides C5, X6, and A7.
Figure 9.
Base stacking orientations of the R-N1-BDO-(61,2) duplex and the S-N1-BDO-(61,2) duplexes at the lesion site as predicted by rMD calculations, as compared to the unmodified ras61 oligodeoxynucleotide duplex. A. The R-N1-BDO-(61,2) duplex detailing base stacking of the X6 and A7 base pairs. B. The R-N1-BDO-(61,2) duplex detailing base stacking of the C5 and X6 base pairs. C. The modified S-N1-BDO-(61,2) duplex detailing base stacking of the X6 and A7 base pairs. D. The S-N1-BDO-(61,2) duplex detailing base stacking of the C5 and X6 base pairs. E. The unmodified ras61 oligodeoxynucleotide duplex detailing base stacking of the A6 and A7 base pairs. F. The unmodified ras61 oligodeoxynucleotide duplex detailing base stacking of the C5 and A6 base pairs.
Discussion
The R-N1-BDO-(61,2) adduct was significantly mutagenic. Like the S-N1-BDO-(61,2) adduct of opposite stereochemistry, it strongly coded for incorporation of dCTP, both in Escherichia coli (26) and in COS-7 cells (27). Studies of the S-N1-BDO-(61,2) adduct revealed the rotation of the glycosyl bond into the high syn conformation. This placed the butadiene moiety into the major groove (28). The earlier studies suggested that butadiene-induced A→G mutations might accrue from three coupled events: (1) alkylation of the N1 imine nitrogen of adenine by butadiene mono- or di-epoxides, (2) deamination of the initially formed N1-dA adducts, and (3) the rotation of the resulting N1-dI adducts about the glycosyl bond into the syn conformation, facilitating Hoogsteen hydrogen bonding with incoming dCTP (27) (Scheme 3). Significantly, this model predicted that deamination was essential to generating A→G mutations, since the N1-dA adduct itself would not be expected to form a Hoogsteen pairing interaction with incoming dCTP. It also predicted that the conformation of the glycosyl bond was the key determinant of mutagenic outcome, as opposed to adduct stereochemistry, thus providing a rationale for the observation that both the R- and S-N1-BDO-(61,2) adducts coded for incorporation of dCTP (26, 27). Consequently, it was of interest to examine the structure of the R-N1-BDO-(61,2) adduct.
Structure of the R-N1-BDO-(61,2) Adduct
The present results indicate, that like the S-N1-BDO-(61,2) adduct, the R-N1-BDO-(61,2) adduct also undergoes rotation about the glycosyl bond, into the syn conformation. The cross-peak intensity between X6 H8 and X6 H1′ was large, indicative of the close proximity of the anomeric and imidazole protons in the syn conformation. The pattern of NOEs between the R-N1-BDO-(61,2) adduct and the DNA firmly established the major groove orientation of the adduct. NOEs were observed between the BD moiety and major groove protons at C5, X6, A7, T16, and T17. The failure to observe NMR resonances from the imino protons at nucleotides T16 and T17, indicative of rapid exchange with solvent (Figure 2) corroborated the predicted loss of Watson-Crick hydrogen bonding at base pairs X6•T17 and A7•T16. The reduction in the melting temperature for the R-N1-BDO-(61,2) adduct to 33° C (50), less than the observed 57° C melting temperature of the unmodified ras61 duplex, was consistent with this conclusion. The altered base stacking at nucleotides C5, X6, and A7 (Figure 10) was observed spectroscopically in the form of chemical shift perturbations clustered at these three base pairs (Figure 6).
Structure-Activity Relationships
(a) Conformation of the Glycosyl Torsion Angle
The finding that for both the R-N1-BDO-(61,2) and S-N1-BDO-(61,2) adducts (28), the N1-adducted dI nucleotide rotated about the glycosyl bond, into the syn (this work) or high syn (S-N1-BDO-(61,2) adduct) conformations supports the notion that the conformation of the glycosyl bond is the key determinant of mutagenic outcome for these N1-dI adducts. Placing the glycosyl bond in the syn or high syn conformation would allow the adducted base to pair with a protonated dCTP during trans-lesion replication (Scheme 3). Simultaneously, rotation about the glycosyl bond places the BD stereocenter at Cβ into the major groove, such that it would not be expected to interfere with Hoogsteen-type templating involving O6 and N7 of the inosine nucleobase. This would presumably yield the observed A→G mutations, irrespective of adduct stereochemistry, consistent with observation (27). Structural studies of the R-N1-BDO-(61,2) and S-N1-BDO-(61,2) adducts opposite dCTP to further examine this hypothesis should now be of considerable interest.
(b) Stereospecific processing
Although the present results suggest that stereochemistry of the R-N1-BDO-(61,2) and S-N1-BDO-(61,2) adducts is not the primary determinant of mutagenic outcome, the two stereoisomeric adducts showed differential levels of mutations. Comparable studies using the R-N1-BDO-(61,2) and S-N1- BDO-(61,2) adducts gave rise to A→G transitions with overall mutagenic frequencies of 60 and 90%, respectively (27). When ligated into 51-mer templates, and examined as to trans-lesion replication in vitro, the R-N1-BDO-(61,2) or the S-N1-BDO-(61,2) adducts posed significant blocks to replication by repair-deficient E. coli polymerase III (26). Replication efficiency of the adduct-containing DNAs were found to be reduced to 10% of the wild type for the S-N1-BDO-(61,2) adduct and 25% for the R-N1-BDO-(61,2) adduct. Collectively, these observations suggest that while conformation of the glycosyl bond is the primary determinant of A→G mutations, adduct stereochemistry provides a secondary contribution with regard to differences in the biological processing of these N1-dI adducts.
One significant structural difference between the R- and S-N1-BDO-(61,2) adducts was the orientation of the BD moiety. In the R-N1-BDO-(61,2) adduct the vinyl group was oriented in the 5′ direction of the adducted strand. In contrast, it was oriented in the 3′ direction in the S-N1-BDO-(61,2) oligodeoxynucleotide (28) (Figure 8). This had several consequences, which may be significant with regard to stereospecific differences in the processing of the N1 lesions. For the R-N1-BDO-(61,2) duplex the vinyl group bridged the floor of the major groove between the adducted nucleotide X6 and its complement, T17. In contrast, for the S-N1-BDO-(61,2) duplex, the vinyl group was oriented away from the floor of the major groove and into solution. The major consequence of this was that in the R-N1-BDO-(61,2) duplex there was less opening of the duplex as compared to the S-N1-BDO-(61,2) duplex. In Figure 8, the large solvent accessible "hole" created in the major groove of the S-N1-BDO-(61,2) duplex can be clearly observed. We speculate that the location of the vinyl group spanning the floor of the major groove in the R-N1-BDO-(61,2) duplex somehow sterically "backstops" the incoming nucleotide triphosphate during trans-lesion replication, the lack of which might increase the propensity for incorporation of dCTP opposite the S-N1-BDO-(61,2) lesion.
The role of stereochemistry in modulating biological processing has been established in studies of adducts arising from polycyclic aromatic hydrocarbons (55). Mao et al. (56) observed stereoselective resistance to digestion of stereoisomeric N2-dG benzo[a]pyrene adducts by phosphodiesterases I and II. Moriya et al. demonstrated that the fidelity of translesion synthesis of such adducts depends upon chirality (57), as does mutagenic potential (58). Choi et al. (59, 60) noted differences in T7 polymerase processing. Likewise, Perlow et al. (61) proposed stereospecific differences for the interactions of these adducts with RNA polymerase II. Overall, stereochemistry appears to modulate the processing of adducts by a number of enzymes (62).
Biological Implications
Mutagenesis studies carried out in vivo with BD using B6c3F1 lacI transgenic mice showed point mutations at both dG and dA, with approximately 20% occurring at A•T base pairs, suggesting that adenine adducts of butadiene are mutagenically significant. The observation that site-specific mutagenesis of these N1-dI adducts in both Escherichia coli (26) and in COS-7 cells (27) yielded A→G transitions was surprising because adenine-specific mutations observed in mice were primarily A→T transversions (63–67), although it was reported that butadiene induced A→G transitions in H-ras codon 61 (68). The present results provide an attractive hypothesis as to the genesis of A→G mutations, i.e., that they are induced by highly mutagenic N1-(1-hydroxy-3-buten-2-yl)-2′-deoxyinosine adducts. The failure to observe high levels of A→G mutations in mice suggests the possibility that these adducts are not present at high levels in vivo. In this regard, it is interesting to note that the more commonly observed A→T mutations could be generated similarly, via incorporation of protonated dATP opposite an N1-dI adduct in the syn conformation about the glycosyl bond (Scheme 3). In this regard, the recent observation of a Hoogsteen-type pair between template dA and incoming TTP in a ternary crystal structure of the human Y-family DNA polymerase ι is of particular interest (69), and is consistent with the notion that the pol ι enzyme might exploit Hoogsteen-type interactions to bypass a variety of bulky DNA adducts.
Supplementary Material
The Supporting Information contains Table S1, showing the 1H chemical shift assignments for the R-N1-BDO-(61,2) adduct, Table S2, showing the NOE restraints utilized in the rMD calculations for the R-N1-BDO-(61,2) adduct, and Table S3, showing force field parameterization and partial charges for the R-N1-BDO-(61,2) adduct. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
Dr. Jaison Jacob assisted with NMR spectroscopy and Dr. Jarrod Smith assisted with molecular modeling.
Footnotes
This work was supported by NIH grant ES-05509 (M.P.S.). Funding for the NMR spectrometers was supplied by Vanderbilt University; by NIH grant RR-05805, and the Vanderbilt Center in Molecular Toxicology, ES-00267. The Vanderbilt-Ingram Cancer Center is supported by NIH grant CA-68485.
Abbreviations: BD, butadiene; BDE, 3,4-epoxy-1,2-butanediol; BDO, butadiene monoepoxide (1,2-epoxy-3-butene); BDO2, butadiene diepoxide (1,2:3,4-diepoxybutane); CPK, Corey-Pauling-Koltun space-filling models; DQF-COSY, double-quantum filtered correlation spectroscopy; HMBC, heteronuclear multiple bond correlation spectroscopy; R1x, sixth root residual; rMD, restrained molecular dynamics; rmsd, root mean square deviation; SBR, styrene-butadiene rubber; TOCSY, total correlation spectroscopy; TPPI, time-proportional phase increment.
The definitions of the prochiral protons at Cα of the BD moiety are based upon the Cahn, Ingold, and Prelog nomenclature. The proton Hα′ is defined as the pro-R proton at Cα′·Hα″ is defined as the pro-S proton at Cα.
References
- 1.Himmelstein MW, Acquavella JF, Recio L, Medinsky MA, Bond JA. Toxicology and epidemiology of 1,3-butadiene. Crit. Rev. Toxicol. 1997;27:1–108. doi: 10.3109/10408449709037482. [DOI] [PubMed] [Google Scholar]
- 2.Jackson MA, Stack HF, Rice JM, Waters MD. A review of the genetic and related effects of 1,3-butadiene in rodents and humans. Mutat. Res. 2000;463:181–213. doi: 10.1016/s1383-5742(00)00056-9. [DOI] [PubMed] [Google Scholar]
- 3.Pelz N, Dempster NM, Shore PR. Analysis of low molecular weight hydrocarbons including 1,3-butadiene in engine exhaust gases using an aluminum oxide porous-layer open-tubular fused-silica column. J. Chromatogr. Sci. 1990;28:230–235. doi: 10.1093/chromsci/28.5.230. [DOI] [PubMed] [Google Scholar]
- 4.Brunnemann KD, Kagan MR, Cox JE, Hoffmann D. Analysis of 1,3-butadiene and other selected gas-phase components in cigarette mainstream and sidestream smoke by gas chromatography-mass selective detection. Carcinogenesis. 1990;11:1863–1868. doi: 10.1093/carcin/11.10.1863. [DOI] [PubMed] [Google Scholar]
- 5.Huff JE, Melnick RL, Solleveld HA, Haseman JK, Powers M, Miller RA. Multiple organ carcinogenicity of 1,3-butadiene in B6c3F1 mice after 60 weeks of inhalation exposure. Science. 1985;227:548–549. doi: 10.1126/science.3966163. [DOI] [PubMed] [Google Scholar]
- 6.Melnick RL, Huff J, Chou BJ, Miller RA. Carcinogenicity of 1,3-butadiene in C57BL/6 x C3H F1 mice at low exposure concentrations. Cancer Res. 1990;50:6592–6599. [PubMed] [Google Scholar]
- 7.Melnick RL, Huff JE, Roycroft JH, Chou BJ, Miller RA. Inhalation toxicology and carcinogenicity of 1,3-butadiene in B6c3F1 mice following 65 weeks of exposure. Environ. Health Perspect. 1990;86:27–36. doi: 10.1289/ehp.908627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owen PE, Glaister JR. Inhalation toxicity and carcinogenicity of 1,3-butadiene in sprague- dawley rats. Environ. Health Perspect. 1990;86:19–25. doi: 10.1289/ehp.908619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.United States Environmental Protection Agency. 1,3-Butadiene. Carcinogenicity assessment for lifetime exposure: Weight-of-evidence characterization. 2002 available at http://www.epa.gov/iris/subst/0139.htm.
- 10.International Agency for Research on Cancer. Re-evaluation of some organic chemicals, hydrazine and hydrogen peroxide, IRC Monographs on the Evaluation of Carcinogenic Risks to Humans. IARC Sci. Publ. 1999;71:109–125. [PMC free article] [PubMed] [Google Scholar]
- 11.Ward JB, Jr, Ammenheuser MM, Bechtold WE, Whorton EB, Jr, Legator MS. hprt mutant lymphocyte frequencies in workers at a 1,3-butadiene production plant. Environ. Health Perspect. 1994;102 Suppl 9:79–85. doi: 10.1289/ehp.94102s979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ward JB, Jr, Ammenheuser MM, Whorton EB, Jr, Bechtold WE, Kelsey KT, Legator MS. Biological monitoring for mutagenic effects of occupational exposure to butadiene. Toxicology. 1996;113:84–90. doi: 10.1016/0300-483x(96)03431-2. [DOI] [PubMed] [Google Scholar]
- 13.Sram RJ, Rossner P, Peltonen K, Podrazilova K, Mrackova G, Demopoulos NA, Stephanou G, Vlachodimitropoulos D, Darroudi F, Tates AD. Chromosomal aberrations, sister-chromatid exchanges, cells with high frequency of SCE, micronuclei and comet assay parameters in 1,3-butadiene-exposed workers. Mutat. Res. 1998;419:145–154. doi: 10.1016/s1383-5718(98)00135-1. [DOI] [PubMed] [Google Scholar]
- 14.Delzell E, Sathiakumar N, Hovinga M, Macaluso M, Julian J, Larson R, Cole P, Muir DC. A follow-up study of synthetic rubber workers. Toxicology. 1996;113:182–189. doi: 10.1016/0300-483x(96)03443-9. [DOI] [PubMed] [Google Scholar]
- 15.Meinhardt TJ, Lemen RA, Crandall MS, Young RJ. Environmental epidemiologic investigation of the styrene-butadiene rubber industry. Mortality patterns with discussion of the hematopoietic and lymphatic malignancies. Scand. J. Work Environ. Health. 1982;8:250–259. doi: 10.5271/sjweh.2469. [DOI] [PubMed] [Google Scholar]
- 16.Matanoski G, Francis M, Correa-Villasenor A, Elliott E, Santos-Burgoa C, Schwartz L. Cancer epidemiology among styrene-butadiene rubber workers. IARC Sci. Publ. 1993;127:363–374. [PubMed] [Google Scholar]
- 17.Santos-Burgoa C, Matanoski GM, Zeger S, Schwartz L. Lymphohematopoietic cancer in styrene-butadiene polymerization workers. Am. J. Epidemiol. 1992;136:843–854. doi: 10.1093/aje/136.7.843. [see comments]. [DOI] [PubMed] [Google Scholar]
- 18.Macaluso M, Larson R, Delzell E, Sathiakumar N, Hovinga M, Julian J, Muir D, Cole P. Leukemia and cumulative exposure to butadiene, styrene and benzene among workers in the synthetic rubber industry. Toxicology. 1996;113:190–202. doi: 10.1016/0300-483x(96)03444-0. [DOI] [PubMed] [Google Scholar]
- 19.Matanoski GM, Schwartz L. Mortality of workers in styrene-butadiene polymer production. J. Occup. Med. 1987;29:675–680. [PubMed] [Google Scholar]
- 20.Santos-Burgoa C, Eden-Wynter RA, Riojas-Rodriguez H, Matanoski GM. Living in a chemical world. Health impact of 1,3-butadiene carcinogenesis. Ann. N. Y. Acad. Sci. 1997;837:176–188. doi: 10.1111/j.1749-6632.1997.tb56874.x. [DOI] [PubMed] [Google Scholar]
- 21.Matanoski G, Elliott E, Tao X, Francis M, Correa-Villasenor A, Santos-Burgoa C. Lymphohematopoietic cancers and butadiene and styrene exposure in synthetic rubber manufacture. Ann. N. Y. Acad. Sci. 1997;837:157–169. doi: 10.1111/j.1749-6632.1997.tb56872.x. [DOI] [PubMed] [Google Scholar]
- 22.Albertini R, Clewell H, Himmelstein MW, Morinello E, Olin S, Preston J, Scarano L, Smith MT, Swenberg J, Tice R, Travis C. The use of non-tumor data in cancer risk assessment: Reflections on butadiene, vinyl chloride, and benzene. Regul. Toxicol. Pharmacol. 2003;37:105–132. doi: 10.1016/s0273-2300(02)00019-3. [DOI] [PubMed] [Google Scholar]
- 23.Csanady GA, Guengerich FP, Bond JA. Comparison of the biotransformation of 1,3-butadiene and its metabolite, butadiene monoepoxide, by hepatic and pulmonary tissues from humans, rats and mice [published erratum appears in Carcinogenesis 1993 Apr;14(4):784] Carcinogenesis. 1992;13:1143–1153. doi: 10.1093/carcin/13.7.1143. [DOI] [PubMed] [Google Scholar]
- 24.Duescher RJ, Elfarra AA. Human liver microsomes are efficient catalysts of 1,3-butadiene oxidation: evidence for major roles by cytochromes P450 2A6 and 2E1. Arch. Biochem. Biophys. 1994;311:342–349. doi: 10.1006/abbi.1994.1246. [DOI] [PubMed] [Google Scholar]
- 25.Selzer RR, Elfarra AA. Characterization of N1- and N6-adenosine adducts and N1-inosine adducts formed by the reaction of butadiene monoxide with adenosine: Evidence for the N1-adenosine adducts as major initial products. Chem. Res. Toxicol. 1996;9:875–881. doi: 10.1021/tx960039a. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez DA, Kowalczyk A, Ward JBJ, Harris CM, Harris TM, Lloyd RS. Point mutations induced by 1,2-epoxy-3-butene N1 deoxyinosine adducts. Environ. Mol. Mutagen. 2001;38:292–296. doi: 10.1002/em.10026. [DOI] [PubMed] [Google Scholar]
- 27.Kanuri M, Nechev LV, Tamura PJ, Harris CM, Harris TM, Lloyd RS. Mutagenic spectrum of butadiene-derived N1-deoxyinosine adducts and N6,N6-deoxyadenosine intrastrand cross-links in mammalian cells. Chem. Res. Toxicol. 2002;15:1572–1580. doi: 10.1021/tx025591g. [DOI] [PubMed] [Google Scholar]
- 28.Scholdberg TA, Merritt WK, Dean SM, Kowalczyk A, Harris CM, Harris Tm, Lloyd RS, Stone MP. Structure of an oligodeoxynucleotide containing a butadiene oxide-Derived N1-hydroxyalkyl deoxyinosine adduct in the human N-ras codon 61 sequence. Biochemistry. 2005;44 doi: 10.1021/bi0482452. in press. [DOI] [PubMed] [Google Scholar]
- 29.Schmitz U, James TL. How to generate accurate solution structures of double-helical nucleic acid fragments using nuclear magnetic resonance and restrained molecular dynamics. Meth. Enzymol. 1995;261:3–44. doi: 10.1016/s0076-6879(95)61003-0. [DOI] [PubMed] [Google Scholar]
- 30.Borer PN. Handbook of Biochemistry and Molecular Biology. Cleveland, OH: CRC Press; 1975. [Google Scholar]
- 31.Feng B, Stone MP. Solution structure of an oligodeoxynucleotide containing the human N-ras codon 61 sequence refined from 1H NMR using molecular dynamics restrained by nuclear Overhauser effects. Chem. Res. Toxicol. 1995;8:821–832. doi: 10.1021/tx00048a002. [DOI] [PubMed] [Google Scholar]
- 32.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 33.James TL. Relaxation matrix analysis of two-dimensional nuclear Overhauser effect spectra. Curr. Opin. Struct. Biol. 1991;1:1042–1053. [Google Scholar]
- 34.Keepers JW, James TL. A theoretical study of distance determination from NMR. Two-dimensional nuclear Overhauser effect spectra. J. Magn. Reson. 1984;57:404–426. [Google Scholar]
- 35.Borgias BA, James TL. Two-dimensional nuclear Overhauser effect: Complete relaxation matrix analysis. Meth. Enzymol. 1989;176:169–183. doi: 10.1016/0076-6879(89)76011-0. [DOI] [PubMed] [Google Scholar]
- 36.Borgias BA, James TL. MARDIGRAS--a procedure for matrix analysis of relaxation for discerning geometry of an aqueous structure. J. Magn. Reson. 1990;87:475–487. [Google Scholar]
- 37.Liu H, Spielmann HP, Ulyanov NB, Wemmer DE, James TL. Interproton distance bounds from 2D NOE intensities: effect of experimental noise and peak integration errors. J. Biomol. NMR. 1995;6:390–402. doi: 10.1007/BF00197638. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Zuiderweg ERP, Glick GD. Solution structure of a disulfide cross-linked DNA hairpin. J. Am. Chem. Soc. 1995;117:2981–2991. [Google Scholar]
- 39.Geen H, Freeman R. Band-selective radiofrequency pulses. J. Magn. Reson. 1991;93:93–141. [Google Scholar]
- 40.Lankhorst PP, Haasnoot AG, Erkelens C, Altona C. Carbon-13 NMR in conformational analysis of nucleic acid fragments. 3. The magnitude of torsional angle in d(TpA) from CCOP and HCOP NMR coupling constants. Nucleic Acids Res. 1984;12:5419–5428. doi: 10.1093/nar/12.13.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnott S, Hukins DWL. Optimised parameters for A-DNA and B-DNA. Biochem. Biophys. Res. Comm. 1972;47:1504–1509. doi: 10.1016/0006-291X(72)90243-4. [DOI] [PubMed] [Google Scholar]
- 42.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer RJ, Duan Y, Pitera J, Massova I, Seibel GL, Singh UC, Weiner PK, Kollman PA. AMBER 7.0. San Francisco, CA: University of California; 2002. [Google Scholar]
- 43.Clore GM, Brunger AT, Karplus M, Gronenborn AM. Application of olecular dynamics with interproton distance restraints to three-dimensional protein structure determination. J. Mol. Biol. 1986;191:523–551. doi: 10.1016/0022-2836(86)90146-4. [DOI] [PubMed] [Google Scholar]
- 44.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comp. Phys. 1977;23:327–341. [Google Scholar]
- 45.Bashford D, Case DA. Generalized Born models of macromolecular solvation effects. Ann. Rev. Phys. Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 46.Tsui V, Case DA. Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers. 2000;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 47.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179. [Google Scholar]
- 48.Keepers J, Kollman PA, James TL. Molecular mechanical studies of base-pair opening in d(CGCGC):d(GCGCG), dG5:dC5, d(TATAT):d(ATATA), and dA5:dT5 in the B and Z forms of DNA. Biopolymers. 1984;23:2499–2511. doi: 10.1002/bip.360231124. [DOI] [PubMed] [Google Scholar]
- 49.Lu XJ, Olson WK. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kowalczyk A, Harris CM, Harris TM. Synthesis and characterization of oligodeoxynucleotides containing an N1 beta-hydroxyalkyl adduct of 2′-deoxyinosine. Chem. Res. Toxicol. 2001;14:746–753. doi: 10.1021/tx010025r. [DOI] [PubMed] [Google Scholar]
- 51.Patel DJ, Shapiro L, Hare D. DNA and RNA: NMR studies of conformations and dynamics in solution. Q. Rev. Biophys. 1987;20:35–112. doi: 10.1017/s0033583500004224. [DOI] [PubMed] [Google Scholar]
- 52.Reid BR. Sequence-specific assignments and their use in NMR studies of DNA structure. Q. Rev. Biophys. 1987;20:2–28. doi: 10.1017/s0033583500004212. [DOI] [PubMed] [Google Scholar]
- 53.Weisz K, Shafer RH, Egan W, James TL. Solution structure of the octamer motif in immunoglobulin genes via restrained molecular dynamics calculations. Biochemistry. 1994;33:354–366. doi: 10.1021/bi00167a046. [DOI] [PubMed] [Google Scholar]
- 54.Nikonowicz EP, Gorenstein DG. Two-dimensional 1H and 31P NMR spectra and restrained molecular dynamics structure of a mismatched GA decamer oligodeoxyribonucleotide duplex. Biochemistry. 1990;29:8845–8858. doi: 10.1021/bi00489a048. [DOI] [PubMed] [Google Scholar]
- 55.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisomeric covalent polycyclic aromatic carcinogen-DNA adducts: Principles, Patterns and diversity. Chem. Res. Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 56.Mao B, Li B, Amin S, Cosman M, Geacintov NE. Opposite stereoselective resistance to digestion by phosphodiesterases I and II of benzo[a]pyrene diol epoxide-modified oligonucleotide adducts. Biochemistry. 1993;32:11785–11793. doi: 10.1021/bi00095a006. [DOI] [PubMed] [Google Scholar]
- 57.Moriya M, Spiegel S, Fernandes A, Amin S, Liu T, Geacintov N, Grollman AP. Fidelity of translesional synthesis past benzo[a]pyrene diol epoxide-2′-deoxyguanosine DNA adducts: Marked effects of host cell, sequence context, and chirality. Biochemistry. 1996;35:16646–16651. doi: 10.1021/bi9608875. [DOI] [PubMed] [Google Scholar]
- 58.Fernandes A, Liu T, Amin S, Geacintov NE, Grollman AP, Moriya M. Mutagenic potential of stereoisomeric bay region (+)- and (−)-cis-anti-benzo[a]pyrene diol epoxide-N2-2′-deoxyguanosine adducts in Escherichia coli and simian kidney cells. Biochemistry. 1998;37:10164–10172. doi: 10.1021/bi980401f. [DOI] [PubMed] [Google Scholar]
- 59.Choi DJ, Marino-Alessandri DJ, Geacintov NE, Scicchitano DA. Site-specific benzo[a]pyrene diol epoxide-DNA adducts inhibit transcription elongation by bacteriophage T7 RNA polymerase. Biochemistry. 1994;33:780–787. doi: 10.1021/bi00169a020. [DOI] [PubMed] [Google Scholar]
- 60.Choi DJRR, Liu T, Geacintov NE, Scicchitano DA. Incorrect base insertion and prematurely terminated transcripts during T7 RNA polymerase transcription elongation past benzo[a]pyrenediol epoxide-modified DNA. J. Mol. Biol. 1996;264:213–219. doi: 10.1006/jmbi.1996.0635. [DOI] [PubMed] [Google Scholar]
- 61.Perlow RAKA, Hingerty BE, Geacintov NE, Broyde S, Scicchitano DA. DNA adducts from a tumorigenic metabolite of benzo[a]pyrene block human RNA polymerase II elongation in a sequence- and stereochemistry-dependent manner. J. Mol. Biol. 2002;321:29–47. doi: 10.1016/s0022-2836(02)00593-4. [DOI] [PubMed] [Google Scholar]
- 62.Shukla R, Jelinsky S, Liu T, Geacintov NE, Loechler EL. How stereochemistry affects mutagenesis by N2-deoxyguanosine adducts of 7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene: Configuration of the adduct bond is more important than those of the hydroxyl groups. Biochemistry. 1997;36:13263–13269. doi: 10.1021/bi971195z. [DOI] [PubMed] [Google Scholar]
- 63.Recio L, Saranko CJ, Steen AM. 1,3-butadiene: Cancer, mutations, and adducts. Part II: Roles of two metabolites of 1,3-butadiene in mediating its in vivo genotoxicity. Res. Rep. Health Eff. Inst. 2000:49–87. discussion 141–149. [PubMed] [Google Scholar]
- 64.Recio L, Steen AM, Pluta LJ, Meyer KG, Saranko CJ. Mutational spectrum of 1,3-butadiene and metabolites 1,2-epoxybutene and 1,2:3,4-diepoxybutane to assess mutagenic mechanisms. Chem. Biol. Interact. 2001;135–136:325–341. doi: 10.1016/s0009-2797(01)00220-4. [DOI] [PubMed] [Google Scholar]
- 65.Sisk SC, Pluta LJ, Bond JA, Recio L. Molecular analysis of lacI mutants from bone marrow of B6c3F1 transgenic mice following inhalation exposure to 1,3-butadiene. Carcinogenesis. 1994;15:471–477. doi: 10.1093/carcin/15.3.471. [DOI] [PubMed] [Google Scholar]
- 66.Recio L, Meyer KG. Increased frequency of mutations at A:T base pairs in the bone marrow of B6c3F1 laci transgenic mice exposed to 1,3-butadiene. Environ. Mol. Mutagen. 1995;26:1–8. doi: 10.1002/em.2850260102. [DOI] [PubMed] [Google Scholar]
- 67.Steen AM, Meyer KG, Recio L. Analysis of hprt mutations occurring in human TK6 lymphoblastoid cells following exposure to 1,2:3,4-diepoxybutane. Mutagenesis. 1997;12:61–67. doi: 10.1093/mutage/12.2.61. [DOI] [PubMed] [Google Scholar]
- 68.Goodrow TL, Nichols WW, Storer RD, Anderson MW, Maronpot RR. Activation of H-ras is prevalent in 1,3-butadiene-induced and spontaneously occurring murine Harderian gland tumors. Carcinogenesis. 1994;15:2665–2667. doi: 10.1093/carcin/15.11.2665. [DOI] [PubMed] [Google Scholar]
- 69.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase ι occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supporting Information contains Table S1, showing the 1H chemical shift assignments for the R-N1-BDO-(61,2) adduct, Table S2, showing the NOE restraints utilized in the rMD calculations for the R-N1-BDO-(61,2) adduct, and Table S3, showing force field parameterization and partial charges for the R-N1-BDO-(61,2) adduct. This material is available free of charge via the Internet at http://pubs.acs.org.