Abstract
The major virulence cluster of Listeria monocytogenes harbors six virulence genes that encode proteins critical for the intracellular life cycle of this human and animal pathogen. In this study, we determined the sequence (8,709 nt) of the virulence gene cluster (including the six main virulence genes) in 40 L. monocytogenes isolates from different source populations (human clinical cases, animal clinical cases, foods, and natural environments). An alignment of the full length cluster as well as individual gene alignments and alignments of intragenic regions were used for phylogenetic, recombination, and positive selection analyses. Initial phylogenetic analyses showed that the sequences represented two main clusters, consistent with previously defined L. monocytogenes phylogenetic lineages. The 40 sequences represented 25 distinct allelic types and the overall alignment included 592 polymorphic sites. Overall, our data show that (i) virulence genes in the main L. monocytogenes virulence gene cluster include highly conserved genes (i.e., hly, prfA) as well as diverse genes that appear to have evolved by positiveselection (mpl, actA, plcA), (ii) recombination has played an important role in the evolution of the virulence gene cluster, but is limited to lineage II isolates, and (iii) the promoter region driving the transcription of virulence genes transcribed early in intracellular infection (i.e., hly, plcA) has evolved by positive selection. The genes and intragenic regions in the L. monocytogenes virulence gene cluster thus have evolved independently, despite their close physical linkage, likely reflecting distinct selective pressures associated with expression and function of the proteins encoded in this region.
1. Introduction
Listeria monocytogenes is a Gram-positive facultative intracellular pathogen that can cause gastroenteritis as well as invasive disease in humans and animals (Vazquez-Boland et al., 2001). Human listeriosis is typically foodborne and L. monocytogenes can be found in many different environments, including food processing plants, farms, and urban and natural environments (Lappi et al., 2004; Sauders et al., 2006). A number of virulence genes important in the intracellular life cycle of L. monocytogenes have been identified. Six key L. monocytogenes virulence genes (i.e., prfA, plcA, hly, mpl, actA, and plcB) that are critical for the intracellular life cycle, including vacuolar escape and cell-to-cell spread, are located in a virulence gene island, which is often referred to as the “prfA virulence gene cluster (pVGC)“ (Ward et al., 2004) or the “Listeria pathogenicity island 1 (LIPI-1)” (Vazquez-Boland et al., 2001). hly encodes listeriolysin O (LLO), which facilitates L. monocytogenes escape from the host cell vacuole. plcA and plcB encode the two phospholipases, (PI-PLC and PC-PLC), which also contribute to disruption of the host cell vacuole membrane (Gaillard et al., 1987; Smith et al., 1995), while mpl encodes a zinc metalloproteinase needed to activate PC-PLC. actA encodes the ActA protein, which is responsible for host actin accumulation and motility. prfA, which can be transcribed from at least three different promoters (Camilli et al., 1993; Freitag et al., 1993), encodes the transcriptional factor PrfA; PrfA positively regulates transcription of the other genes in the virulence cluster as well as of some other virulence genes (e.g., inlA, inlB) (Chakraborty et al., 1992; Domann et al., 1992; Leimeister-Wachter et al., 1990; Scortti et al., 2007). PrfA-dependent transcription of virulence genes appears to be hierarchical, such that virulence genes needed early in the intracellular life cycle (e.g. hly, plcA) are more rapidly activated than genes required later (e.g., actA) (Sheehan et al., 1995). While hly and plcA are transcribed in opposite directions using a shared PrfA binding site that overlaps with the −35 site for both promoters, hly can also be transcribed from a second putatively PrfA-independent promoter (Mengaud et al.,1989). A PrfA-dependent promoter upstream of mpl generate a monocistronic mpl transcript as well as a polycistronic mpl-actA-plcB transcript, while another PrfA-dependent promoter upstream of actA can generate an actA as well as an actA-plcB transcript (reviewed in Kreft and Vazquez-Boland, 2001). In addition to the coding sequences, the prfA virulence gene cluster thus includes intergenic regions with complex regulatory elements that are needed for virulence of L. monocytogenes.
L. monocytogenes is a diverse organism with a structured population that includes at least three phylogenetic lineages (Rasmussen et al., 1995). While two lineages (I and II) are common, lineage III appears to be uncommon (Jeffers et al., 2001; Nightingale et al., 2004; Sauders et al., 2006). Although isolates from all lineages seem to be capable of causing disease, epidemiological and phenotypic differences among these lineages have been observed. Lineage II strains, although the most prevalent in foods, are underrepresented among human sporadic listeriosis cases and are seldom associated with human listeriosis outbreaks (De Cesare et al.,2007; Gray et al., 2004; Jeffers et al., 2001; McLauchlin et al., 2004; Norton et al., 2001). Conversely, lineage I isolates, and in particular lineage I serotype 4b strains, have been associated with most human outbreaks and the majority of human listeriosis cases in most countries despite their lower prevalence in foods (McLauchlin et al., 2004). These epidemiological findings suggested that all or some lineage I strains could be more virulent than lineage II strains, a hypothesis that has been supported by some in vitro assays, including tissue culture assays that found that lineage I isolates, on average, seem to form more, and larger plaques in mouse L cells (Gray et al., 2004; Norton et al., 2001; Wiedmann et al., 1997). Moreover, several lineage II isolates were shown to carry nonsense or frameshift mutations in virulence genes (particularly in inlA), which may lead to virulence attenuation (Rousseaux et al.,2004; Nightingale et al., 2005a; Orsi et al., 2007; Roche et al., 2005; Velge et al., 2007).
The goal of this study was to use evolutionary analyses (including positive selection and recombination analyses) of both coding and intergenic regions in the prfA cluster to improve our understanding of the evolution and adaptation of this virulence gene cluster in L. monocytogenes, using isolates representing different lineages and obtained from different source population (i.e., human and animal clinical cases, foods, and natural environments). While one study (Ward et al., 2004) has previously used DNA sequence data for the prfA cluster for a phylogenetic analyses, comprehensive analyses on the contributions of recombination and positive selection to the evolution of this virulence gene cluster have been missing so far. In particular, positive selection analyses have only been reported for a fragment of one virulence gene (i.e., actA) in the prfA cluster (Nightingale et al., 2005b) and no positive selection analyses have been reported for the non-coding and regulatory regions in the prfA cluster.
2. Methods
2.1. Listeria monocytogenes isolates
The 40 L. monocytogenes isolates used here were selected from a larger set of 132 isolates as previously described (Orsi et al., 2007; Tsai et al., 2006); DNA sequences for selected internalin genes had previously been reported and analyzed for these isolates (Orsi et al., 2007; Tsai et al., 2006). Briefly, the original set of 132 isolates represented isolates from four source populations, including human clinical cases (n = 60), animal clinical cases (n = 30), foods (n = 30), and pristine environment (n = 12); these isolates had been obtained from two previous studies, which provide details on these isolates (Nightingale et al., 2005b; Sauders et al., 2006). For each of the four source populations, ten isolates were randomly selected (using a random number table) to yield the set of 40 isolates used here (Supplemental Table 1; all supplemental are available at http://www.foodscience.cornell.edu/cals/foodsci/research/labs/wiedmann/links/orsi_2008.cfm). Serotypes were previously reported for 30 isolates (Nightingale et al., 2005b) and were determined for 10 isolates using the methods previously described (Nadon et al., 2001); serotype data were used to determine clustering of serotypes within a lineage.
2.2. DNA amplification and sequencing
The L. monocytogenes virulence gene cluster (see Figure 1 for a schematic) was amplified using a total of eight separate PCRs (see Supplemental Table 2 for primers). PCR products were purified using the Qiaquick Purification kit (Qiagen) and sequenced using PCR primers and internal primers (Supplemental Table 2); sequencing was performed (at the Biotechnology Resource Center, Cornell University) using Big Dye Terminator chemistry and AmpliTaq-FS DNA Polymerase. Sequences were proofread and assembled using Seqman (Lasergene). The sequences used for the full virulence gene cluster alignment have been deposited in GenBank (accession numbers EU372018 to EU372057).
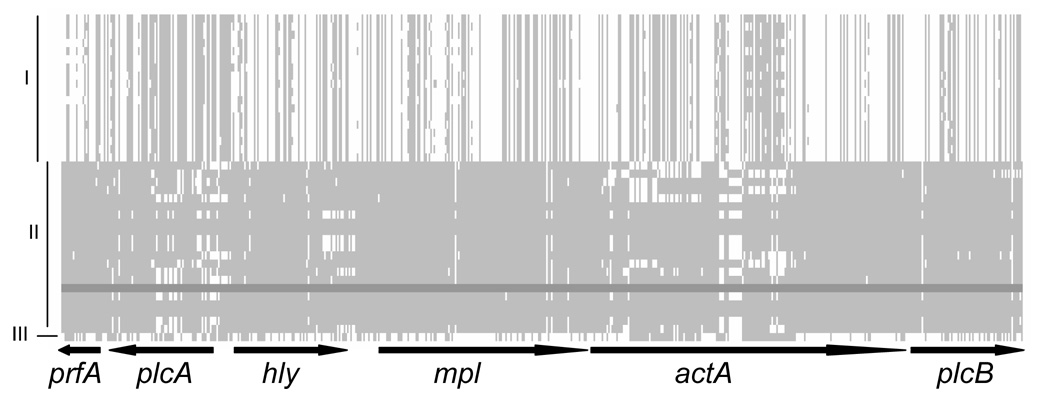
Figure 1.
Alignment of polymorphic sites in the virulence cluster. Gaps are not considered. Lineages are labeled as I, II and III. A lineage II isolate (FSL S4–497) was used as reference (dark gray shade). Lightly shaded areas represent nucleotides in other isolates that match the nucleotide present in FSL S4–497 for a given site. Regions in white within lineage II sequences may represent recombination events. Schematic of the coding regions are shown below the alignment. The lengths of the coding regions are proportional to the number of polymorphic sites within each coding region.
2.3. Descriptive analyses
Clustal W implemented in Megalign (Lasergene) was used to create alignments of each gene and of the whole virulence cluster. DNASP version 4.0 (Rozas and Rozas, 1999) was used to assess the number of allelic types, number of polymorphic sites (S), number of nonsynonymous (NNS) and synonymous substitutions (NSS), and nucleotide diversity (π) in each alignment.
2.4. Phylogenetic analysis
The model of DNA substitution that best fit each data was identified using MODELTEST (Posada and Crandall, 1998) and was used to create phylogenetic trees using the maximum likelihood method implemented in PAUP (http://paup.csit.fsu.edu/). The phylogeny for the full virulence gene cluster was rooted using the L. ivanovii subsp. londoniensis NRRL 33021 sequence (GenBank accession number AY510073) previously published (Ward et al., 2004). Support for each branch was assessed by the bootstrap procedure in PAUP (100 replicates).
2.5. Positive selection analyses of coding regions
Alignments and trees generated for each of the six coding regions within the virulence gene cluster were used to assess whether a given gene had evolved by positive selection; only the sequences for the 40 L. monocytogenes isolates were used for these analyses. Trees were generated using the maximum likelihood method as described above. Positive selection in coding regions can be assessed by comparing the rate of nonsynonymous substitutions (dN) to the rate of synonymous substitutions (dS) in that region. By assuming that the rate of synonymous mutation is constant and not targeted by natural selection, a dN/dS > 1 indicates that nonsynonymous mutations were more likely to be fixed in the population than synonymous mutations suggesting positive selection of nonsynonymous mutations. While some studies use an estimation of a single average dN/dS for a whole gene to identify genes under positive selection, the approaches implemented “Phylogenetic Analysis using Maximum Likelihood” (PAML) provide an advancement over these methods by allowing for detection of positive selection at specific sites and in specific lineages (Lefebure and Stanhope, 2007; Yang et al., 2000; Zhang et al., 2005). In PAML, three null models, in which positive selection is not allowed, and three alternative models, in which positive selection is allowed, can be compared using a Likelihood Ratio Test (LRT) to assess whether the alternative model is significantly better than the null model. If an alternative model is accepted over the null model, one can conclude that a given gene evolved by positive selection (Yang et al., 2000). Specifically, the null models M0, M1a, and M7 are compared to the alternative models M3, M2a, and M8, respectively. M0 assumes only one class of ω along the sequence whereas M3 assumes three classes of ω; the comparison between these two models can be used to determine whether selection differed along a given sequence (suggesting, for example, that regions in a gene differ in their selection). M1a assumes two classes of codon sites each with a different value of ω (i.e., ω0 < 1 and ω1 = 1), while M2a assumes one additional class of sites (i.e., ω2) with an ω > 1. Similarly, M7 assumes ten classes of sites with ω following a beta distribution that approximate a continuous distribution constrained to values between 0 and 1, while M8 assumes an extra class of sites with ω1 > 1. Hence, comparisons between M1a and M2a as well as comparisons between M7 and M8 assess whether a gene has evolved by positive selection; acceptance of an alternative model (i.e., M2a or M8) suggests that a given gene evolved by positive selection. However, the test between M7 and M8 is less conservative than the test between M1a and M2a as models M7 and M8 do not assume a class of sites with ω = 1. A Bayes Empirical Bayes (BEB) approach implemented in PAML was used to further identify specific codon sites with a significant posterior probability of having evolved by positive selection.
In addition to the tests described above, branch-site models as described by Zhang et al. (2005) were used to assess whether a given gene evolved by positive selection within specific branches in a tree; we specifically tested branches within L. monocytogenes lineage I or II for positive selection in each gene. Similar to the tests described above, the branch-site test relies on the comparison of a null model with an alternative model. While both the null model and the alternative model have four site classes, only the alternative model allows positive selection (i.e., ω classes with ω > 1) in the branch(es) of interest (Zhang et al., 2005). Therefore, if the alternative model is accepted over the null model, one can conclude that a given gene evolved under positive selection in the branch(es) of interest. A BEB approach was also used to identify specific codon sites evolving by positive selection in the branches of interest. All analyses for assessing evolution by positive selection were carried out using PAML version 3.15.
2.6. Positive selection analyses of the non-coding regions
Six separate alignments (each including a given gene and its upstream and downstream non-coding regions) were used to construct phylogenetic trees; these alignments and phylogenetic trees were then used to identify non-coding fragments that have evolved by positive selection using the program EvoNC (Wong and Nielsen, 2004). Like PAML, EvoNC assumes that synonymous mutations are neutral and happen in a constant rate within the coding fragment. Hence, calculating the ratio of the estimated nucleotide substitution rate in the non-coding region to the estimated synonymous substitution rate in the coding region ratio (this ratio is designated as ζ) provides a numerical approach to assess whether a non-coding region has evolved by positive selection. Three models implemented in EvoNC can be used in two different tests to determine whether a non-coding region is under positive selection; the neutral model assumes two classes of nucleotide sites with ζ0 < 1 and ζ1 = 1; the two-category model assumes two classes with ζ0 < 1 and ζ1 ≥ 1; and the three-category model assumes three classes of nucleotides with: ζ0 < 1, ζ1 = 1, and ζ2 > 1. The test comparing the neutral model and the three-category model is called Test 1; Test 1 is more conservative than Test 2, which compares the neutral model and the two-category model (Wong and Nielsen, 2004).
2.7. Recombination analyses
The Sawyer’s test implemented in GENECONV (http://www.math.wustl.edu/~sawyer) was initially used to identify recombinant fragments within the virulence cluster, using the default settings of the program. As previously described (Nightingale et al., 2005b), GENECONV fragments that shared the same 5’ or 3’ ends were considered to represent the same recombination event.
Clonal Frame v1.1 (Didelot and Falush, 2007) was used to identify isolates that have recently imported external DNA fragments in the virulence cluster. Two independent runs of the program were used to estimate a 95% consensus tree. Branches supported by the tree were analyzed for recombination. Only recombination events with probability higher than 90% were considered. The program was run with 100,000 burn-in iteration and 100,000 sampling iteration.
3. Results
3.1. Descriptive analysis
The alignment of the virulence gene cluster for the 40 L. monocytogenes isolates was 8,709 bp long with an average G + C content of 36.3 %. A total of 7,984 sites were monomorphic; 133 sites in the alignment represented indels (insertions or deletions). Overall, 592 polymorphic sites (S) were identified (excluding the sites with indels), including 117 singletons (i.e., a polymorphic sites with substitution in only one isolate) and 475 informative sites (451 and 24 sites with two and three nucleotide variants, respectively). The 40 sequences represented 25 unique virulence gene cluster alleles. The overall nucleotide diversity (π) was estimated as 0.0246; π was significantly higher for lineage II isolates (21 isolates; πII = 0.0064) than lineage I isolates (18 isolates, πI = 0.0030) (Table 1). Lineage I and II isolates shared nine substitutions whereas 267 were fixed in either lineage I or II.
Table 1.
Descriptive analysis of the virulence gene cluster nt sequence among 40 L. monocytogenes isolates.
| Fragment (length in nt)a | Hb | Sc | PISd | Substitutions | NSSe | NNSf | πg | π (NSS) | π (NNS) |
|---|---|---|---|---|---|---|---|---|---|
| Cluster (8709), all | 25 | 592 | 475 | 616 | NA | NA | 0.0246 | NA | NA |
| Lineage I | 12 | 75 | 64 | 75 | NA | NA | 0.0030 | NA | NA |
| Lineage II | 12 | 178 | 147 | 181 | NA | NA | 0.0064 | NA | NA |
| prfA (711), all | 9 | 25 | 23 | 25 | 23 | 2 | 0.0143 | 0.0684 | 0.0003 |
| Lineage I | 4 | 6 | 6 | 6 | 6 | 0 | 0.0028 | 0.0129 | 0 |
| Lineage II | 4 | 3 | 0 | 3 | 1 | 2 | 0.0004 | 0.0006 | 0.0003 |
| plcA (951), all | 18 | 66 | 55 | 67 | 50 | 17 | 0.0211 | 0.0729 | 0.0072 |
| Lineage I | 8 | 7 | 6 | 7 | 5 | 2 | 0.0028 | 0.0098 | 0.0009 |
| Lineage II | 9 | 35 | 33 | 35 | 25 | 10 | 0.0138 | 0.0431 | 0.0056 |
| hly (1587), all | 19 | 71 | 55 | 73 | 67 | 6 | 0.0160 | 0.0685 | 0.0020 |
| Lineage I | 8 | 8 | 7 | 8 | 7 | 1 | 0.0017 | 0.0061 | 0.0004 |
| Lineage II | 10 | 23 | 20 | 23 | 20 | 3 | 0.0042 | 0.0159 | 0.0008 |
| mpl (1530), all | 18 | 126 | 91 | 130 | 100 | 30 | 0.0293 | 0.1084 | 0.0096 |
| Lineage I | 10 | 19 | 13 | 19 | 14 | 5 | 0.0035 | 0.0114 | 0.0013 |
| Lineage II | 7 | 7 | 6 | 7 | 4 | 3 | 0.0015 | 0.0036 | 0.0009 |
| actA (1917), all | 20 | 184 | 161 | 197 | 98 | 90 | 0.0383 | 0.0977 | 0.0226 |
| Lineage I | 9 | 24 | 23 | 24 | 13 | 10 | 0.0052 | 0.0123 | 0.0030 |
| Lineage II | 10 | 82 | 74 | 85 | 47 | 35 | 0.0149 | 0.0371 | 0.0090 |
| plcB (867), all | 11 | 62 | 46 | 64 | 45 | 19 | 0.0254 | 0.0868 | 0.0097 |
| Lineage I | 5 | 5 | 5 | 5 | 2 | 3 | 0.0017 | 0.0035 | 0.0012 |
| Lineage II | 5 | 13 | 3 | 13 | 7 | 6 | 0.0023 | 0.0054 | 0.0015 |
the nt positions for the six coding regions in the alignment include: prfA, 1 – 711 (located in the opposite strand); plcA, 988 – 1938 (located in the opposite strand); hly, 2181 – 3767; mpl, 4105 – 5634; actA, 5837 – 7753; plcB, 7793 – 8659; data are shown for all 40 isolates as well as for lineage I and II separately (18 and 21 isolates, respectively; the data for the one lineage III isolate are not shown separately)
H: number of haplotypes
S: number of polymorphic sites (excluding polymorphic sites within indels)
PIS: number of parsimonious informative sites;
NSS: number of synonymous substitutions;
NNS: number of nonsynonymous substitutions;
π = nucleotide diversity (average pairwise differences per site); the π values for lineage I and II strains (for the whole cluster as well as for all genes) are statistically significantly different (P < 0.0001, T test).
Descriptive analyses of the six genes in the virulence gene cluster showed considerable variation in nucleotide diversity and other parameters among these genes (Table 1). actA was the most diverse gene (π = 0.0383) with a total of 184 polymorphic sites and 197 substitutions (excluding polymorphic sites in the indel regions). Thirteen polymorphic sites had three different nucleotides. Twelve isolates (11 in lineage I and 1 in lineage II) showed a single large deletion of 105 nucleotide in actA (representing the majority of the indel sites observed in the virulence gene cluster), representing a previously reported deletion (Smith et al., 1996; Wiedmann et al., 1997; Moriishi et al., 1998; Jacquet et al., 2002) of one of the four Proline-Rich Repeats (PRR) in ActA. On average, lineage I isolates showed significantly lower (P < 0.0001) nucleotide diversity in actA (πI = 0.0052) as compared to lineage II isolates (πII = 0.0149) (Table 1). prfA was the most conserved gene (π= 0.0143) with only 25 substitutions, including only two nonsynonymous substitutions; one of these substitutions (K197N) is present in one lineage II isolate (FSL S4–304) and the lineage III isolate (FSL F2–695). While prfA and mpl showed higher nucleotide diversity (π) in lineage I isolates, all other genes in the virulence gene cluster showed higher nucleotide diversity in lineage II isolates (Table 1).
3.2. Phylogenetic analysis
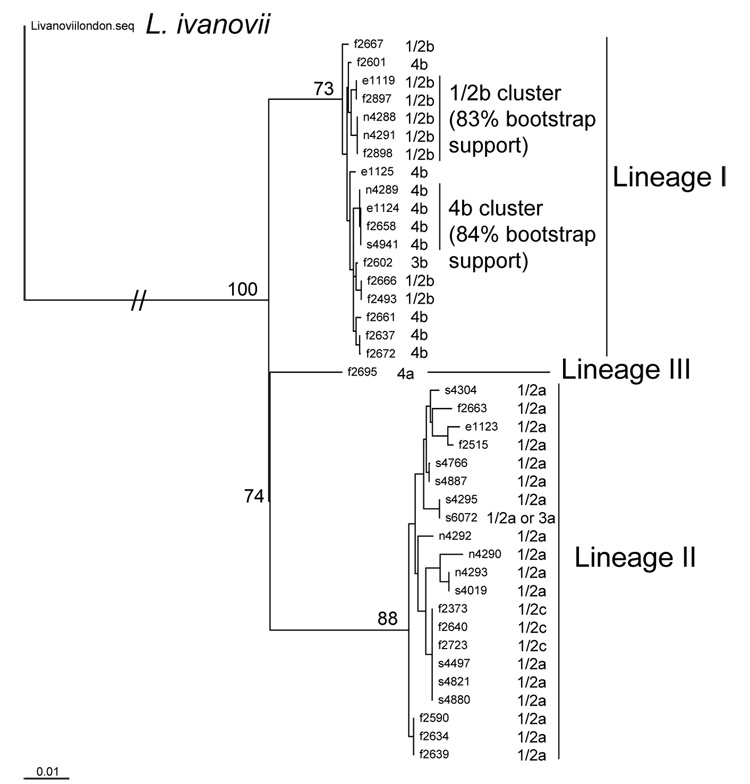
The maximum likelihood tree constructed using the complete virulence gene cluster sequence showed two main clusters (Fig. 2). Isolate grouping into these clusters is consistent with previous lineage classification of these isolates (Nightingale et al., 2005b; Orsi et al., 2007; Sauders et al., 2006; Tsai et al., 2006) and also consistent with serotype classification (i.e., serotype 1/2a and 1/2c isolates group into lineage II and serotypes 1/2b, 3b and 4b cluster into lineage I) (Nadon et al., 2001). The one serotype 4a, lineage III isolate included in our isolate set was clearly separate from the lineage I and II clusters. Long branch lengths were observed among lineage II isolates but not among lineage I isolates. Lineage I isolates formed two main clusters, one containing 1/2b isolates (83% bootstrap support) and one containing 4b isolates (84% bootstrap support; Fig. 2), even though 1/2b and 4 b isolates sometimes clustered together, consistent with previous reports (Call et al., 2003; Ward et al., 2004; Nightingale et al., 2005b). Most lineage II isolates belonged to serotype 1/2a, one lineage II isolate (FSL S6–072) could not be serotyped unambiguously and belongs to either serotype 1/2a or 3a; for this isolate the H antigen was identified as AB and the O antigen was identified as II, despite repeated serotyping attempts this isolate reacted neither with the O: IV or the O:I antibodies indicating that this isolate is either serotype 3a or 1/2a. The virulence gene cluster sequences for all three serotype 1/2c isolates (lineage II; all three isolated from foods) were identical to each other, consistent with previous studies that found 1/2c isolates to be highly clonal (Zhang et al., 2004; Orsi et al., 2007), and were also indistinguishable from three serotype 1/2a isolates obtained from the pristine environment (allelic type 18, Supplemental Table 1). Clustering of serotype 1/2a and 1/2c isolates has also been observed previously (Mereghetti et al., 2002; Zhang et al., 2004; Orsi et al., 2007) and suggests that serotype 1/2c emerged from a serotype 1/2a ancestor.
Figure 2.
Maximum likelihood tree of the virulence gene cluster for the 40 L. monocytogenes isolates in the study. The virulence gene cluster of a L. ivanovii subspecies londoniensis (NRRL 33021) was used as outgroup. Bootstrap values are shown at the branches that define the lineage of the isolates.
3.3. Positive selection analysis of the coding regions
Three of the six coding regions in the virulence gene cluster (i.e., mpl, plcA, and actA) showed evidence for positive selection. For all three genes M0 was rejected in favor of the alternative model M3 (Table 2), suggesting that selection is not homogeneous across these genes. For mpl and plcA, the branch-specific analyses showed evidence for positive selection among branches within lineage I and II, respectively (Table 2). For mpl, 1.5% of sites were classified into the site category for positively selected sites (ω = 7.28) in the branch specific model for lineage I; one mpl aa site (aa 103) showed a posterior probability > 95% of being under positive selection (Table 2). For plcA, 3.8% of sites were classified into the site category for positively selected sites (the ω for this site category was estimated to be 2.82) in the branch-specific model for lineage II; no plcA sites were identified as having a posterior probability > 95% of being under positive selection.
Table 2.
Positive selection in coding regions.
| LRT and parameters | prfA | plcA | hly | mpl | actA | plcB |
|---|---|---|---|---|---|---|
| M0 vs M3 LRTa | 6.5852 | 52.3129* | 5.0478 | 18.3828* | 153.0975* | 0.0 |
| ωb | 1.20 | 2.12 | 0.61 | 1.05 | 2.39 | 0.105 |
| pc | 0.0169 | 0.0696 | 0.0347 | 0.0917 | 0.0978 | 0.3034 |
| M1a vs M2a LRTa | 0.02886 | 5.2502 | 0.0 | 0.0019 | 12.3971* | 0.0005 |
| ωb | 1.20 | 2.12 | 1.00 | 1.04 | 2.55 | 1.00 |
| pc | 0.0169 | 0.0696 | 0.0109 | 0.0917 | 0.0771 | 0.00 |
| aa sites under pos. selectiond | - | - | - | - | 461 (97%) | - |
| M7 vs M8 LRTa | 2.1510 | 5.2529 | 0.7376 | 0.0097 | 13.7596* | 0.0 |
| ωb | 1.20 | 2.12 | 1.00 | 1.07 | 2.40 | 1.00 |
| pc | 0.0169 | 0.0696 | 0.0175 | 0.0882 | 0.0955 | 0.00 |
| aa sites under pos. selectiond | - | aa 119 | - | - | aa 112 (97%) | - |
| (95%) | aa 123 (96%) | |||||
| aa 291 (97%) | ||||||
| aa 449 (97%) | ||||||
| aa 461 (99%) | ||||||
| aa 522 (95%) | ||||||
| Lineage I LRTa | NAe | 0.0 | 0.0 | 6.2440* | 1.4834 | 0.0 |
| ωb | NAe | 1.00 | 1.00 | 7.28 | 6.77 | 1.00 |
| pc | NAe | 0.00 | 0.0056 | 0.0151 | 0.174 | 0.00 |
| aa sites under pos. selectiond | NAe | - | - | aa 103 | - | - |
| (98%) | ||||||
| Lineage II LRTa | 0.0 | 4.6409* | 1.1946 | 0.0001 | 5.0142* | 0.0 |
| ωb | 1.00 | 2.82 | 3.95 | 1.00 | 3.59 | 1.00 |
| pc | 0.00 | 0.0381 | 0.0044 | 0.0802 | 0.0245 | 0.2064 |
| aa sites under pos. selectiond | - | - | - | - | - | - |
This row reports the results for the likelihood ratio test (LRT) for different models (e.g., M0 versus M3; lineage specific tests are indicated as “Lineage I LRT” and “Lineage II LRT”); the value shown represents the likelihood ratio between the two models;
a indicates that the null model can be rejected in favor to the alternative model (P < 0.05; significance is calculated using a χ2-distribution with degrees of freedom equals the difference in the number of parameters estimated for the alternative model in comparison to the null model
ω = dN/dS for the site category that allows ω to be larger than 1 (i.e., ω2 in M2, M3, and the branch specific models; ω in M8)
p = the proportion of sites under positive selection
aa site identified by Bayes Empirical Bayes (BEB) as having probability > 95% of being under positive selection are shown; posterior probabilities are given in parenthesis; “−“ indicates that no sites showed probability ≥ 95% of having evolved by positive selection
NA, not assessed because no amino acid changes were observed within lineage I in prfA
For actA, both overall tests (i.e., M1a vs M2a and M7 vs. M8, Table 2) showed evidence for positive selection (Table 2). In addition, the branch-specific analyses for actA showed evidence for positive selection among lineage II branches (Table 2). M8 estimated that 9.6% of the sites evolved by positive selection (ω = 2.40) and identified six aa sites (aa 112, 123, 291, 449, 461 and 522) with posterior probabilities > 95% of being under positive selection. The more conservative M2a model estimated that 7.7% of actA aa sites evolved by positive selection (ω = 2.55) and identified one aa site (aa 461) with posterior probability > 95% of being under positive selection. In the branch specific analysis, 2.5% of the sites were estimated to have evolved by positive selection (ω = 3.59), but no sites presented a posterior probability > 95% of being under positive selection. ActA aa site 498, which had previously been reported, using a different isolate set, to be under positive selection (Roberts and Wiedmann, 2006) was classified into the positively selected sites, although with a posterior probability < 95 % (i.e., 63 %).
Among lineage II isolates, the nucleotide diversity among nonsynonymous (πNNS) and synonymous (πNSS) sites were higher for the genes identified as being under positive selection in this lineage, i.e., plcA and actA (Table 1), suggesting that plcA and actA evolved faster than the other virulence genes in this lineage. Moreover, actA showed the highest πNNS among all isolates (0.0226) while prfA and hly showed the lowest πNNS among all isolates (Table 1).
3.4. Positive selection and polymorphism analysis for non-coding regions
Analysis in non-coding regions was performed separately using each gene including the upstream and downstream non-coding regions, except that no sequence downstream of prfA (i.e., on the 5’ end of the virulence cluster) and only 47 nucleotides downstream of plcB (i.e., on the 3’ end of the virulence gene cluster) were available for analyses. Both, the analysis with plcA and the analysis with hly (representing two adjacent genes), found significant evidence for positive selection in the non-coding areas; five out of the six sites identified as being under positive selection (with a posterior probability > 95%; Table 3) fall in the intergenic region between plcA and hly, which was included in both analyses (as this region is located 5’ of both of these genes, which are adjacent to each other and divergently transcribed). One site downstream of hly was also identified as being under positive selection (Table 3), but this site was only identified in the analysis with hly, but not in the analysis with mpl (where this site would be included in the non-coding region upstream of mpl). Our analyses thus provide strong evidence for positive selection specifically in the intergenic region between plcA and hly; this region harbors the promoter sequences for both hly and plcA, which share a PrfA box and are transcribed in opposite directions. None of the sites under positive selection falls within the PrfA box or a −10 promoter region. However, the nucleotide variation at the positively selected site 92 (Table 3, Fig. 3) generates a putative new −10 site, which matches the consensus −10 for a promoter recognized by the alternative sigma factor σB; a search for σB promoter regions using the genome of the lineage I strain F2365 (Nelson et al., 2004) and a Hidden Markov Model (HMM) constructed from known σB promoters in L. monocytogenes (Raengpradub et al., 2008), identified the putative promoter generated by the nucleotide 92 polymorphism, suggesting that this may indeed represent a σB-dependent promoter for hly, which is present in some lineage I isolates (i.e., 16/18 lineage I isolates sequenced here). In addition, the nucleotide variation at the positively selected site 42 creates a possible −35 region (GTTA, which is present in all lineage I and 10/21 lineage II isolates), which may represent a σB-dependent promoter combined with a −10 region (GGCAT), even though the spacing between the −35 and −10 region would be unusually short [12 nucleotides rather than the typical 13 to 17 nucleotide spacing for a σB-dependent promoter (Kazmierczak et al., 2003)].
Table 3.
Positive selection in non-coding regions.
| Alignmenta | LRT M2 vs M1b | LRT M3 vs M1b | Zeta(ζ) | Frequency | Sites under positive selectionc |
|---|---|---|---|---|---|
| plcA | 16.63* | 16.55* | 16.24 | 0.01 | 42**; 53**; 180** |
| hly | 10.93* | 10.92* | 5.70 | 0.10 | 42*; 53*; 92*; 195*; 410* |
| mpl | 0.13 | 0.12 | 1.49 | 0.14 | - |
| actA | 0.14 | 0.13 | 1.48 | 0.11 | - |
| plcB | 0.90 | 0.86 | 3.58 | 0.14 | - |
Alignments contained the coding regions for the listed genes as well as the noncoding regions upstream and downstream of each gene (e.g. the plcA alignment contained the intergenic region between plcA and prfA, the plcA coding region, and the intergenic region between plcA and hly). See Fig. 1 for the relative position of the genes in the virulence gene cluster
These two columns report the results for the likelihood ratio test (LRT) for different models (M2 versus M1 [test 2]; M3 versus M1 [test 1]); the value shown represents the likelihood ratio between the two models;
indicates that the null model can be rejected in favor to the alternative model (P < 0.05)
Sites with posterior probability > 95% of being under positive selection as estimated by M3; “−“ indicates that no sites were identified as evolving by positive selection;
indicates sites identified with posterior probabilities > 95% and > 99%, respectively. Sites 42, 53, 92, 180, and 195 are located between plcA and hly (see Fig. 3); site 410 is located between hly and mpl (i.e., 168 bp downstream of the hly stop codon)
Figure 3.
Noncoding region between plcA and hly. The first 200 nucleotides upstream plcA are shown for selected isolates representing sequences found among lineage I (I), lineage II (II) and lineage III (III). Arrows indicate sites with high probability (≥95%) of having evolved by positive selection. The hly P1 −35 region (nucleotides 84–89), P1 −10 region (nucleotides 108–113), PrfA box (nucleotides 60–73), and P2 −10 region (nucleotides 97–102) are italicized. The transcriptional start sites (+1) of the hly P1 (nucleotide 120) and P2 (nucleotide 99) promoters are in bold. The putative σB-dependent promoter identified by HMM in the lineage I strain F2365 (−35 region, nucleotides 75–78; −10 region, nucleotides 93–97) is shown in bold.
In addition to the positively selected sites in the hly-plcA intergenic region, we also identified a number of polymorphic sites (which were not specifically identified as being under positive selection), which may affect promoter elements. Specifically, we identified three polymorphic sites within the −35 region (GGGACA) of the putatively PrfA-independent hly P1 promoter (Mengaud et al., 1989) (Fig. 3); one mutation in the lineage III isolate FSL F2–695 changes this −35 region to GAGACA, while one mutation in the lineage II isolate FSL F2–515 changes this region to AGGACA. In three other lineage II isolates (FSL S4–304, FSL F2–663, and FSL E1–123), the hly P1 promoter −35 region is AGGATA and thus differs by two nucleotides from P1 promoter −35 region of most other L. monocytogenes isolates in our study.
3.5. Recombination analysis
Initial GENECONV analyses identified 91 unique fragments, which could be grouped into 26 independent events (i.e., events with distinct 5’ and 3’ breakpoints). actA showed the most recombination events, while prfA showed the least recombination events (Supplemental Table 3).
We subsequently used ClonalFrame to further assess recombination events and to identify the recipient strains in the recombination events. ClonalFrame detected no recombination events where lineage I isolates served as recipients, while it detected 25 events where lineage II isolates served as recipients in a recombination event (Fig. 4; Table 4); these events occurred in 18 branches (in each of five branches two recombination events; in one branch [branch F; Table 4] three events occurred). Recombination events were identified in isolates from all four source populations (natural environment, foods, animal and human clinical cases). While ClonalFrame identified a number of recombination events in the single lineage III isolate (FSL F2–695) and in the ancestral branch between lineage II and lineage I/III, these data are not reported as only a single highly divergent lineage III isolate was included in our data. The majority of the recombination events seem to fall within the coding sequences of actA (11 events) and plcA (5 events); four recombination events fall in the hly-plcA intergenic region (Fig.1 and Fig. 4; Table 4).
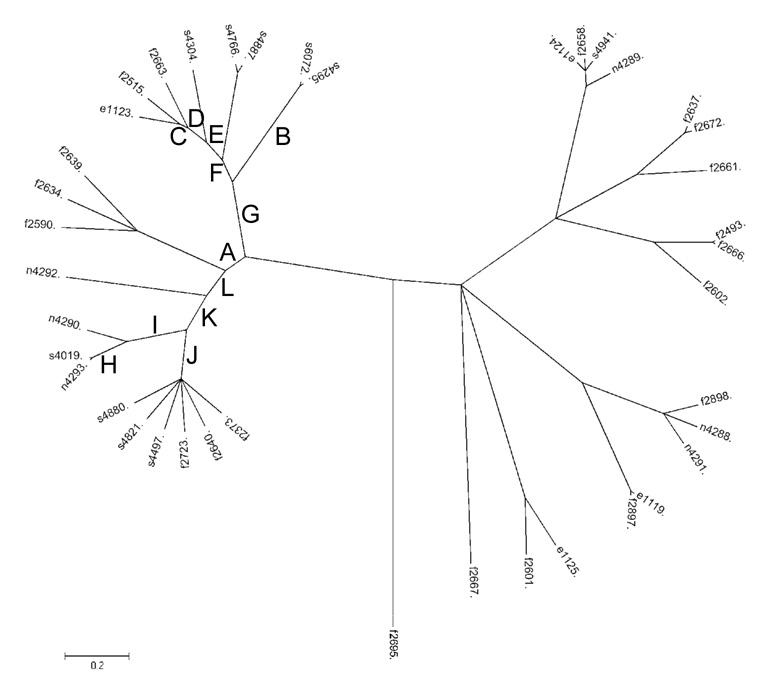
Figure 4.
Phylogeny of the 40 L. monocytogenes isolates inferred by ClonalFrame based on the virulence cluster sequence data. The tree represents a 95 % consensus between two independent runs of the program with the same initial parameters. In order to construct the tree, ClonalFrame takes into account the possibility of recombination. Internal branches where recombination events were identified are marked with letters (see Table 4 for details regarding the recombination events).
Table 4.
Recombination events identified by ClonalFrame.
| Brancha | Lineage of recipientb | Regionc |
|---|---|---|
| A | II | plcA ↔ plcA |
| B | II | actA ↔ actA |
| FSL S4–304 | II | actA ↔ actA |
| FSL F2–663 | II | actA ↔ actA |
| FSL F2–515 | II | plcA ↔ plcA; actA ↔ actA |
| FSL E1–123 | II | actA ↔ actA |
| C | II | actA ↔ actA |
| D | II | plcA ↔ plcA |
| E | II | plcA-hly ↔ plcA-hly |
| F | II | plcA-hly ↔ plcA-hly; hly ↔ hly-mpl |
| G | II | plcA ↔ plcA-hly |
| FSL N4–292 | II | actA ↔ actA |
| FSL N4–290 | II | actA ↔ actA; plcB ↔ plcB |
| H | II | actA ↔ actA |
| I | II | actA ↔ actA |
| J | II | plcA ↔ plcA |
| K | II | plcA-hly ↔ plcA-hly; actA ↔ actA |
| L | II | hly ↔ hly-mpl |
For internal branches, letters are shown (A to L; corresponding to the branch labels in Fig. 4); for external branches, the isolate name is shown (e.g., FSL S4–304 indicates that the recombination even was identified in the branch leading to the FSL S4–304 sequence).
Indicates the lineage of the recipient sequence.
“gene-gene” designations denote intergenic regions (e.g., plcA-hly denotes the intergenic region between plcA and hly); “↔” indicates the extent of the recombinant fragment (e.g., in branch G, one event occurred with the recombinant fragment extending from the coding region of plcA to the intergenic region between plcA and hly). Regions are approximate and were determined based on Clonalframe outputs.
4. Discussion
We determined and analyzed the DNA sequences for six virulence genes and intergenic regions in the prfA cluster in 40 L. monocytogenes isolates representing diverse source populations. Initial phylogenetic analyses confirmed that the 40 sequences group into two main clusters representing the two common L. monocytogenes lineages that have first been described by Piffaretti et al. (1989) and subsequently been confirmed by phylogenetic analyses based on various virulence and housekeeping genes (Nightingale et al., 2005b; Orsi et al., 2007; Tsai et al., 2006) as well as a previous phylogenetic study of the prfA virulence gene cluster (Ward et al., 2004); these two main clusters have also been defined by all other subtyping studies that included cluster analyses, including PFGE and ribotyping studies (as reviewed by Wiedmann, 2002). Subsequent positive selection and recombination analyses showed that (i) virulence genes in the main L. monocytogenes virulence gene cluster includes highly conserved genes (i.e., hly, prfA) as well as diverse genes that appear to have evolved by positive selection (mpl, actA, plcB), (ii) recombination has played an important role in the evolution of the virulence gene cluster, but is limited to lineage II isolates, and (iii) the promoter region driving the transcription of virulence genes transcribed early in intracellular infection (i.e., hly, plcA) appears to have evolved by positive selection and recombination.
4.1. Virulence genes in the main L. monocytogenes virulence gene cluster include highly conserved genes (i.e., hly, prfA) as well as diverse genes that appear to have evolved by positive selection (mpl, actA, plcB)
The genetic diversity for the six genes in the virulence cluster ranged from 0.0143 (prfA) to 0.0383 (actA) with an average diversity for the whole virulence gene cluster of 0.0246. Using the same 40 isolates, similar diversity was previously observed for another virulence gene, inlA (0.02134) (Orsi et al., 2007), while other internalin genes found in most or all of these isolates (i.e., inlB, inlC, inlC2, inlD, inlE) (Tsai et al., 2006) showed a wider range of diversity (0.0167 to 0.0702) with some highly diverse genes (e.g., inlE; π = 0.0702). Nucleotide diversity within actA was not limited to single nucleotide polymorphisms, but also included a previously reported (Wiedmann et al., 1997; Moriishi et al., 1998; Jacquet et al., 2002) deletion of a fragment encoding a 35 aa Proline-Rich Repeats (PRR) in a number of lineage I isolates. This deletion yields an ActA protein with three rather than four PRRs; while Smith et al. (1996) previously reported a linear relationship between the number of PRRs and the rate of bacterial movement in the host cell cytoplasm, the effects of PRR variation on virulence (Smith et al., 1996) and in natural strains remain unclear (Roberts and Wiedmann, 2006).
The three most diverse genes in the virulence gene island (i.e., plcA, actA and mpl) also showed evidence for positive selection in their coding regions, although with different lineage-specific patterns of positive selection. While positive selection in plcA and mpl were restricted to lineage II and lineage I isolates, respectively, positive selection in actA was identified in the overall analysis, suggesting that this gene evolved by positive selection in different branches. Interestingly, ActA region C (aa 50–126), which is necessary for continuous actin filament elongation (Lasa et al., 1997), was very diverse with ten polymorphic sites, including two sites (aa 112 and 123) that were identified by M8 as being under positive selection and where three amino acid variants were observed in each site. Another site identified as being under positive selection (aa 291) falls within the first PRR domain. On the other hand, the site with highest probability of being under positive selection (aa 461) as well as other two sites (aa 449 and 522) fall in regions with no described function. Overall, these findings suggest that actA may be adapting to certain functions in addition to cell-to-cell spread [e.g., a role in host cell invasion (Suarez et al., 2001)] that require specific aa sequences in regions that have not yet been assigned specific biological functions. This hypothesis is consistent with experimental data, which showed that an isogenic mutation in a positively selected site in ActA did not affect cell-to-cell spread phenotype (Roberts and Wiedmann, 2006).
Although mpl showed evidence for positive selection and considerable nucleotide and aa diversity, the Mpl aa 84–92 region, which has been identified as a CTL epitope in mice (Busch et al., 1997) was conserved in all isolates analyzed, possibly because this region may represent the active domain of Mpl. Similarly, the major murine CTL epitope region in LLO (aa 91 and 99) (Pamer et al., 1991; Vijh and Pamer, 1997) was conserved among all 40 isolates, possibly because conservation of this region appears to be important for virulence (Lety et al., 2006). These findings illustrate conservation of certain sequence features, including some that may be expected to be under selection for diversification. Overall, the analyses reported provide important new data on specific aa sites under positive selection in mpl, actA, and plcA; identification of these sites provides an opportunity for mutational analyses of specific polymorphic sites in order to probe the contributions of diversification in these virulence genes to niche adaptation in L. monocytogenes.
4.2. Recombination has played an important role in the evolution of the virulence gene cluster, but is limited to lineage II isolates
Interestingly, our data suggest that recombination events within the virulence gene cluster commonly involve lineage II strains as recipients, but rarely involve lineage I isolates as recipients (recombination in lineage III isolates was not characterized as only a single lineage III isolate was included in our isolate set). These findings are in agreement with previous studies that have shown considerable recombination in other virulence genes, such as inlA, inlC, inlC2,inlD, inlE, inlF and inlG among lineage II isolates (Orsi et al., 2007; Tsai et al., 2006), with recombination generally more frequently in lineage II than lineage I isolates. Similar observations (i.e., more common recombination among lineage II strains than among lineage I strains) have also been reported for L. monocytogenes housekeeping genes (Meinersmann et al., 2004). Recombination thus appears to be critical for the diversification of lineage II isolates. While lineage II has generally been reported as showing considerably higher nucleotide diversity than lineage I isolates, our data suggest that this higher diversity in lineage II strains may be largely due to recombination. Specifically, this hypothesis is supported by the observation that the two virulence genes with no evidence for recombination (i.e., prfA and mpl) show higher nucleotide diversity among lineage I strains than among lineage II strains. Similarly, sigB, the only gene which showed no evidence for recombination among 120 L. monocytogenes isolates in previous study, showed identical nucleotide diversity among lineage I and II isolates (Nightingale et al., 2005b). Additional evolutionary analyses in L. monocytogenes genes with no evidence for recombination will be necessary to further characterize the evolutionary history of lineages I and II and to test the current hypothesis that only lineage I has experienced a recent bottleneck (Meinersmann et al., 2004). Our data reported here specifically suggest as a possible alternative hypothesis that lineage II simply diversified more rapidly, e.g., through more frequent recombination than lineage I. Possible reasons for more common recombination in lineage II may involve molecular (e.g. different restriction systems in each lineage), epidemiological (e.g. abundance of different isolates in environments where horizontal gene transfer might be facilitated) or evolutionary (e.g. different selective pressures to maintain recombinant fragments in the population) factors.
4.3. The promoter region driving the transcription of virulence genes transcribed early in intracellular infection (i.e., hly, plcA) appears to have evolved by positive selection and recombination
While analyses for positive selection in coding regions are increasingly recognized as a powerful tool for identifying functionally important genes or residues within a gene, few studies have used emerging evolutionary analyses tools (Wong and Nielsen, 2004) to identify intergenic regions under positive selection, including the specific nucleotide sites that may be under positive selection. Interestingly, the positive selection analyses on non-coding regions in the L. monocytogenes virulence gene cluster provided initial evidence that the intergenic region between hly and plcA includes nucleotide sites that are under positive selection; Clonalframe also provided evidence for recombination events in this region. While the PrfA binding site in this region (as well as the other PrfA binding sites upstream of prfA, mpl, and actA) was perfectly conserved, a nucleotide change in at least one of the positively selected sites in hly and plcA intergenic region (nucleotide 92, Figure 3) appears to generate a putative σB-dependent promoter upstream of hly; this mutation was only found in lineage I isolates. While there is no evidence that the transcription of hly is σB-dependent in a lineage II strain (Chaturongakul and Boor, 2006), no data are currently available on σB-dependent hly transcription in lineage I strains. Future experimental validation of our findings is thus still necessary. Diversification of virulence gene regulation among L. monocytogenes lineages I and II has recently been demonstrated by comparative transcriptome analyses though (Severino et al., 2007). These analyses (Severino et al., 2007) also reported evidence for differences in transcription of σB- and σB-dependent genes among lineages, supporting the potential importance of our finding.
Supplementary Material
Acknowledgements
This work was partially supported by USDA Special Research Grant 2005-34459-15625 (to MW) and National Institutes of Health (NIH) Grant No. R01GM63259 (to MW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Busch DH, Bouwer HG, Hinrichs D, Pamer EG. A nonamer peptide derived from Listeria monocytogenes metalloprotease is presented to cytolytic T lymphocytes. Infect. Immun. 1997;65:5326–5329. doi: 10.1128/iai.65.12.5326-5329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call DR, Borucki MK, Besser TE. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J. Clin. Microbiol. 2003;41:632–639. doi: 10.1128/JCM.41.2.632-639.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli A, Tilney LG, Portnoy DA. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 1993;8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T, Leimeister-Wachter M, Domann E, Hartl M, Goebel W, Nichterlein T, Notermans S. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 1992;174:568–574. doi: 10.1128/jb.174.2.568-574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul S, Boor KJ. SigmaB activation under environmental and energy stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 2006;72:5197–5203. doi: 10.1128/AEM.03058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cesare A, Mioni R, Manfreda G. Prevalence of Listeria monocytogenes in fresh and fermented Italian sausages and ribotyping of contaminating strains. J. Food. Microbiol. 2007;120:124–130. doi: 10.1016/j.ijfoodmicro.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domann E, Wehland J, Rohde M, Pistor S, Hartl M, Goebel W, Leimeister-Wachter M, Wuenscher M, Chakraborty T. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 1992;11:1981–1990. doi: 10.1002/j.1460-2075.1992.tb05252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag NE, Rong L, Portnoy DA. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 1993;61:2537–2544. doi: 10.1128/iai.61.6.2537-2544.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard JL, Berche P, Mounier J, Richard S, Sansonetti P. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect. Immun. 1987;55:2822–2829. doi: 10.1128/iai.55.11.2822-2829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Zadoks RN, Fortes ED, Dogan B, Cai S, Chen Y, Scott VN, Gombas DE, Boor KJ, Wiedmann M. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 2004;70:5833–5841. doi: 10.1128/AEM.70.10.5833-5841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet C, Gouin E, Jeannel D, Cossart P, Rocourt J. Expression of ActA, Ami, InlB, and listeriolysin O in Listeria monocytogenes of human and food origin. Appl. Environ. Microbiol. 2002;68:616–622. doi: 10.1128/AEM.68.2.616-622.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers GT, Bruce JL, McDonough PL, Scarlett J, Boor KJ, Wiedmann M. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiol. 2001;147:1095–1104. doi: 10.1099/00221287-147-5-1095. [DOI] [PubMed] [Google Scholar]
- Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 2003;185:5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft J, Vazquez-Boland JA. Regulation of virulence genes in Listeria. J. Med. Microbiol. 2001;291:145–157. doi: 10.1078/1438-4221-00111. [DOI] [PubMed] [Google Scholar]
- Lappi VR, Thimothe J, Nightingale KK, Gall K, Scott VN, Wiedmann M. Longitudinal studies on Listeria in smoked fish plants: impact of intervention strategies on contamination patterns. J. Food Prot. 2004;67:2500–2514. doi: 10.4315/0362-028x-67.11.2500. [DOI] [PubMed] [Google Scholar]
- Lasa I, Gouin E, Goethals M, Vancompernolle K, David V, Vandekerckhove J, Cossart P. Identification of two regions in the N-terminal domain of ActA involved in the actin comet tail formation by Listeria monocytogenes. Embo J. 1997;16:1531–1540. doi: 10.1093/emboj/16.7.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebure T, Stanhope MJ. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 2007;8:R71. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimeister-Wachter M, Haffner C, Domann E, Goebel W, Chakraborty T. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. U.S.A. 1990;87:8336–8340. doi: 10.1073/pnas.87.21.8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lety MA, Frehel C, Raynaud C, Dupuis M, Charbit A. Exploring the role of the CTL epitope region of listeriolysin O in the pathogenesis of Listeria monocytogenes. Microbiol. 2006;152:1287–1296. doi: 10.1099/mic.0.28754-0. [DOI] [PubMed] [Google Scholar]
- McLauchlin J, Mitchell RT, Smerdon WJ, Jewell K. Listeria monocytogenes and listeriosis: a review of hazard characterisation for use in microbiological risk assessment of foods. Int. J. Food Microbiol. 2004;92:15–33. doi: 10.1016/S0168-1605(03)00326-X. [DOI] [PubMed] [Google Scholar]
- Meinersmann RJ, Phillips RW, Wiedmann M, Berrang ME. Multilocus sequence typing of Listeria monocytogenes by use of hypervariable genes reveals clonal and recombination histories of three lineages. Appl. Environ. Microbiol. 2004;70:2193–2203. doi: 10.1128/AEM.70.4.2193-2203.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengaud J, Vicente MF, Cossart P. Transcriptional mapping and nucleotide sequence of the Listeria monocytogenes hlyA region reveal structural features that may be involved in regulation. Infect. Immun. 1989;57:3695–3701. doi: 10.1128/iai.57.12.3695-3701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereghetti L, Lanotte P, Savoye-Marczuk V, Marquet-Van Der Mee N, Audurier A, Quentin R. Combined ribotyping and random multiprimer DNA analysis to probe the population structure of Listeria monocytogenes. Appl. Environ. Microbiol. 2002;68:2849–2857. doi: 10.1128/AEM.68.6.2849-2857.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriishi K, Terao M, Koura M, Inoue S. Sequence analysis of the actA gene of Listeria monocytogenes isolated from human. Microbiol. Immunol. 1998;42:129–132. doi: 10.1111/j.1348-0421.1998.tb02261.x. [DOI] [PubMed] [Google Scholar]
- Nadon CA, Woodward DL, Young C, Rodgers FG, Wiedmann M. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 2001;39:2704–2707. doi: 10.1128/JCM.39.7.2704-2707.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, Kolonay JF, Rasko DA, Angiuoli SV, Gill SR, Paulsen IT, Peterson J, White O, Nelson WC, Nierman W, Beanan MJ, Brinkac LM, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Haft DH, Selengut J, Van Aken S, Khouri H, Fedorova N, Forberger H, Tran B, Kathariou S, Wonderling LD, Uhlich GA, Bayles DO, Luchansky JB, Fraser CM. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 2004;32:2386–2395. doi: 10.1093/nar/gkh562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale KK, Schukken YH, Nightingale CR, Fortes ED, Ho AJ, Her Z, Grohn YT, McDonough PL, Wiedmann M. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 2004;70:4458–4467. doi: 10.1128/AEM.70.8.4458-4467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale KK, Windham K, Martin KE, Yeung M, Wiedmann M. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 2005a;71:8764–8772. doi: 10.1128/AEM.71.12.8764-8772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale KK, Windham K, Wiedmann M. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 2005b;187:5537–5551. doi: 10.1128/JB.187.16.5537-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton DM, Scarlett JM, Horton K, Sue D, Thimothe J, Boor KJ, Wiedmann M. Characterization and pathogenic potential of Listeria monocytogenes isolates from the smoked fish industry. Appl. Environ. Microbiol. 2001;67:646–653. doi: 10.1128/AEM.67.2.646-653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi RH, Ripoll DR, Yeung M, Nightingale KK, Wiedmann M. Recombination and positive selection contribute to evolution of Listeria monocytogenes inlA. Microbiol. 2007;153:2666–2678. doi: 10.1099/mic.0.2007/007310-0. [DOI] [PubMed] [Google Scholar]
- Pamer EG, Harty JT, Bevan MJ. Precise prediction of a dominant class I MHC-restricted epitope of Listeria monocytogenes. Nature. 1991;353:852–855. doi: 10.1038/353852a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffaretti JC, Kressebuch H, Aeschbacher M, Bille J, Bannerman E, Musser JM, Selander RK, Rocourt J. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. U.S.A. 1989;86:3818–3822. doi: 10.1073/pnas.86.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Raengpradub S, Wiedmann M, Boor KJ. Comparative analysis of the sigma B-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 2008;74:158–171. doi: 10.1128/AEM.00951-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen OF, Skouboe P, Dons L, Rossen L, Olsen JE. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiol. 1995;141:2053–2061. doi: 10.1099/13500872-141-9-2053. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Wiedmann M. Allelic exchange and site-directed mutagenesis probe the contribution of ActA amino-acid variability to phosphorylation and virulence-associated phenotypes among Listeria monocytogenes strains. FEMS Microbiol. Lett. 2006;254:300–307. doi: 10.1111/j.1574-6968.2005.00041.x. [DOI] [PubMed] [Google Scholar]
- Roche SM, Gracieux P, Milohanic E, Albert I, Virlogeux-Payant I, Temoin S, Grepinet O, Kerouanton A, Jacquet C, Cossart P, Velge P. Investigation of specific substitutions in virulence genes characterizing phenotypic groups of low-virulence field strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2005;71:6039–6048. doi: 10.1128/AEM.71.10.6039-6048.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux S, Olier M, Lemaitre JP, Piveteau P, Guzzo J. Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl Environ. Microbiol. 2004;70:2180–2185. doi: 10.1128/AEM.70.4.2180-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Sauders BD, Durak MZ, Fortes E, Windham K, Schukken Y, Lembo AJ, Jr, Akey B, Nightingale KK, Wiedmann M. Molecular characterization of Listeria monocytogenes from natural and urban environments. J. Food Prot. 2006;69:93–105. doi: 10.4315/0362-028x-69.1.93. [DOI] [PubMed] [Google Scholar]
- Scortti M, Monzo HJ, Lacharme-Lora L, Lewis DA, Vazquez-Boland JA. The PrfA virulence regulon. Microbes. Infect. 2007;9:1196–1207. doi: 10.1016/j.micinf.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Severino P, Dussurget O, Vencio RZ, Dumas E, Garrido P, Padilla G, Piveteau P, Lemaitre JP, Kunst F, Glaser P, Buchrieser C. Comparative transcriptome analysis of Listeria monocytogenes strains of the two major lineages reveals differences in virulence, cell wall and stress response. Appl. Environ. Microbiol. 2007;73:6078–6088. doi: 10.1128/AEM.02730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan B, Klarsfeld A, Msadek T, Cossart P. Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J. Bacteriol. 1995;177:6469–6476. doi: 10.1128/jb.177.22.6469-6476.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Marquis H, Jones S, Johnston NC, Portnoy DA, Goldfine H. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Theriot JA, Portnoy DA. The tandem repeat domain in the Listeria monocytogenes ActA protein controls the rate of actin-based motility, the percentage of moving bacteria, and the localization of vasodilator-stimulated phosphoprotein and profilin. J. Cell Biol. 1996;135:647–660. doi: 10.1083/jcb.135.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez M, Gonzalez-Zorn B, Vega Y, Chico-Calero I, Vazquez-Boland JA. A role for ActA in epithelial cell invasion by Listeria monocytogenes. Cell Microbiol. 2001;3:853–864. doi: 10.1046/j.1462-5822.2001.00160.x. [DOI] [PubMed] [Google Scholar]
- Tsai YH, Orsi RH, Nightingale KK, Wiedmann M. Listeria monocytogenes internalins are highly diverse and evolved by recombination and positive selection. Infect. Genet. Evol. 2006;6:378–389. doi: 10.1016/j.meegid.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velge P, Herler M, Johansson J, Roche SM, Temoin S, Fedorov AA, Gracieux P, Almo SC, Goebel W, Cossart P. A naturally occurring mutation K220T in the pleiotropic activator PrfA of Listeria monocytogenes results in a loss of virulence due to decreasing DNA-binding affinity. Microbiol. 2007;153:995–1005. doi: 10.1099/mic.0.2006/002238-0. [DOI] [PubMed] [Google Scholar]
- Vijh S, Pamer EG. Immunodominant and subdominant CTL responses to Listeria monocytogenes infection. J. Immunol. 1997;158:3366–3371. [PubMed] [Google Scholar]
- Ward TJ, Gorski L, Borucki MK, Mandrell RE, Hutchins J, Pupedis K. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 2004;186:4994–5002. doi: 10.1128/JB.186.15.4994-5002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann M, Bruce JL, Keating C, Johnson AE, McDonough PL, Batt CA. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 1997;65:2707–2716. doi: 10.1128/iai.65.7.2707-2716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedmann M. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 2002;85:524–531. [PubMed] [Google Scholar]
- Wong WS, Nielsen R. Detecting selection in noncoding regions of nucleotide sequences. Genetics. 2004;167:949–958. doi: 10.1534/genetics.102.010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zhang W, Jayarao BM, Knabel SJ. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl. Environ. Microbiol. 2004;70:913–920. doi: 10.1128/AEM.70.2.913-920.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.