Abstract
Aminoacyl-tRNA synthetases (aaRSs) join amino acids to 1 of 2 terminal hydroxyl groups of their cognate tRNAs, thereby contributing to the overall fidelity of protein synthesis. In class II histidyl-tRNA synthetase (HisRS) the nonbridging Sp-oxygen of the adenylate is a potential general base for aminoacyl transfer. To test for conservation of this mechanism in other aaRSs and the role of terminal hydroxyls of tRNA in aminoacyl transfer, we investigated the class II Escherichia coli threonyl-tRNA synthetase (ThrRS). As with other class II aaRSs, the rate-determining step for ThrRS is amino acid activation. In ThrRS, however, the 2′-OH of A76 of tRNAThr and a conserved active-site histidine (His-309) collaborate to catalyze aminoacyl transfer by a mechanism distinct from HisRS. Conserved residues in the ThrRS active site were replaced with alanine, and then the resulting mutant proteins were analyzed by steady-state and rapid kinetics. Nearly all mutants preferentially affected the amino acid activation step, with only a modest effect on aminoacyl transfer. By contrast, H309A ThrRS decreased transfer 242-fold and imposed a kinetic block to CCA accommodation. His-309 hydrogen bonds to the 2′-OH of A76, and substitution of the latter by hydrogen or fluorine decreased aminoacyl transfer by 763- and 94-fold, respectively. The proton relay mechanism suggested by these data to promote aminoacylation is reminiscent of the NAD+-dependent mechanisms of alcohol dehydrogenases and sirtuins and the RNA-mediated catalysis of the ribosomal peptidyl transferase center.
Keywords: proton relay, threonine, translation, aminoacyl-tRNA synthetase, transient kinetics
The attachment of specific amino acids to their cognate tRNAs by aminoacyl tRNA synthetases (aaRSs) enables ribosomes to assemble amino acids into proteins accurately, as dictated by the sequence of codons in the mRNA. Two distinct classes of aaRSs catalyze aminoacylation, which occurs in 2 steps. During amino acid activation, the cognate amino acid is condensed with ATP to form an enzyme-bound adenylate, with release of pyrophosphate. This is followed by aminoacyl transfer, where the amino acid is transferred to either the 2′ (class I aaRSs) or 3′ (class II aaRSs) hydroxyl of A76 to generate aminoacylated tRNA, along with AMP (1, 2).
The active sites of aaRSs from both classes are remarkably devoid of candidate residues to serve as general bases for aminoacyl transfer. On the basis of structural information in the GlnRS system (3, 4) and later rapid kinetics studies using histidyl-tRNA synthetase (HisRS), the pro-S nonbridging oxygen of the adenylate was proposed to serve as a general base for aminoacyl transfer (5). The latter study also highlighted a role for tRNA in modulating amino acid activation, marking it as the putative rate-determining step for overall aminoacylation. The generality of these proposals has not yet been investigated in detail, even among aaRSs in the same class.
Among class II aaRSs, threonyl-tRNA synthetase (ThrRS) shares the canonical class IIa catalytic and anticodon-binding domains with HisRS but also possesses a dedicated N-terminal domain to hydrolyze Ser-tRNAThr (6, 7). Although the molecular basis of cognate amino acid and tRNA recognition in the aminoacyl synthetic site is clear (6–9), a rigorous kinetic analysis of the role of active-site residues and putative catalytic groups of substrates has not been reported.
Here, the mechanism of the aminoacylation reaction catalyzed by Escherichia coli ThrRS and the role of conserved residues in the ThrRS active site were investigated by mutagenesis, functional group analysis, and rapid kinetics. By these approaches, the contributions of conserved active-site residues, the nonbridging oxygens of the adenylate, and the 2′-OH of A76 to catalysis were investigated. A proton relay model of tRNA-mediated catalysis by an aaRS is suggested by these results that highlights the role of the 2′-OH of A76 and an active-site histidine in aminoacyl transfer. Similar proton relay models have been proposed for enzymes employing nicotinamide-based cofactors, underscoring a similar catalytic role for tRNAs and extant cofactors that themselves may have originally been derived from RNA.
Results
Mutational Analysis of the ThrRS Active Site.
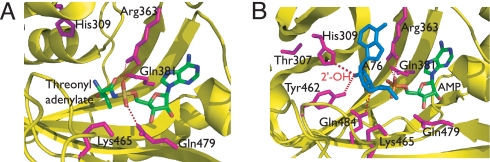
In the ThrRS active site, residues located on conformationally mobile regions provide substrate recognition (6–9). Highly conserved residues [Fig. 1 A and B and supporting information (SI) Fig. S1] were substituted with alanine, and the resulting mutant proteins were characterized functionally. The initial analysis featured the pyrophosphate exchange and aminoacylation assays, which provide the Michaelis constants for threonine and tRNA, respectively, and the kcat parameters. As measured by pyrophosphate exchange (Table S1), the most severe decreases (700-fold and 35-fold, respectively) were seen with mutants of class II conserved Arg-363 and ThrRS family conserved Lys-465, which contact the pro-R and pro-S nonbridging oxygen atoms of the adenylate. The kcat values for steady-state aminoacylation by R363A and K465A were decreased 1,005- and 574-fold, respectively, relative to the wild-type ThrRS, commensurate with the decreases observed in the pyrophosphate exchange assay. The kcat for aminoacylation by K465A was an order of magnitude less than that of pyrophosphate exchange. Notably, neither R363A nor K465A significantly altered the Km for threonine. Increases in the Km (100- and 10-fold, respectively) for threonine were seen with both class II conserved Q381A (removes contact to the carbonyl oxygen of adenylate) and ThrRS family conserved H309A (removes contact to the 2′-OH of A76) mutants. By contrast, Q479A, which eliminates a stabilizing interaction with K465, had very little effect on both reactions. Among all of the mutants, H309A was the only mutant with a significant (33-fold) decrease in aminoacylation and a small 2-fold effect on the kcat for pyrophosphate exchange.
Fig. 1.
Adenylate and tRNA recognition by E. coli ThrRS. Close-up views of the active site of the ThrRS–adenylate complex [Protein Data Bank (PDB) ID code 1EVL] (A) and ThrRS–AMP–tRNAThr (PDB ID code 1QF6) (B). The active-site secondary structures are rendered in yellow, the active site residues in magenta, and the adenylate in CPK coloring, and A76 in blue. The dotted lines represent H bonds.
To dissect the contribution of the side chains discussed above to individual steps in aminoacylation, rapid chemical quench experiments were performed. Multiple-turnover experiments were performed under conditions of excess ATP, tRNA, and threonine over enzyme and followed product formation for several turnovers. The multiple-turnover rate is equivalent to the steady-state kcat and reflects contributions from both the amino acid activation (adenylation) and aminoacyl transfer half-reactions. Aminoacylated tRNA formation was followed by using [14C]Thr as labeled substrate, whereas AMP formation was followed by using [α-32P]ATP. Single-turnover experiments were performed by rapidly mixing preformed spin column-purified enzyme: adenylate complex (in excess) with tRNA (4–5 times Km) to derive the rate of aminoacyl transfer. The resulting parameter, ktrans, may represent the chemical rate of aminoacyl transfer or a rate-limiting conformational change that precedes or follows the chemistry step.
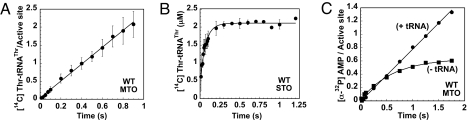
Measurement of the multiple-turnover rate of aminoacylation for wild-type ThrRS gave a rate of 2.4 ± 0.4 s−1, in good agreement with the steady-state kcat of 2.01 ± 0.1 s−1 reported above (Fig. 2A, Table 1, and Table S1). The absence of a burst in the multiple-turnover progress curve indicates that aminoacyl-tRNA release is unlikely to limit overall aminoacylation. The ktrans determined from a single-turnover experiment (14.5 ± 2.5 s−1, Fig. 2B) was 6-fold faster than the multiple-turnover rate of aminoacylation but less than the 29 s−1 measured by stopped-flow fluorometry for the forward rate of amino acid activation in the absence of tRNA (10). Thus, both half-reactions of aminoacylation appear to be paradoxically faster than the complete aminoacylation reaction. However, a direct comparison of the rate of [α-32P]AMP formation in the presence (0.8 s−1) and absence (1.6 s−1) of tRNA under presteady-state conditions showed that tRNA caused a slight inhibition of the rate of amino acid activation (Fig. 2C). Accordingly, the rate-determining step for the complete aminoacylation reaction summing the contributions of amino acid activation and aminoacyl transfer is likely to be amino acid activation in the presence of tRNA, as it is for HisRS (5).
Fig. 2.
Presteady-state kinetics of aminoacylation by ThrRS wild-type (WT) enzyme. (A) Multiple-turnover (MTO) of aminoacylation monitoring the [14C]Thr-tRNAThr formation. The data were analyzed by linear regression. (B) Single-turnover (STO) aminoacyl transfer reaction. The curve was fit to a single exponential equation. (C) The influence of tRNA on adenylation. The latter was studied under MTO conditions, monitoring the production of [α-32P]AMP in the presence (filled circles) or absence (filled squares) of tRNA.
Table 1.
Presteady-state kinetics of ThrRS wild type and mutants
| Enzymes | Multiple turnover (aminoacylation), kcat, s−1 | Single turnover (aminoacyl transfer), ktrans, s−1 |
|---|---|---|
| ThrRSWT | 2.4 ± 0.4 | 14.5 ± 2.5 |
| K465A | 0.0094 ± 0.002 (255) | 3.6 ± 1.2 (4) |
| R363A | 0.0009 ± 0.00003 (2,666) | ND* |
| Q381A | 0.066 ± 0.013 (36) | 7.58 ± 1.76 (2) |
| Q479A | 0.31 ± 0.03 (8) | 6.34 ± 0.27 (2) |
| H309A | 0.07 ± 0.02† (34) | 0.06 ± 0.002 (242) |
All experiments were performed at 37 °C (pH 8.0) as described in SI Methods. The values represent the mean ± SD of three independent experiments. Values in parentheses in the kcat and ktrans columns show the fold decrease with respect to ThrRS enzyme.
*Not determined.
†H309A mutant exhibited complex kinetics with an initial lag followed by linear phase. The value reported is the rate of the linear phase.
The ktrans obtained from single-turnover experiments using Q381A, Q479A, and K465A ThrRS mutants were all within a factor of 2–4 relative to wild-type ThrRS, suggesting that the decreases in aminoacylation measured for these mutants in the steady state resulted principally from decreases in the rate of the amino acid activation (Table 1, Table S1, and Fig. S2 a–c). Accordingly, the contacts made by these residues to the pro-S nonbridging oxygen and carbonyl of the adenylate do not contribute significantly to aminoacyl transfer. The contact to the pro-R oxygen could not be evaluated because of difficulties in purifying sufficient quantities of the R363A protein for single-turnover assays.
Unique Contribution of His-309 to ThrRS Aminoacyl Transfer.
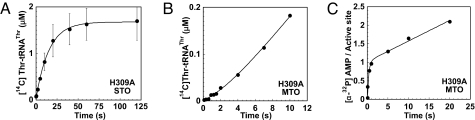
The most significantly compromised mutant with respect to aminoacyl transfer was H309A, with a 242-fold decrease (Fig. 3A and Table 1). The magnitude of this decrease suggests that H309A is the only ThrRS variant whose overall rate of aminoacylation is limited by aminoacyl transfer. When the single-turnover kinetics of the H309A mutant were analyzed under different tRNA concentrations (5 and 10 μM), the amplitudes increased in proportion to the increased tRNA concentration, but the rate of aminoacyl transfer did not increase from 0.06 ± 0.002 s−1 (Fig. S2d). The apparent half-maximal tRNA concentration for maximal rate of transfer (i.e., the [S]0.5) was similar to that for wild-type ThrRS, suggesting that H309A only slightly affects the overall affinity of tRNA for ThrRS, in agreement with the Km of tRNA for H309A (Table S1). Moreover, the catalytic contribution of His-309 depends on the fixed position of its side-chain imidazole because supplementation of free imidazole did not affect ktrans (Fig. S2e). The in vivo function of the H309A mutant was tested in a complementation experiment employing a thrS-null strain of bacteria (11). Compared with the control, the H309A strain exhibited a pronounced lag phase before log phase growth and transitioned into stationary phase at significantly lower A600 than the wild type (Fig. S3).
Fig. 3.
Transient kinetics of H309A ThrRS. (A) Single-turnover (STO) aminoacyl transfer by H309A ThrRS, monitoring the [14C]Thr-tRNAThr formation. (B) Multiple-turnover (MTO) conditions monitoring [14C]Thr-tRNAThr formation, illustrating the distinct lag phase kinetics, followed by a linear phase. (C) Adenylation reaction of H309A was studied under MTO conditions monitoring the formation of [α-32P]AMP.
The H309A mutant enzyme was also notable for a burst of AMP formation in the first turnover that was 50-fold faster (3.06 vs. 0.048 s−1) than the subsequent linear rate (Fig. 3C). The burst amplitude of AMP production for H309A was equal to a stoichiometry of 0.9 mol of AMP per active site rather than the 0.5 reported for HisRS (12). His-309 contacts the 2′-OH of A76, and the coupling of an increased rate of amino acid activation to a decreased rate of aminoacyl transfer suggests that both processes are associated with CCA positioning.
Multiple-turnover rapid-quench experiments highlighted an additional difference between H309A and K465A, R363A, Q479A, and Q381A. These latter mutants all exhibited simple linear progress curves, with multiple-turnover rates within a factor of 2 of the steady-state kcat values (Table 1 and Fig. S4 a–d), indicating that none is limited by the rate of product release. The progress curve for H309A, by contrast, was described more accurately by an exponential lag phase followed by a linear phase (Fig. 3C). This lag phase gradually disappeared with an increasing range (8–32 μM) of tRNA concentrations (data not shown). A fit of these data to a global kinetic model required an additional step either before or after tRNA binding, but before the chemical step of aminoacyl transfer. The H309A mutant may therefore form an initially unproductive complex with cognate tRNA that subsequently isomerizes to an active form. Based on the crystallographic analysis of heterologous aaRS–tRNA complexes (13), this is likely to involve repositioning of the CCA end.
The 2′-OH of tRNA A76 Is a Significant Contributor to Aminoacyl Transfer.
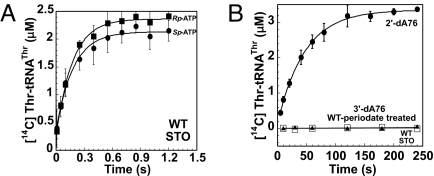
In HisRS, a thio substitution at the pro-S oxygen of the adenylate reduced aminoacyl transfer by 10,000-fold, whereas thio substitution of the pro-R oxygen reduced transfer 50-fold. The pro-S oxygen of the adenylate therefore represents the likely general base for aminoacyl transfer (5). Single-turnover rapid-quench experiments employing Sp and Rp-α-phosphorothioate-substituted ATPs were performed on ThrRS to test the generality of this observation. However, no stereospecific effect was observed, and the aminoacyl transfer rates for both substituted adenylates were no more than 3-fold slower (5.5 s−1 for Sp and 5.7 s−1 for Rp, vs. 14.5 s−1) than the rate using standard ATP (Fig. 4A). For ThrRS, therefore, neither of the nonbridging oxygens of the adenylate is likely to serve as general base for aminoacyl transfer.
Fig. 4.
Single-turnover progress curves for α-phosphorothioate-substituted adenylate and 2′- and 3′-substituted variants at tRNA A76 using ThrRS wild-type enzyme. (A) Rp (filled squares) and Sp (filled circles) α-phosphorothioate-substituted ATP analogs. (B) Modified A76 tRNAs (2′-dA76-, 3′-dA76-, and tRNAThr-periodate-treated). The 3′-dA76 (open squares) and tRNAThr-periodate treated (filled triangles) served as the controls for the reaction.
In the co-crystal structure of the ThrRS–tRNAThr complex (6), Gln-484 makes the closest approach to the 3′-OH of A76, but its side chain does not readily ionize. We therefore considered the adjacent 2′-OH as a candidate for general base and generated 2′-dA76 and 2′-fluoro-A76 tRNA derivatives of tRNAThr by pyrophosphorolysis and nucleotide exchange reactions (14, 15). Control experiments used 3′-dA76 and wild-type periodate-treated tRNAThr molecules. Consistent with the determined regiospecificity of ThrRS (16), aminoacyl transfer was abolished in the 3′-dA76 tRNA. The 2′-dA76 and 2′-fluoro-A76 tRNAs were also significantly, but not completely, diminished in their ability to support aminoacyl transfer, on the order of 763-fold for the former and 94-fold for the latter (Fig. 4B and Fig. S5a). Notably, the decrease in ktrans observed with 2′-dA76 was significantly greater than any of the decreases observed with the single mutants in the ThrRS active site.
The decrease in ktrans associated with conversion of the 2′-OH to 2′-dA76 and 2′-fluoro and His-309 to H309A derivatives (Table 2 and Fig. S5 b and c) was analyzed by a double mutant thermodynamic free energy cycle, as described in Fig. S6 a and b. The ΔΔΔG° free energy of interaction for H309A and 2′-dA76 is −2.8 kcal/mol, whereas the ΔΔΔG° free energy of interaction for H309A and 2′-fluoro-A76 is −1.35 kcal/mol. The algebraic sign of these interaction terms indicates that the interaction between His-309 and the 2′-OH is synergistic and thermodynamically coupled.
Table 2.
Role of the 2′-OH of A76 in aminoacyl transfer
| ThrRSWT or mutant | tRNAThr A76 variant | Rate of aminoacyl transfer, ktranss−1 | Free energy change ΔΔG, kcal/mol |
|---|---|---|---|
| Wild type | Wild type | 14.5 ± 2.5 (1) | 0 |
| Wild type | 2′H-A76 | 0.019 ± 0.003 (763) | 4.1 |
| Wild type | 2′F-A76 | 0.154 ± 0.004 (94) | 2.8 |
| H309A | Wild type | 0.06 ± 0.002 (242) | 3.4 |
| H309A | 2′H-A76 | 0.008 ± 0.001 (1,813) | 4.6 |
| H309A | 2′F-A76 | 0.006 ± 0.002 (2,417) | 4.8 |
All values represent the mean ± SD of three independent experiments. Values in parentheses represent the fold reductions (ktrans-WT/ktrans-mut) in ktrans with respect to ThrRS WT. The change in the free energy (kcal/mol) was computed from ΔΔG = −RT ln (krel), where krel = (ktrans-WT/ktrans-mut).
Discussion
Similarities in the Overall Class II aaRSs Catalytic Cycle Do Not Preclude Differences in the Mechanisms of Amino Acid Activation and Aminoacyl Transfer.
As seen for HisRS (5, 12), the rates of amino acid activation (10) and aminoacyl transfer (Fig. 2B) catalyzed by ThrRS are both faster than the overall rate of the aminoacylation cycle when both half-reactions proceed in sequence (Fig. 2A). The rate of amino acid activation was modestly attenuated by the presence of the tRNA (Fig. 2C), an effect that could be uncoupled by mutations in the enzyme (Fig. 3C). Accordingly, amino acid activation in the presence of tRNA appears to be the rate-determining step for both HisRS and ThrRS.
Despite these similarities, conserved active site residues make different contributions in the 2 systems. Although interactions with the α-PO4 of ATP were found to be important for both ThrRS and HisRS, the degree of stabilization of the adenylation transition state provided by interactions to the pro-R and pro-S nonbridging oxygens differed. In ThrRS, the largest contributions to adenylate stabilization were provided by Arg-363 and Lys-465, which contact the nonbridging pro-R and pro-S oxygens, respectively. The contacts made by Arg-363 to both the substrate threonine carboxylate and the α-phosphate (6, 8) were found to be particularly significant (Table S1). By contrast, the loss of the Lys-465 adenylate pro-S oxygen interaction had a much a smaller effect on pyrophosphate exchange reaction than did loss of the corresponding contact made by Arg-363 (Table S1). This is also in contrast to the 1,000-fold drop in amino acid activation associated with R259H, which substitutes a contact to the pro-S oxygen of the adenylate in HisRS (5).
The functional distinctions between ThrRS and HisRS were even more significant with respect to aminoacyl transfer. Although HisRS and PheRS exhibit stereospecificity with respect to their sensitivity to phosphorothioates and disfavor the use of Rp-ATPαS as substrate (17), use of phosphorothioate-substituted ATP had only a modest effect on ThrRS (compare Fig. 2B with Fig. 4A). The interaction between Lys-465 and the pro-S oxygen also contributed virtually nothing to aminoacyl transfer (Table 1 and Fig. S2c). AaRSs within the same family can thus share broad similarities with respect to their overall catalytic cycle (i.e., the rate-determining step) and yet still exhibit pronounced differences in how active-site residues and substrate groups contribute to the mechanism of aminoacylation.
Involvement of the 2′-OH of A76 in a Proton Relay for Aminoacyl Transfer.
The unsuitability of both the nonbridging oxygens of the adenylate and Gln-484 as general bases served to focus attention on the 2′-OH of A76, an attractive candidate because experiments employing 2′-deoxy- and 2′-fluoro-tRNA molecules revealed that both produced significant decreases in transfer rates, 763-fold for the former and 94-fold for the latter (Table 2). Although both substitutions effectively eliminate the possibility of proton transfer between vicinal hydroxyl groups, the 2′-deoxy- and the 2′-fluoro groups differ with respect to their structural and hydrogen bonding features. The 2′-deoxy replacement abolishes the capacity for hydrogen bonding and promotes the C2′-endo conformation found in the riboses of DNA. By contrast, the fluoro substitution retains a modest ability to form hydrogen bonds as an acceptor and promotes the C3′-endo conformation characteristic of RNA (18). The combination of the structural consequences of the ribose conformation and the potential for hydrogen bonding may thus rationalize the higher activity of the fluoro substitution.
The recovery of aminoacyl transfer function by the fluoro group was, however, dependent on the presence of His-309. Without the His-309 side chain, the fluoro substitution was more deleterious than the deoxy (Table 2 and Fig. S5 b and c). A notable consequence of the H309A mutation was to produce a lag in the accumulation of aminoacylated tRNA (Fig. 3B). As noted above, H309A may decrease the rate of isomerization ThrRS–tRNAThr from a nonproductive to productive complex, which in turn may depend on the correct orientation of the CCA moiety. If so, one function of the hydrogen bond between His-309 and the 2′-OH may be to establish the position and orientation of A76 for transfer.
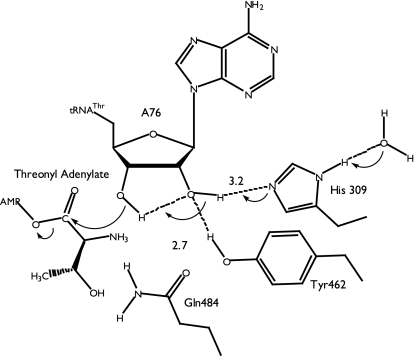
The residual 93-fold drop in aminoacyl transfer activity of the 2′-fluoro relative to wild type suggests that positioning alone is insufficient to explain the requirement for the 2′-OH. The 2′-OH may also contribute to aminoacyl transfer by a proton transfer mechanism involving His-309 (Fig. 5). In this mechanism, the imidazole ring of His-309 can either activate the 2′-OH directly, or indirectly, via a water molecule. In either scenario, the activated 2′-OH serves to activate the 3′-OH, leading to aminoacyl transfer. The protonated His-309 would then subsequently lose the proton to solvent.
Fig. 5.
The mechanism of aminoacyl transfer mediated by the 2′-OH of tRNA A76 in E. coli ThrRS. Dotted lines represent H-bonds, and the arrows indicate the proton relay. See Discussion for the detailed explanation of the mechanism.
The model proposed above accounts for the absence of a general base in the ThrRS active site and the mutational and functional group substitution data. In principle, the proton on the 2′-OH could be transferred either to His-309 or Tyr-462, as phenolate. There is a strong precedent for histidine serving as general base in other systems, particularly ribonuclease A (19). Although substitution of His-309 had the largest impact of any side-chain mutation tested, the effect was not as great as the corresponding substitution mutants in ribonuclease A, which are decreased nearly 3 orders of magnitude (20). In contrast, proton relay mechanisms involving tyrosine are less well established (21, 22), and an initial presteady-state characterization of Y462F showed no more than a 6-fold drop in activity (unpublished data).
Precedents for the proton relay model are provided by the histidine-NAD+ dependent horse liver alcohol dehydrogenase (ADH) and sirtuins. In ADH, a relay mechanism serves to transfer the proton from the substrate alcoholic OH group to the β-OH of Ser-48, then to 2′-OH of NAD+, to the imidazole moiety of His-51, and then to water (23). This activation step facilitates the oxidation process and prepares the system for subsequent hydride transfer. Replacement of His-51 with Gln in yeast ADH is associated with a 30-fold decrease in catalytic efficiency (24). His-51 is not universally conserved in ADH orthologs, but can be infrequently replaced with Thr, Tyr, or Asn (24). This is reminiscent of the replacements of His-309 by Tyr in the ThrRSs of some archaeal organisms (Fig. S1). The sirtuins are protein deacetylases for which a proton relay mechanism has also been proposed. In the currently favored peptidyl-imidate mechanism, the attack of the 2′-OH of NAD+ on the imidate of the acetyllysine is activated by proton transfer to the adjacent 3′-OH and then to nearby His-16 (25). This is strikingly similar to the mechanism for aminoacyl transfer proposed here. Replacement of His-16 with Ala is similarly associated with decreased deacetylation function (26).
The proton relay model accounts for the relatively larger effect of the 2′-OH substitution in tRNAThr on aminoacyl transfer than that seen for other related tRNA systems under steady-state aminoacylation conditions and for the relatively minor effect of H309A on Km for tRNA (Table S1). In other tRNA systems, removal of the vicinal hydroxyl decreases aminoacylation by factors that range from a low of only 2-fold (16) to a moderate 35-fold (27), to a high of 100-fold (28). Loss of adjacent hydroxyl groups on A76 has also been shown to decrease tRNA-dependent editing (29). In class I IleRS, the deacylation of Val-tRNAIle is decreased by >500-fold when the adjacent 3′-OH is replaced with hydrogen (30), whereas in class II PheRS, substitution of the 3′-OH similarly reduces activity (31). The latter observation led to a model in which the 3′-OH assists in the activation of an essential hydrolytic water molecule during the deacylation of Tyr-tRNAPhe (31). Although the work here reports a system in which a contribution of the adjacent vicinal hydroxyl of A76 has been specifically assigned to the transfer step, the possibility that other systems may feature a role of the adjacent hydroxyl in aminoacyl transfer and/or editing remains to be investigated.
Catalytic Role of the 2′-OH of tRNAThr Compared with Other RNA Systems, Including Peptidyl Transferase.
Substitution of the 2′-OH groups at the 5′ end of the exon of Tetrahymena group I intron with either hydrogen or fluorine reduced rates of splicing by ≈600- and ≈10-fold, respectively (32), whereas ribosomal peptidyl transferase function is decreased ≈106-fold in single-turnover assays with 2′-H- and 2′-fluoro-substituted P-site tRNA substrates (33). Consistent with the crystal structure of the peptidyl transferase center (34), the 2′-OH of the P-site tRNA was therefore proposed to serve as a general base to activate the α-NH3 nucleophile (33). Alternatively, the ribosome lowers the entropic barrier to activation, aided by well-defined short-range hydrogen bonds between active-site RNA functional groups and critical substrate functional groups (35). Such interactions may facilitate proton transfer in the peptidyl transferase center (36, 37). For ThrRS, interactions between His-309, Tyr-462, and the 2′-OH that position A76 relative to the scissile bond of the adenylate may represent a similar strategy to increase catalytic power by combination of a delocalized proton on the 3′-OH and reduced entropy.
The 102 to 103 rate enhancement provided by the 2′-OH of A76 to the aminoacylation of tRNAThr is several orders of magnitude less than the contribution of tRNA-assisted catalysis to peptidyl transferase (18). Both systems, however, illustrate how catalytic features of primordial RNA-based aminoacylating enzymes (38) may have been partially retained to enhance protein-based catalysis. As part of the evolution of a modern translation apparatus with greater speed and accuracy, genetic selection might have driven early RNA-based aminoacylation catalysts to partner with simple protein-based enzymes, with functional groups on both partners contributing to the overall catalytic mechanism. Such a model could explain why in the case of the ribosome and the aaRSs, obvious protein-derived general acid/general base catalysts are missing and why the catalytic contribution of ionizable ribose groups is still significant. Along these lines, the parallel roles of the 2′-OH in tRNAThr and the 2′-OH of the NAD+ cofactor of ADH in their respective proton relay mechanisms is striking and recalls the early proposal from H. B. White (39) that contemporary coenzymes may share an evolutionary history with catalytic RNAs.
Methods
Material Preparation.
The wild-type E. coli ThrRS, ThrRS mutants, and CCA-adding enzymes were expressed and purified by Ni-nitrilotriacetate affinity chromatography as described in ref. 10. The tRNA was expressed from pWFW1015 (40) in XL-1 Blue (Stratagene) cells and then purified as described in ref. 41. The tRNA A76 modification reactions were performed by using the in vivo produced full-length tRNAThr according to optimized protocols (15, 30). Details of these procedures and their modifications are provided in SI Methods.
Kinetic Assays.
The pyrophosphate exchange reaction (8) and the steady-state aminoacylation assays were conducted according to published protocols (41). Single-turnover and presteady-state kinetic assays were performed as described for HisRS (5, 12). The bacterial complementation assay was performed as described (6, 11). All details, including modifications and data analysis, are presented in SI Methods.
Supplementary Material
Acknowledgments.
We thank Ya-Ming Hou (Thomas Jefferson University, Philadelphia) for providing overexpressing CCA-adding enzyme strain and Paul Schimmel (Scripps Research Institute, La Jolla, CA) for tRNAThr overexpression plasmid; Christopher Berger, Charles W. Carter, Jr., Eugene G. Mueller, and Scott Strobel, for the critical reading of the manuscript; Ethan Guth for helpful discussions and assistance in Kintek global fitting; and Mathias Springer (Institut de Biologie Physico-Chimique, Paris, France) for providing the thrS-null bacterial strain. This work was supported by the National Institute of General Medical Sciences, National Institutes of Health Grant GM54899.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804247105/DCSupplemental.
References
- 1.Arnez JG, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem Sci. 1997;22:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 2.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 3.Perona JJ, Rould MA, Steitz TA. Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry. 1993;32:8758–8771. doi: 10.1021/bi00085a006. [DOI] [PubMed] [Google Scholar]
- 4.Cavarelli J, et al. The active site of yeast aspartyl-tRNA synthetase: Structural and functional aspects of the aminoacylation reaction. EMBO J. 1994;13:327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guth E, Connolly SH, Bovee M, Francklyn CS. A substrate-assisted concerted mechanism for aminoacylation by a class II aminoacyl-tRNA synthetase. Biochemistry. 2005;44:3785–3794. doi: 10.1021/bi047923h. [DOI] [PubMed] [Google Scholar]
- 6.Sankaranarayanan R, et al. The structure of threonyl-tRNA synthetase-tRNATyr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 7.Dock-Bregeon A, et al. Transfer RNA-mediated editing in threonyl-tRNA synthetase: The class II solution to the double discrimination problem. Cell. 2000;103:877–884. doi: 10.1016/s0092-8674(00)00191-4. [DOI] [PubMed] [Google Scholar]
- 8.Sankaranarayanan R, et al. Zinc ion-mediated amino acid discrimination by threonyl-tRNA synthetase. Nat Struct Biol. 2000;7:461–465. doi: 10.1038/75856. [DOI] [PubMed] [Google Scholar]
- 9.Torres-Larios A, et al. Conformational movements and cooperativity upon amino acid, ATP, and tRNA binding in threonyl-tRNA synthetase. J Mol Biol. 2003;331:201–211. doi: 10.1016/s0022-2836(03)00719-8. [DOI] [PubMed] [Google Scholar]
- 10.Bovee ML, Pierce MA, Francklyn CS. Induced fit and kinetic mechanism of adenylation catalyzed by Escherichia coli threonyl-tRNA synthetase. Biochemistry. 2003;42:15102–15113. doi: 10.1021/bi0355701. [DOI] [PubMed] [Google Scholar]
- 11.Caillet J, et al. Mutations in residues involved in zinc binding in the catalytic site of Escherichia coli threonyl-tRNA synthetase confer a dominant lethal phenotype. J Bacteriol. 2007;189:6839–6848. doi: 10.1128/JB.00439-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guth E, Francklyn CS. Kinetic discrimination of tRNA identity by the conserved motif 2 loop of a class II aminoacyl-tRNA synthetase. Mol Cell. 2007;25:531–542. doi: 10.1016/j.molcel.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moulinier L, et al. The structure of an AspRS-tRNA(Asp) complex reveals a tRNA-dependent control mechanism. EMBO J. 2001;20:5290–5301. doi: 10.1093/emboj/20.18.5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrenfeld GM, Francis TA, Hecht SM. Loss of positional specificity in the aminoacylation of Escherichia coli tRNAGly. J Biol Chem. 1983;258:11745–11750. [PubMed] [Google Scholar]
- 15.Francis TA, Ehrenfeld GM, Gregory MR, Hecht SM. Transfer RNA pyrophosphorolysis with CTP(ATP):tRNA nucleotidyltransferase: A direct route to tRNAs modified at the 3′ terminus. J Biol Chem. 1983;258:4279–4284. [PubMed] [Google Scholar]
- 16.Sprinzl M, Cramer F. Site of aminoacylation of tRNAs from Escherichia coli with respect to the 2′- or 3′-hydroxyl group of the terminal adenosine. Proc Natl Acad Sci USA. 1975;72:3049–3053. doi: 10.1073/pnas.72.8.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von der Haar F, Cramer F, Eckstein F, Stahl KW. On the stereochemistry of activation of phenylalanine by phenylalanyl-tRNA synthetase from baker's yeast. Eur J Biochem. 1977;76:263–267. doi: 10.1111/j.1432-1033.1977.tb11591.x. [DOI] [PubMed] [Google Scholar]
- 18.Weinger JS, Strobel SA. Participation of the tRNA A76 hydroxyl groups throughout translation. Biochemistry. 2006;45:5939–5948. doi: 10.1021/bi060183n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herschlag D. Ribonuclease revisited: Catalysis via the classical general acid–base mechanism or a triester-like mechanism. J Am Chem Soc. 1994;116:11631–11635. [Google Scholar]
- 20.Thompson JE, Raines RT. Value of general acid–base catalysis to ribonuclease A. J Am Chem Soc. 1994;116:5467–5468. doi: 10.1021/ja00091a060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong B, et al. Hydride transfer versus hydrogen radical transfer in thymidylate synthase. J Am Chem Soc. 2006;128:5636–5637. doi: 10.1021/ja060196o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang YH, Chuang LY, Hwang CC. Mechanism of proton transfer in the 3α-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni. J Biol Chem. 2007;282:34306–34314. doi: 10.1074/jbc.M706336200. [DOI] [PubMed] [Google Scholar]
- 23.Hennecke M, Plapp BV. Involvement of histidine residues in the activity of horse liver alcohol dehydrogenase. Biochemistry. 1983;22:3721–3728. doi: 10.1021/bi00285a001. [DOI] [PubMed] [Google Scholar]
- 24.LeBrun LA, Park DH, Ramaswamy S, Plapp BV. Participation of histidine-51 in catalysis by horse liver alcohol dehydrogenase. Biochemistry. 2004;43:3014–3026. doi: 10.1021/bi036103m. [DOI] [PubMed] [Google Scholar]
- 25.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 26.Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog–NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 27.Vortler CS, et al. Determination of 2′-hydroxyl and phosphate groups important for aminoacylation of Escherichia coli tRNAAsp: A nucleotide analogue interference study. RNA. 1998;4:1444–1454. doi: 10.1017/s1355838298980967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pleiss JA, Wolfson AD, Uhlenbeck OC. Mapping contacts between Escherichia coli alanyl tRNA synthetase and 2′ hydroxyls using a complete tRNA molecule. Biochemistry. 2000;39:8250–8258. doi: 10.1021/bi0001022. [DOI] [PubMed] [Google Scholar]
- 29.von der Haar F, Cramer F. Hydrolytic action of aminoacyl-tRNA synthetases from baker's yeast: “Chemical proofreading” preventing acylation of tRNA(I1e) with misactivated valine. Biochemistry. 1976;15:4131–4138. doi: 10.1021/bi00663a034. [DOI] [PubMed] [Google Scholar]
- 30.Nordin BE, Schimmel P. Plasticity of recognition of the 3′ end of mischarged tRNA by class I aminoacyl-tRNA synthetases. J Biol Chem. 2002;277:20510–20517. doi: 10.1074/jbc.M202023200. [DOI] [PubMed] [Google Scholar]
- 31.Ling J, Roy H, Ibba M. Mechanism of tRNA-dependent editing in translational quality control. Proc Natl Acad Sci USA. 2007;104:72–77. doi: 10.1073/pnas.0606272104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herschlag D, Eckstein F, Cech TR. Contributions of 2′-hydroxyl groups of the RNA substrate to binding and catalysis by the Tetrahymena ribozyme: An energetic picture of an active site composed of RNA. Biochemistry. 1993;32:8299–8311. doi: 10.1021/bi00083a034. [DOI] [PubMed] [Google Scholar]
- 33.Weinger JS, et al. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat Struct Mol Biol. 2004;11:1101–1106. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- 34.Schmeing TM, et al. Structural insights into the roles of water and the 2′-hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell. 2005;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Sievers A, Beringer M, Rodnina MV, Wolfenden R. The ribosome as an entropy trap. Proc Natl Acad Sci USA. 2004;101:7897–7901. doi: 10.1073/pnas.0402488101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trobro S, Aqvist J. Analysis of predictions for the catalytic mechanism of ribosomal peptidyl transfer. Biochemistry. 2006;45:7049–7056. doi: 10.1021/bi0605383. [DOI] [PubMed] [Google Scholar]
- 37.Beringer M, Rodnina MV. The ribosomal peptidyl transferase. Mol Cell. 2007;26:311–321. doi: 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 38.Lee N, et al. Ribozyme-catalyzed tRNA aminoacylation. Nat Struct Biol. 2000;7:28–33. doi: 10.1038/71225. [DOI] [PubMed] [Google Scholar]
- 39.White HB., III Coenzymes as fossils of an earlier metabolic state. J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- 40.Waas WF, Schimmel P. Evidence that tRNA synthetase-directed proton transfer stops mistranslation. Biochemistry. 2007;46:12062–12070. doi: 10.1021/bi7007454. [DOI] [PubMed] [Google Scholar]
- 41.Francklyn CS, First EA, Perona JJ, Hou Y-M. Methods for kinetic and thermodynamic analysis of aminoacyl-tRNA synthetases. Methods. 2008;44:100–118. doi: 10.1016/j.ymeth.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.