Abstract
A highly efficient selection method for enhanced enzyme enantioselectivity based on yeast surface display and fluorescence-activated cell sorting (FACS) is developed and validated. Its application to horseradish peroxidase has resulted in enzyme variants up to 2 orders of magnitude selective toward either substrate enantiomer at will. These marked improvements in enantioselectivity are demonstrated for the surface-bound and soluble enzymes and rationalized by computational docking studies.
Keywords: directed evolution, enzyme design, molecular modeling, redox enzymes, stereoselectivity
There is an ever-growing demand for enantiopure chemical compounds, in particular for new pharmaceuticals (1). Although enzymes, being chiral molecules, offer undeniable benefits as asymmetric catalysts in organic synthesis, their enantioselectivity for desired unnatural substrates is often insufficient for practical applications (2). Improving enzymatic enantioselectivity toward a given substrate is thus a practically important but arduous task. Established strategies for achieving this goal without genetically modifying the enzyme include solvent engineering (3), bioimprinting (4), optimization of reaction conditions (5), and coupling biocatalysis with chemical catalysis (6).
Recently, much emphasis has been placed on protein engineering, particularly by directed evolution, as an effective approach to create enzymes with improved properties (7, 8). Although effective in improving such enzyme properties as catalytic activity and thermal stability, this approach has been far less successful in evolving enzymes with higher enantioselectivity (9). In particular, an efficient search of protein sequence space with respect to enantioselectivity and development of high-throughput selection methods for assaying enantioselectivity remain major challenges (10, 11). Consequently, enzyme enantioselectivities achieved thus far by using directed evolution typically have been quite modest (11).
These problems are particularly severe for such a complex (albeit catalytically powerful and versatile) enzyme as horseradish peroxidase (HRP). Among other reactions, HRP catalyzes oxidation of numerous phenols with hydrogen peroxide but typically does that with low enantioselectivity (12). It contains multiple disulfides, N-linked glycosylations, and a catalytically essential heme moiety, making the enzyme refractory to expression in prokaryotes (13). Therefore, to screen large libraries of HRP variants, a eukaryotic system, such as yeast, must be used (14).
In the present study, we have developed and validated a highly efficient selection method based on yeast surface display and fluorescence-activated cell sorting (FACS) that has led to raising HRP's enantioselectivity up to 2 orders of magnitude toward either substrate enantiomer at will. These marked improvements in enantioselectivity have been demonstrated and rationalized for both the surface-bound and soluble enzymes.
Results and Discussion
We recently demonstrated that the enantioselectivity of HRP displayed on the cell surface of yeast can be readily determined by using fluorescent phenolic substrates (15). Employing this methodology in the present study, we determined the enantioselectivity of wild-type HRP toward a representative chiral phenol, tyrosinol, linked to 2 different positional isomers of the Alexa Fluor 488 fluorescent dye (1 and 2 in Fig. 1). The enantioselectivity, E(l/d) (defined herein as the initial rate of the enzymatic oxidation of the l enantiomer divided by that of the d enantiomer), of wild-type yeast-surface-bound HRP was negligible for both substrates: 1.6 ± 0.5 and 0.8 ± 0.1 for 1 and 2, respectively (the first 2 entries in Table 1), in agreement with that for other chiral phenolic substrates (12). We then endeavored to enhance it toward both enantiomers of the substrates 1 and 2 by means of directed evolution.
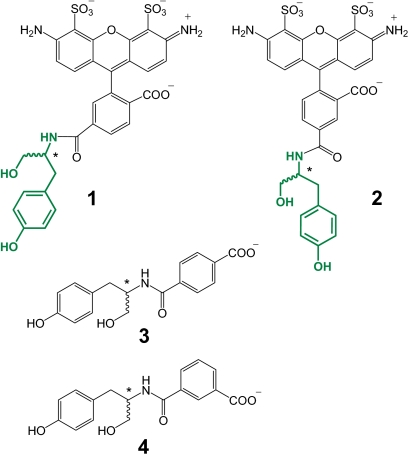
Fig. 1.
Chemical structures of the reducing substrates used in the present study to assess HRP's enantioselectivity (asterisks designate stereogenic centers); the tyrosinol (chiral) portion is shown in green.
Table 1.
Enantioselectivities of l and d selective yeast-bound HRP variants toward 1 and 2 discovered in 3 rounds of directed evolution
| HRP variant | Substrate | vl* | vd* | E(l/d)† |
|---|---|---|---|---|
| Wild-type | 1 | 2.6 ± 0.6 | 1.6 ± 0.7 | 1.6 ± 0.5 |
| Wild-type | 2 | 21 ± 3 | 25 ± 4 | 0.8 ± 0.1 |
| LIII‡ | 1 | 85 ± 6 | 1.8 ± 0.5 | 49 ± 1 |
| LIII‡ | 2 | 4.2 ± 0.6 | 0.4 ± 0.1 | 10 ± 1 |
| DIII§ | 1 | 0.8 ± 0.3 | 2.4 ± 0.8 | 0.3 ± 0.4 |
| DIII§ | 2 | 2.0 ± 0.4 | 154 ± 4 | 0.013 ± 0.003 |
* vl and vd are the initial rates of oxidation of the l enantiomer and d enantiomer, respectively, reported in MFU/min. All experiments were conducted at least in triplicate with the mean and standard deviation values given in the table. See Methods for details.
†Enantioselectivity, E(l/d), is defined as vl/vd. Note that E(l/d) × E(d/l) = 1.
‡The l selective variant discovered in 3 rounds of directed evolution.
§The d selective variant discovered in 3 rounds of directed evolution.
Despite some progress in the first main step of the directed evolution methodology, creation of genetic diversity in the target gene in the form of gene libraries (16), the development of an effective high-throughput selection method for enzymatic enantioselectivity remains daunting (11, 17). Although FACS has shown much promise as a high-throughput selection method (18), it requires a stable link between genotype (the DNA encoding a particular enzyme variant) and phenotype (the enzyme's enantioselectivity) if the selection is to be carried out on the entire gene pool at once (19).
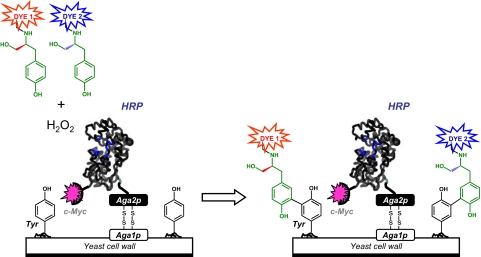
We reasoned that such a link could be created when 2 fluorescent enantiomeric substrates are simultaneously oxidized by HRP that is displayed on the surface of yeast. In this scheme, an enantiomeric pair of chiral phenolic HRP substrates is conjugated to 2 different fluorescent dyes (Dye 1 and Dye 2 in Fig. 2). The enzymatic oxidation of these conjugates yields phenolic free radicals that are captured by the cell surface, thereby creating yeast cells stained with 2 different colored dyes. The ratio of the fluorescence intensities (Dye 1/Dye 2) of these cells should correlate to the enzyme's enantioselectivity (to be exact, the enzyme's selectivity toward the same chiral fragment of the substrate because the dyes are different), thus establishing the genotype–phenotype link required for FACS analysis. Furthermore, this 2-color selection method affords a “dual selection”—selecting enzyme variants with reactivity for the desired enantiomer, while simultaneously excluding those with reactivity toward the undesired enantiomer, a key attribute of an ultrahigh-throughput selection method.
Fig. 2.
Schematic representation of the ultrahigh-throughput selection method for yeast-surface-bound HRP variants with enhanced l or d enantioselectivity. HRP, expressed as a fusion protein to the c-Myc tag and Aga2p mating agglutinin protein, is displayed on the yeast surface via disulfide bridges between the Aga2p and Aga1p proteins. Enzymatic oxidation of the l and d enantiomers of tyrosinol (shown in green) conjugated to fluorescent dyes (Dye 1 and Dye 2) yields phenoxyl radicals that then nonenzymatically react with Tyr residues of membrane-bound proteins; this reaction leads to labeled cells with fluorescence intensity that depends on the enantioselectivity of HRP. The enzymatic activity is normalized via fluorescently labeled antibodies against the c-Myc tag (magenta star). Multiparameter FACS is used to isolate cells with the highest ratio of fluorescence intensities (Dye 1/Dye 2 or Dye 2/Dye 1) encoding l or d selective HRP variants.
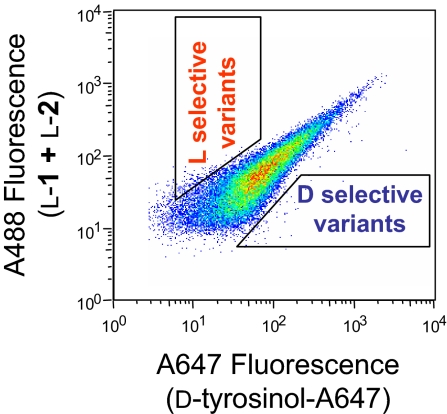
To test this idea, we covalently attached 1 enantiomer of tyrosinol to the Alexa Fluor 488 dye (A488, obtained commercially as a mixture of 2 positional isomers) and the other to the Alexa Fluor 647 dye (A647, used by us herein for screening purposes only). We then determined HRP's activity and selectivity toward these fluorescent conjugates by incubating yeast cells displaying wild-type HRP on their surface with the oxidizing substrate H2O2 and an equimolar mixture of l-tyrosinol conjugated to A488 and d-tyrosinol to A647, followed by FACS analysis. The resultant dual-parameter dot plot of A488 (y axis) vs. A647 (x axis) fluorescence intensity exhibits a clustered signal in the middle (Fig. 3) indicating that (i) the enzyme was active toward both the enantiomeric substrates and (ii) the cells were labeled with both dyes to an equal intensity. The same pattern of fluorescence intensities (i.e., a clustered signal) was observed with d-tyrosinol attached to A488 and l-tyrosinol to A647 (data not shown) under otherwise identical conditions, indicating that the identity of the dye had little effect on the enzyme's enantiopreference. Consequently, using the aforementioned substrate pairs along with FACS, one could screen HRP-based libraries and select HRP variants with higher l or d enantioselectivity by isolating cells above or below the diagonal cluster, respectively, as depicted schematically in Fig. 3.
Fig. 3.
Multiparameter FACS analysis of surface-bound, wild-type HRP incubated with l-1 + l-2, d-tyrosinol-A647, and H2O2. The regions outlined by trapezoids schematically represent library sort gates, used to isolate l selective (cells with high A488 and low A647 fluorescence) and d selective (cells with low A488 and high A647 fluorescence) HRP variants.
To validate our high-throughput selection method, in the first 2 rounds of evolution, we examined the effectiveness of 2 types of libraries, those produced by active-site-targeted saturated mutagenesis and random mutagenesis, in creating both l and d selective HRP variants. Each round of evolution included a round of mutagenesis of the variant with highest enantioselectivity, followed by screening and selection. In the third round of evolution, we used a library that was created by randomizing an area of the gene at, or near, the enzyme active site. This modification of the experimental approach was prompted by the findings of the first 2 rounds of evolution, namely that (i) the mutations that impacted enantioselectivity discovered through random mutagenesis were located close to the active site and (ii) the locations of these mutations did not seem to be obvious targets for saturated mutagenesis, underscoring the difficulty in identifying residues whose alteration would affect the enantioselectivity.
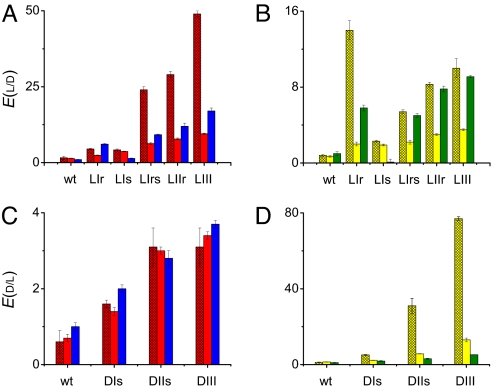
The foregoing libraries assayed by our selection method systematically yielded HRP variants with enhanced l and d enantioselectivity for both substrates 1 and 2 (Fig. 4). As seen in Fig. 4A, the l enantioselectivity of surface-bound HRP variants toward 1 improved steadily and markedly with each round of evolution giving rise to E(l/d) values of 4.5 ± 0.2, 29 ± 1, and 49 ± 1, respectively. Interestingly, the most enantioselective variants produced by random mutagenesis in the 1st and 2nd rounds (LIr and LIIr, respectively) both have single mutations near the active site with a high impact on enantioselectivity. By modifying our strategy to create genetic diversity as explained in the preceding paragraph, in the 3rd round of evolution we discovered a highly l selective variant, LIII, with a total of 8 mutations (listed in the legend to Fig. 4). Inspection of the initial rates of oxidation of 1 catalyzed by this variant reveals that a high E(l/d) value, 49 ± 1, was achieved by increasing the reactivity of the l enantiomer (85 ± 6 mean fluorescence units MFU/min for LIII vs. 2.6 ± 0.6 MFU/min for the wild-type enzyme; Table 1), whereas the oxidation rate of its d counterpart remained similarly low (1.8 ± 0.5 and 1.6 ± 0.7 MFU/min, respectively; Table 1). Likewise, the enantioselectivities of the l selective HRP variants toward 2 rise steadily with each consecutive round of evolution (Fig. 4B). The LIII variant exhibits a 13-fold increase in E(l/d) toward 2 compared with the wild-type enzyme (Table 1). However, because our selection strategy centered on simultaneously isolating variants with high enantiopreference toward both l-1 and l-2, the LIII variant has a lower E(l/d) value toward 2 compared to that of LIr (Fig. 4B).
Fig. 4.
Enantioselectivities of l selective variants toward 1 and 3 (A) and 2 and 4 (B) as well as d selective variants toward 1 and 3 (C) and 2 and 4 (D), discovered in each round of evolution. Red, yellow, blue, and green bar colors represent substrates 1, 2, 3, and 4, respectively; hatched and solid bars designate surface-bound and soluble HRP, respectively. The l and d letters designate the direction of enantiopreference; the Roman numerals after the letters define the round of directed evolution; the r and/or s letters after the Roman numerals indicate whether these variants were isolated from the random or saturated mutagenesis libraries, respectively. Mutations: LIr (Arg178Gln), LIs (Phe68Leu, Gly69Ala, Asn72Glu, Ser73Leu, Ala74Tyr) (14), LIrs (LIr + LIs), LIIr (LIrs + Gln147Arg), LIII (LIIr + Asn158Asp), DIs (Phe68Glu, Gly69Pro, Asn72Lys) (14), DIIs (DIs + Asn137Arg, Ala140His, Phe142Lys, Phe143Met), and DIII (DIIs + Ser167Ile).
We also discovered HRP variants whose d enantioselectivity rose consistently with each round of evolution for both 1 (Fig. 4C) and 2 (Fig. 4D). The most enantioselective variant identified, DIII, has an E(d/l) value of 77 ± 1 toward 2, i.e., a 64-fold improvement compared with the wild-type enzyme (Table 1). Like LIII, this variant also has 8 mutations, all near the active site (listed in the legend to Fig. 4). As seen in Table 1, the enhancement in enantioselectivity of DIII toward 2 stems from both a faster oxidation of the d enantiomer and a slower oxidation of the l enantiomer. The enantioselectivity of DIII toward 1 is enhanced in the same way but to a lesser degree (Table 1). These findings validate the notion that our experimental methodology affords a “dual selection,” i.e., accelerating the evolution of a new function while eliminating the native one. Moreover, our results differ favorably from those with other enzymes evolved for higher enantioselectivity that typically exhibit lower specific activities relative to their parents (20, 21). In contrast, all but one of our most l and d selective variants exhibit higher specific activities than the wild-type enzyme (Table 1).
To determine (i) how the enantioselectivity of the discovered HRP variants depends on cell-surface immobilization and (ii) the effect of the fluorescent dye on the enantioselectivity of these variants, we expressed and purified the corresponding soluble enzyme species. To maintain the glycosylation pattern of the surface-bound HRP, we used the same yeast host for heterologous expression of the soluble enzyme. Because HRP is expressed poorly in this yeast (14, 22), we used a strain of Saccharomyces cerevisiae wherein the folding chaperones protein disulfide isomerase (PDI) and Ig heavy-chain-binding protein (BiP) are overexpressed. This strain, in combination with an optimized induction medium (see Methods) and secretory leader sequence (23), dramatically elevated the secretion of HRP from micrograms to several milligrams of functional enzyme per liter of culture.
After purification of the soluble enantioselective HRP variants, their E values were measured. As seen in Fig. 4 A and B, the soluble wild-type enzyme has the same low enantioselectivity as its predecessor displayed on the yeast surface: E(l/d) values of 1.4 ± 0.1 and 0.7 ± 0.1 for 1 and 2, respectively. Moreover, the enantioselectivities of the soluble variants, as of the surface-bound ones, increase with each round of directed evolution, although the E values of the soluble enzyme are several times lower (Fig. 4). This phenomenon is not uncommon (24) and consistent with the basic rule of directed evolution “one gets what one selects for.” Specifically, our selection method was applied to the HRP enzyme fused to a large, highly glycosylated, Aga2p–Aga1p protein complex (a total of 1,150 aa residues) integrated into the cell wall (as schematically depicted in Fig. 2). The attachment of HRP to the Aga2p–Aga1p protein complex likely restricts the number of conformations that the enzyme can adopt which may affect enantioselectivity. In contrast, in analyzing the enantioselectivity of the soluble enzyme, we used a much smaller protein consisting of HRP linked merely to 2 affinity purification tags (a total of 330 aa residues).
To elucidate the role of the fluorescent dye portions of 1 and 2 in the enantioselectivities of the discovered HRP variants, we measured the E values of the soluble wild-type HRP, as well as LIII and DIII, with tyrosinol. As seen in Table 2, the LIII variant, highly enantioselective toward 1 and 2, is one-half as enantioselective with tyrosinol as the wild-type enzyme. On the other hand, DIII, the variant with the highest preference for d-1 and d-2, is also d enantioselective with tyrosinol (Table 2). As seen in Table 2, however, LIII and DIII exhibit inverted enantioselectivities toward N-acetyl-tyrosinol compared with tyrosinol, suggesting the influence of the positive charge of the substrate. Therefore, attaching the fluorescent dye to tyrosinol plays an important role in determining the enantioselectivity of the discovered HRP variants, once again confirming that one gets what one selects for.
Table 2.
Enantioselectivities of soluble HRP variants toward tyrosinol and N-acetyl-tyrosinol
Separately, we uncovered significant differences in enantioselectivities of the surface-bound enzyme depending on the structural isomer of the dye attached to tyrosinol (1 and 2). For example, DIII is far more enantioselective toward 2 than toward 1 (E(d/l) of 77 ± 1 vs. 3.1 ± 0.5), whereas LIII, on the contrary, strongly prefers 1 to 2 (E(l/d) of 49 ± 1 vs. 10 ± 1) (Table 1). The same trends hold for their soluble counterparts, although the differences in enantioselectivities are more modest (Fig. 4). As is evident from Fig. 1, the structural difference between 1 and 2 arises from the attachment point of tyrosinol: particularly, it is 5 carbon atoms away from the carboxyl group of the benzoate moiety in 1, whereas in 2, it is 4 carbons away. Hence, one explanation of the difference in E values for the regioisomers might be the location of the negatively charged carboxyl group with respect to the stereogenic center. It is also possible, however, that the fused phenyl rings of the A488 dye play a role in enantioselectivity.
To distinguish between these 2 alternatives, we measured the enantioselectivities of the soluble HRP variants toward analogs of 1 and 2 that lack the fused phenyl rings of the dye but still retain the benzoate group (3 and 4, respectively, in Fig. 1). (Our experimental methodology does not allow characterization of the surface-bound enzyme by using these nonfluorescent substrates.) The wild-type enzyme exhibits no enantiodiscrimination with either 3 or 4, as evidenced by the E values of unity for both regioisomers (Fig. 4 A and B). However, as with the substrates 1 and 2, the enantioselectivity toward 3 and 4 is enhanced with each round of evolution for both the l and d selective variants (Fig. 4). Furthermore, the l selective variants are more enantioselective with 3 than with 4, whereas the opposite is true for the d selective ones, consistent with the data obtained for the corresponding variants with 1 and 2 (Fig. 4). These results point to the disposition of the carboxyl group as the main determinant in the enantioselectivity of the discovered variants.
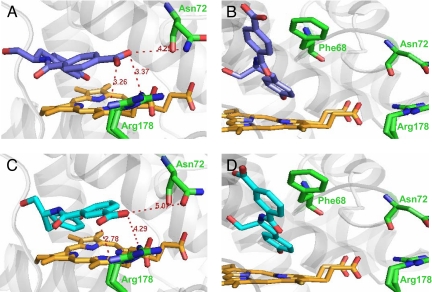
To examine how the location of the negatively charged carboxylate vis-à-vis the stereogenic center can give rise to vastly different enantioselectivities of the isolated variants with 3 and 4 (used instead of 1 and 2, respectively, because of their much simpler structures), we used molecular modeling to simulate complexes of these regioisomers with the wild-type enzyme. As depicted in Fig. 5, the l and d enantiomers of both 3 and 4 exhibit distinct binding modes: in particular, the locations of their carboxyl groups in the active site differ for each enantiomer. The most striking difference in enantioselectivity between 3 and 4 is seen with the LIs variant, which is marginally l selective with 3 (E(l/d) = 1.4 ± 0.1; Fig. 4A) but highly d selective with 4 (E(l/d) = 0.1 ± 0.3; Fig. 4B). Of the 5 mutations of the LIs variant, one seems particularly influential, namely Asn72Glu. Inspection of Fig. 5 A and C reveals that the Asn-72 residue is located close to the carboxyl group of d-4, and even closer to that of d-3 (but not for their l counterparts): the distance between that amino acid residue's amide nitrogen and the carboxylate's oxygens is 5.07 and 4.25 Å, respectively. Therefore, replacing Asn-72 with the negatively charged Glu should weaken the binding of d-3 to the enzyme in the transition state because of electrostatic repulsion, leading to a slower oxidation of d-3 and, in turn, imparting l enantioselectivity. Furthermore, d-4's carboxylate is closer to Arg-178 than d-3's (Fig. 5 A and C). Therefore, the Asn72Glu mutation is more likely to electrostatically repel the d-4 substrate into a new orientation, resulting in a salt bridge with Arg-178 in the transition state, thus enhancing the binding affinity of d-4 and making the LIs variant highly d selective with 4. If this hypothesis is correct, the LIs variant should become l selective with 4 instead when the putative salt bridge is eliminated. Indeed, when the Arg178Gln mutation is introduced, the resultant LIrs variant becomes l selective with 4 (E(l/d) = 5.0 ± 0.2), as well as with 3 (E(l/d) = 9.2 ± 0.2) (Fig. 4 A and B).
Fig. 5.
Modeled complexes of wild-type HRP with d-3 (A), l-3 (B), d-4 (C), and l-4 (D). For clarity, only the active site of the enzyme is shown with the heme moiety in orange, substrate in blue, and some mutated residues in green. Distances indicated are in angstroms. The Arg-178 residue is shown in a double rotamer configuration as it appears in the crystal structure (29); only 1 rotamer configuration was used in docking experiments. See Methods for details of how these models were built.
In the case of the d selective variants, the differences in enantioselectivities with 3 and 4 are more modest compared with the l selective ones: e.g., for DIII the E(d/l) values are 3.7 ± 0.1 with 3 and 5.2 ± 0.1 with 4 (Fig. 4 C and D). Nevertheless, the enantioselectivity enhancement in these variants still could be attributed to the position of the substrate's carboxylate with respect to the enzyme mutations. For example, the DIs variant has 3 mutations, including Phe68Glu and Asn72Lys. The latter one is likely to accelerate the oxidation of d-3 and d-4 by increasing their binding affinities for the enzyme in the transition state through either the establishment of a salt bridge or hydrogen bonding between the positively charged Lys and the carboxylate of d-3 (Fig. 5A) or d-4 (Fig. 5C). On the other hand, the Phe68Glu mutation may lower the oxidation rate of the l enantiomers of 3 and 4 because of electrostatic repulsion of the negatively charged carboxyl group (Fig. 5 B and D, respectively).
In closing, we have developed an ultrahigh-throughput selection method for enzyme enantioselectivity, based on yeast-cell-surface display paired with FACS, validated by discovering highly enantioselective variants of HRP toward tyrosinol conjugated to the A488 fluorescent dye. We have found that the enantioselectivity of the isolated HRP variants depends on the attachment of tyrosinol to the benzoate moiety of this dye and specifically on the position of the carboxylate. The discovered HRP variants are several times more enantioselective when bound to the cell surface than when solubilized. Such surface-bound enzymes with enhanced enantioselectivity toward commercially useful substrates may be used as naturally immobilized asymmetric biocatalysts in chemical reactors.
Materials and Methods
Syntheses.
The tyrosinol-A647 conjugate, 1, 2, 3, 4, and N-acetyl-tyrosinol were synthesized as described in supporting information (SI) Text.
Enantioselectivity of Yeast-Surface-Bound HRP.
The initial rates of substrate oxidation with hydrogen peroxide catalyzed by surface-bound HRP were measured by suspending 1 × 106 HPR-displaying yeast cells in 100 μl of PBS buffer (pH 7.4) containing 15 μM 1 or 2 and 150 μM H2O2, in parallel for both enantiomers. Periodically, 20 μl of the l and d substrate mixtures were withdrawn into 1 ml of PBS containing 0.5% BSA and 10 mM ascorbic acid to quench the reactions. The fluorescently labeled cells were then washed with 0.5 ml of PBS containing 0.1% BSA and labeled with mouse anti-c-Myc monoclonal 9E10 (Covance) and phycoerythrin-goat anti-mouse antibodies (Sigma), as described previously (25). The cells with the same HRP display levels were then analyzed by using a Coulter Epics XL flow cytometer. The mean fluorescence of Alexa Fluor 488 (MFU) for each enantiomer was plotted as a function of time to determine initial reaction rates.
Construction of HRP Libraries. Random Mutagenesis Libraries.
Random mutagenesis libraries were created by using a protocol adapted from ref. 25. Briefly, for both d and l selective enzyme libraries, the HRP gene coding for the best variant was amplified in the presence of nucleotide analogs by using forward (5′-GGTGGAGGAGGCTCTGGTGGAGGCGGTAGCGGAGGCGGAGGGTCGGCTAGC-3′) and reverse (5′-CAGATCTCGAGCTATTACAAGTCCTCTTCAGAAATAAGCTTTTGTTCGGATCC-3′) primers (IDT) under the following conditions. A mixture of 2 ng of pCT2-HRP plasmid (15), 1 μM concentrations of each reverse and forward primer, 0.2 mM dNTPs, 2 mM MgCl2, 2 μM 8-oxo-dGTP, 2 μM dPTP, and 0.05 units·μL−1 Taq polymerase in 1× Taq polymerase reaction buffer was subjected to a thermocycling program which comprised 1 min at 95 °C, followed by 15 cycles of 1 min at 94 °C, 30 s at 60 °C, and 2 min at 72 °C. The mutagenic HRP gene was further amplified 10- to 100-fold in a total volume of 1.5–2 ml by using the aforementioned thermocycling program with 20 additional cycles, and then gel-purified. The libraries were obtained by transforming the mutagenic HRP gene, along with the BamH1-NheI backbone of pCT2con-HRP plasmid, into EBY100 following a published method (25). In the last round of evolution, the libraries were created as described above except that different primers (5′-GTCCTAACGTCTCAAACATAGTACGGGACACTATTGTCAATGAGTTACGATCGGACCC-3′, 5′-GTACGCAGATCGAAGTCGACCAAGGCGCTTAGGTTGCCATTAAGGGGA-CATAGTCC-3′), and the AvrII-PflFI pCT2con-HRP backbone were used. Each library contained ≈107 unique sequences with a mutation frequency of 1–3 mutations per gene.
Saturated Mutagenesis Libraries.
Saturated mutagenesis libraries were constructed by replacing the BsmI-AflII fragment of LIrs and DIs genes with a DNA fragment where Asn-137, Leu-138, Ala-140, Phe-142, and Phe-143 were exhaustively randomized. This DNA fragment was assembled by using oligonucleotides (5′-CAGGAGGTCCCTCTTGGAGGGTTCCTTTGGGACGTCGAGACAGCCTACAAGCATTTTTAGATCTCGCGAATGCG, 5′-CCACCGCTGAGGGCAACGAGATCAGAAGAACGGTTTAAACCAACATTTCTAAAAGAATCCTTAAGTTGTGGAAGTG, 5′-ATTTTTAGATCTCGCGAATGCGNNBNNBCCANNBCCANNBNNBACACTTCCACAACTTAAGGAT) (TriLink) and Phusion High-fidelity DNA polymerase (NEB) under the PCR conditions suggested by the manufacturer of the polymerase following a literature procedure (15). To make LIIs or DIIs libraries, this insert, along with either the pCT2-LIrs or pCT2-DIs plasmid missing the BsmI-AflII fragment, was then used in the EBY100 transformation step as described in ref. 15.
Selection by FACS.
HRP libraries were grown and induced as previously described (15). Freshly induced cells (the number of cells used was at least 10-fold greater than the population diversity) were washed with PBS containing 0.5% BSA, followed by another wash with PBS alone. The cells were resuspended in a PBS solution containing 15 μM l-tyrosinol-A488, 15 μM d-tyrosinol-A647, and 150 μM H2O2, followed by a 30-min shaking at 30 °C. The labeling reaction was then quenched by the addition of ascorbic acid to a final concentration of 10 mM; the cells were washed with PBS containing 0.5% BSA, labeled with mouse anti-c-Myc monoclonal antibody and phycoerythrin-goat anti-mouse antibodies, and sorted as previously described (15). To minimize the influence of the dye on HRP's enantioselectivity, the aforementioned fluorescent substrates were replaced with d-tyrosinol-A488 and l-tyrosinol-A647 in every other round of sorting. After 6 rounds of sorting, plasmid DNA encoding the HRP variants was extracted from the sorted libraries by using the Zymoprep Yeast Plasmid Miniprep kit (Zymo Research) and transformed into XL10-Gold ultracompetent Escherichia coli cells (Stratagene). The plasmids, encoding individual HRP variants were then isolated from E. coli cells and transformed into yeast strain EBY100 for further characterization.
Soluble HRP Purification.
The HPR genes were subcloned into the 4m5.3 plasmid (26) backbone with the Gal1-10 promoter and appS4 leader sequence (23). These HRP-containing plasmids, along with the BiP-overexpressing plasmid pMR-1341 (CEN-URA3) (27), were transformed into a PDI-overexpressing strain of S. cerevisiae, YVH10 (26). The resultant colonies were grown to an OD600 of 5–7 in 1 L of synthetic defined (SD) medium [2% dextrose, 0.34% yeast nitrogen base without (NH4)2SO4, 0.8% casamino acids (VWR), 50 mM sodium phosphate buffer adjusted to pH 6.6] in a 2.5-L fully baffled Tunair flask (Shelton Scientific) by shaking at 250 rpm at 30 °C. To induce expression of the HRP variants, the cells were centrifuged to remove the supernatant and resuspended in 1 L of yeast peptone galactose (YPG) medium (1% Bactoyeast extract, 2% Bactopeptone, 1.8% galactose, 0.2% dextrose, 50 mM sodium phosphate buffer adjusted to pH 6.6) in a 2-L glass Erlenmeyer flask. This medium was supplemented with 50 μg/ml kanamycin, 100 units/ml penicillin G, 200 units/ml streptomycin, 0.034% thiamine HCl, 0.084% δ-aminolevulinic acid, 0.1 mM ferric citrate, and 0.5 μM hemin (100× stock solution was freshly prepared by dissolving hemin in equal parts of ethanol and 0.04 M aqueous NaOH) (Frontier Scientific). The flasks were shaken at 250 rpm at 20 °C for 72 h, with 0.02% δ-aminolevulinic acid added every 24 h. The supernatant was then separated by centrifugation, filter-sterilized, and concentrated to ≈100 ml with a 30K MWCO Amicon Stirred Cell (Millipore). PBS (400 ml) was then added, and the sample was concentrated to a final volume of ≈50 ml. The protein was purified by anti-FLAG (Sigma) affinity chromatography using the protocol provided by the manufacturer. Final protein, digested with EndoH (NEB) to remove glycosylations, was seen as a single band by Coomassie blue staining of an SDS/PAGE gel. Purified yields had RZ (Reinheitszahl) values of 1.5–2.5 and were 1–5 mg/L of initial culture as determined spectrophotometrically by using the extinction coefficient of 100 mM−1cm−1 at 403 nm.
Enantioselectivity of Soluble HRP.
The initial rates of substrate oxidation by soluble HRP were measured by monitoring the rising absorbance of the products as previously described (28). In a typical experiment, the enzyme (0.01–1 μM) was added directly to a spectrophotometric cuvette containing 300 μl of a reaction mixture consisting of H2O2 (0.15 mM for 1 or 2, or 10 mM otherwise) and reducing substrate (15 μM for 1 or 2, or 1 mM otherwise) in a PBS buffer at room temperature. To measure the initial reaction rates, the increases in absorbance as a function of time were recorded at 513 nm for 1 and 2, at 290 nm for 3 and 4, and at 315 nm for tyrosinol and N-acetyl-tyrosinol.
Computational Docking of HRP Variants.
The complexes of wild-type HRP (PDB ID code 7ATJ) (29) with the enantiomers of 3 and 4 were obtained by using a docking method that combines quantum mechanical calculations with Schrödinger's Glide version 4.5. The substrates were geometry-optimized first in molecular mechanics with Macromodel using the OPLS2001 force field and then in quantum mechanics with Jaguar using the Poisson–Boltzmann implicit solvent model of aqueous environment simulation. Quantum mechanics was represented by density functional theory with the B3LYP functional (30) and 6-31G* basis set (31). The presence of iron in the enzyme's heme moiety requires the use of quantum chemical calculations for the complex region that involves electron transfer (32). Therefore, a variation of the previously described QM/MM (quantum mechanical/molecular mechanical) docking algorithm (33, 34) was used. To properly sample binding modes, “restricted docking,” wherein the phenol ring of ferulic acid in the 7ATJ complex served as the restriction point, was performed in addition to standard Glide sampling to generate a total of 10 diverse poses of each enantiomer of 3 and 4. To accurately score these poses, QM/MM single-point energy calculations without geometry optimization were carried out, treating the heme moiety and the substrate as a quantum region. Upon convergence of QM/MM, the atomic charges were fitted for atoms involved in calculations using the ESP (electrostatic potential) method. The poses, with fitted charges, were then ranked by using Glide's score-in-place function, and the lowest binding energy pose was selected.
Supplementary Material
Acknowledgments.
We thank the Massachusetts Institute of Technology Flow Cytometry Core Facility for assistance with flow cytometry and Dr. Daša Lipovšek for making the random mutagenesis library of wild-type HRP. This work was supported by National Institutes of Health Grant GM66712.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809851105/DCSupplemental.
References
- 1.Rouhi M. Chiral chemistry: Traditional methods thrive despite numerous hurdles, including tough luck, slow commercialization of catalytic processes. Chem Eng News. 2004;82:47–62. [Google Scholar]
- 2.Shin H-D, Guo X, Chen R. In: Bioprocessing for Value Added Products from Renewable Resources. Yang ST, editor. Amsterdam: Elsevier; 2006. pp. 351–371. [Google Scholar]
- 3.Klibanov AM. Improving enzymes by using them in organic solvents. Nature. 2001;409:241–246. doi: 10.1038/35051719. [DOI] [PubMed] [Google Scholar]
- 4.Klibanov AM. Enzyme memory—What is remembered and why? Nature. 1995;374:596. doi: 10.1038/374596a0. [DOI] [PubMed] [Google Scholar]
- 5.Phillips RS. Temperature modulation of the stereochemistry of enzymatic catalysis: Prospects for exploitation. Trends Biotechnol. 1996;14:13–16. [Google Scholar]
- 6.Turner NJ. Controlling chirality. Curr Opin Biotechnol. 2003;14:401–406. doi: 10.1016/s0958-1669(03)00093-4. [DOI] [PubMed] [Google Scholar]
- 7.Hult K, Berglund P. Engineered enzymes for improved organic synthesis. Curr Opin Biotechnol. 2003;14:395–400. doi: 10.1016/s0958-1669(03)00095-8. [DOI] [PubMed] [Google Scholar]
- 8.Arnold FH. Combinatorial and computational challenges for biocatalyst design. Nature. 2001;409:253–257. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- 9.Tao HY, Cornish VW. Milestones in directed enzyme evolution. Curr Opin Chem Biol. 2002;6:858–864. doi: 10.1016/s1367-5931(02)00396-4. [DOI] [PubMed] [Google Scholar]
- 10.Reetz MT. Controlling the enantioselectivity of enzymes by directed evolution: Practical and theoretical ramifications. Proc Natl Acad Sci USA. 2004;101:5716–5722. doi: 10.1073/pnas.0306866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reetz MT. Directed evolution of enantioselective enzymes as catalysts for organic synthesis. Adv Catal. 2006;49:1–69. [Google Scholar]
- 12.Gilabert MA, et al. Stereospecificity of horseradish peroxidase. Biol Chem. 2004;385:1177–1184. doi: 10.1515/BC.2004.152. [DOI] [PubMed] [Google Scholar]
- 13.Veitch NC. Horseradish peroxidase: a modern view of a classic enzyme. Phytochemistry. 2004;65:249–259. doi: 10.1016/j.phytochem.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 14.Morawski B, et al. Functional expression of horseradish peroxidase in Saccharomyces cerevisiae and Pichia pastoris. Protein Eng. 2000;13:377–384. doi: 10.1093/protein/13.5.377. [DOI] [PubMed] [Google Scholar]
- 15.Lipovšek D, et al. Selection of horseradish peroxidase variants with enhanced enantioselectivity by yeast surface display. Chem Biol. 2007;14:1176–1185. doi: 10.1016/j.chembiol.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Sen S, Venkata Dasu V, Mandal B. Developments in directed evolution for improving enzyme functions. Appl Biochem Biotechnol. 2007;143:212–223. doi: 10.1007/s12010-007-8003-4. [DOI] [PubMed] [Google Scholar]
- 17.Boersma YL, Droge MJ, Quax WJ. Selection strategies for improved biocatalysts. FEBS J. 2007;274:2181–2195. doi: 10.1111/j.1742-4658.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- 18.Farinas ET. Fluorescence activated cell sorting for enzymatic activity. Comb Chem High Throughput Screen. 2006;9:321–328. doi: 10.2174/138620706776843200. [DOI] [PubMed] [Google Scholar]
- 19.Bershtein S, Tawfik DS. Advances in laboratory evolution of enzymes. Curr Opin Chem Biol. 2008;12:151–158. doi: 10.1016/j.cbpa.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 20.Reetz MT, et al. Learning from directed evolution: Further lessons from theoretical investigations into cooperative mutations in lipase enantioselectivity. ChemBioChem. 2007;8:106–112. doi: 10.1002/cbic.200600359. [DOI] [PubMed] [Google Scholar]
- 21.van Loo B, et al. Directed evolution of epoxide hydrolase from A. radiobacter toward higher enantioselectivity by error-prone PCR and DNA shuffling. Chem Biol. 2004;11:981–990. doi: 10.1016/j.chembiol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 22.Morawski B, Quan S, Arnold FH. Functional expression and stabilization of horseradish peroxidase by directed evolution in Saccharomyces cerevisiae. Biotechnol Bioeng. 2001;76:99–107. doi: 10.1002/bit.1149. [DOI] [PubMed] [Google Scholar]
- 23.Rakestraw AJ, Sazinsky SL, Piatesi A, Antipov E, Wittrup KD. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in S. cerevisiae. Biotechnol Bioeng. 2008 doi: 10.1002/bit.22338. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura Y, et al. Enhancement of activity of lipase-displaying yeast cells and their application to optical resolution of (R,S)-1-benzyloxy-3-chloro-2-propyl monosuccinate. Biotechnol Prog. 2006;22:998–1002. doi: 10.1021/bp060136m. [DOI] [PubMed] [Google Scholar]
- 25.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protocols. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 26.Midelfort KS, et al. Substantial energetic improvement with minimal structural perturbation in a high affinity mutant antibody. J Mol Biol. 2004;343:685–701. doi: 10.1016/j.jmb.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Shusta EV, Raines RT, Pluckthun A, Wittrup KD. Increasing the secretory capacity of Saccharomyces cerevisiae for production of single-chain antibody fragments. Nat Biotechnol. 1998;16:773–777. doi: 10.1038/nbt0898-773. [DOI] [PubMed] [Google Scholar]
- 28.Rojas AM, Gonzalez PA, Antipov E, Klibanov AM. Specificity of a DNA-based (DNAzyme) peroxidative biocatalyst. Biotechnol Lett. 2007;29:227–232. doi: 10.1007/s10529-006-9228-y. [DOI] [PubMed] [Google Scholar]
- 29.Henriksen A, Smith AT, Gajhede M. The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates. J Biol Chem. 1999;274:35005–35011. doi: 10.1074/jbc.274.49.35005. [DOI] [PubMed] [Google Scholar]
- 30.Becke AD. A new mixing of Hartree–Fock and local density-functional theories. J Chem Phys. 1993;98:1372–1377. [Google Scholar]
- 31.Rassolov VA, Pople JA, Ratner MA, Windus TL. 6–31G* basis set for atoms K through Zn. J Chem Phys. 1998;109:1123–1229. [Google Scholar]
- 32.Cho AE. Effect of quantum mechanical charges in binding sites of metalloproteins. BioChip J. 2007;1:70–75. [Google Scholar]
- 33.Cho AE. Quantum mechanical calculations for binding sites of metalloproteins. BioChip J. 2008;2:148–153. [Google Scholar]
- 34.Cho AE, Rinaldo D. Extension of QM/MM docking and its applications to metalloproteins. J Comput Chem. 2008 doi: 10.1002/jcc.21270. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.