Abstract
We have devised a phage display system in which an expanded genetic code is available for directed evolution. This system allows selection to yield proteins containing unnatural amino acids should such sequences functionally outperform ones containing only the 20 canonical amino acids. We have optimized this system for use with several unnatural amino acids and provide a demonstration of its utility through the selection of anti-gp120 antibodies. One such phage-displayed antibody, selected from a naïve germline scFv antibody library in which six residues in VH CDR3 were randomized, contains sulfotyrosine and binds gp120 more effectively than a similarly displayed known sulfated antibody isolated from human serum. These experiments suggest that an expanded “synthetic” genetic code can confer a selective advantage in the directed evolution of proteins with specific properties.
Keywords: directed evolution, phage display, unnatural amino acids
With few exceptions, the genetic codes of all known organisms specify only the 20 canonical amino acids for protein synthesis. Yet it is quite possible that the ability to encode additional amino acids and their corresponding chemical functionalities would be evolutionarily advantageous, especially since nature's choice of 20 could have been arbitrarily fixed at the point of transition between communal and Darwinian evolution paradigms and subsequently sustained by the code's inertia (1). Furthermore, in the limited scope of laboratory-directed evolution, which concerns only one or few specific functions over a short time rather than general organismal fitness over thousands of years, one can easily envision a selective advantage conferred by additional amino acids. Recent developments in our laboratory allow us to explore this possibility. Specifically, orthogonal tRNA/aminoacyl-tRNA synthetase (aaRS) pairs capable of incorporating various unnatural amino acids into proteins in response to unique nonsense and frameshift codons have been added to the translational machinery of Escherichia coli (2). These E. coli (X-E. coli) can now be used for evolution of protein function wherein 21 building blocks, rather than the common 20, are available.
Several unnatural amino acids were initially chosen, on the basis of their unique chemistries, for use in our system. For example, X-E. coli genetically encoding the bidentate metal-chelating amino acid bipyridyl-alanine (3) are well-suited for the evolution of redox and hydrolytic catalysts, as metal ion binding would not require preorganized primary and secondary ligand shells. Similarly, X-E. coli encoding the reactive 4-borono-phenylalanine (4) are well-suited for evolution of receptors specific for glycoproteins or serine protease inhibitors, because the boronate group can form covalent complexes with diols or reactive serine residues. In addition, X-E. coli genetically encoding otherwise posttranslationally modified amino acids, such as sulfotyrosine (5), can be used for evolution of properties that exploit the unique chemical characteristics of the given posttranslational modification, but without any of the host organism and sequence constraints normally limiting such modifications (6). And finally, X-E. coli using keto amino acids, such as para-acetyl-phenylalanine may be advantageous in the evolution of catalysts for reactions involving iminium ion intermediates (e.g., addition, isomerization, or decarboxylation reactions) (7). With this framework in mind, we have developed a system for protein evolution in which unnatural amino acids encoded by X-E. coli are included in phage display libraries. This system is designed such that sequences with unnatural amino acids can be selected based on function from populations containing both sequences with unnatural amino acids and sequences with only the 20 common amino acids. We then used this system for the evolution of anti-gp120 antibodies and found that specific sequences containing sulfotyrosine emerge as winners over all other sequences represented in the population, including those that contain only canonical amino acids. These unique studies demonstrate that an expanded genetic code can confer a selective advantage through the functional contribution of an unnatural amino acid.
Results
Proteins Containing Unnatural Amino Acids Are Correctly Displayed on Phage Coat in a Phagemid Format.
Phage display has proven to be a versatile platform for the directed evolution of various protein functions (8–13). Under the constraints of phage-display evolution, two basic criteria must be met for functional evolution to be successful. First, the phage produced by E. coli must properly and effectively display the protein undergoing evolution; and second, selective advantage (e.g., enrichment) should be as closely linked to functional performance as possible. This requires the mitigation of any systematic biases against certain classes of sequences that are not based on function. Although unnatural amino acids have been displayed on WT M13 phage in single peptides (14), such a system was not amenable to directed evolution experiments within these constraints. We therefore turned to phagemid display, specifically multivalent hyperphage phagemid display (15, 16), which we felt would fulfill these two criteria for both the canonical and unnatural amino acids.
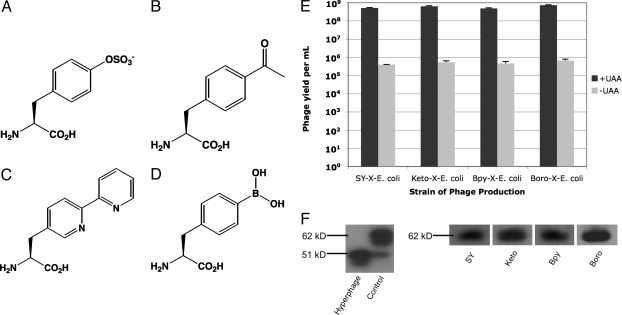
To test whether a phagemid-encoded protein sequence containing an unnatural amino acid could be displayed on the surface of phage, pIII was fused to the C-terminal end of an scFv derived from the common human VH 3–23 and VL A27 germline sequences. An amber codon was substituted at position 111 in the VH CDR3 loop, and this construct was inserted into the pSEX phagemid to create pSEX-GermTAG. This plasmid was subsequently transformed into four different X-E. coli, one encoding sulfotyrosine (SY-X-E. coli) (5), one encoding para-acetyl-phenylalanine (Keto-X-E. coli) (7), one encoding bipyridyl-alanine (Bpy-X-E. coli) (3), and one encoding 4-borono-phenylalanine (Boro-X-E. coli) (4), all in response to the amber codon. Because the only source of pIII for phage packing is phagemid-encoded antibody-pIII fusion (16), phage should be produced only if the amber codon upstream of pIII is suppressed. Hyperphage was then used to generate the respective phage-displayed scFvs from these clones. Phage yield was determined in the presence and absence of the respective unnatural amino acid (Fig. 1 A–D). The addition of the corresponding unnatural amino acid to the growth media resulted in phage yields over 1,000-fold greater than yields in the absence (Fig. 1E), confirming that protein sequences containing unnatural amino acids can be displayed in this system. SDS/PAGE and Western blot analysis of precipitated whole phage was used to verify that the full scFv-pIII fusion was effectively presented (Fig. 1F).
Fig. 1.
Phagemid-display of unnatural amino acids. (A) Structure of sulfotyrosine. (B) Structure of para-acetyl-phenylalanine. (C) Structure of bipyridyl-alanine. (D) Structure of 4-borono-phenylalanine. (E) Yield of phage under optimized conditions where +UAA corresponds to phage produced with the corresponding unnatural amino acid (UAA) supplemented in the media at optimized concentrations and −UAA corresponds to phage produced in the absence of the corresponding unnatural amino acid. Titers were determined in triplicate and error bars correspond to plus standard deviations. (F) Detection of pIII-scFv fusion from whole phage produced using pSEX-GermTAG in the presence of unnatural amino acids. Approximaely 108 phage particles (corresponding to ≈200-μl phage culture precipitated and concentrated to ≈10 μl) per sample were run on a denaturing PAGE gel under reducing conditions and subsequently transferred to a membrane for Western blotting with an anti-pIII antibody. When the same volume of phage similarly produced and prepared from pSEX-GermTAG in the absence of unnatural amino acid was run and Western blotted for pIII, no bands were detected (data not shown). Hyperphage corresponds to phage without displayed scFv. Control corresponds to phage displaying the pIII-scFv fusion from pSEX-GermTAT with only common amino acids; the 51 kDa band is a result of nonspecific proteolysis of the 62-kDa band (pIII-scFv fusion).
Phage-Yield Bias Against Sequences Containing Unnatural Amino Acids Can be Minimized to Allow Directed Evolution.
Although the display of sequences containing unnatural amino acids on the phage coat was successful, early experiments gave low overall phage yields from pSEX-GermTAG when compared to display of sequences from pSEX-GermTAT in which the amber codon was replaced by one specifying tyrosine [supporting information (SI) Table S1]. This was expected because the suppression efficiency of unnatural amino acids using engineered tRNA/aaRS pairs is lower than suppression with the common amino acids using endogenous machinery. Yet to evolve proteins with unnatural amino acids, one must be able to compete sequences containing unnatural amino acids against those containing only the canonical 20 aa over multiple rounds of selection, a scenario that does not tolerate large, systematic expression biases against sequences containing unnatural amino acids. Therefore, we optimized growth conditions (phage production temperature, expression time, and plasmid encoding tRNA/aaRS pairs) and unnatural amino acid concentrations for several X-E. coli (see Table S1) such that yield of phage displaying unnatural amino acids was similar to yield of phage displaying natural sequences. We suspected that optimization could be achieved through growth conditions and amino acid concentrations alone, because it requires only an increase in the rate of full-length fusion-pIII protein expression relative to the rate of the other steps in the phage packaging and assembly process; it does not require increasing amber codon suppression efficiency, likely a much more difficult task. As shown in Table S2, under optimized conditions, the yield/expression bias in favor of sequences containing only the common amino acids was <3-fold for the four X-E. coli. This means that if a sequence containing an unnatural amino acid functionally outperforms the most competitive sequence containing only natural amino acids by at least approximately threefold, it will be enriched despite bias against.
These optimized conditions were then tested with a phagemid library of antibodies to ensure that biases determined for the individual clones extend to the population level. Six consecutive residues completely randomized by site-saturation mutagenesis using NNK were therefore placed into the VH3–23 CDR3 loop, replacing residues 101–106, by overlap PCR using a randomized primer. This was subsequently cloned into the pSEX-GermTAT vector to create pSEX-GermNNK (see SI Text), a library that maintains the same framework as pSEX-GermTAG and pSEX-GermTAT. After transformation into Top10 F', a maximal complexity of 5 × 108 was experimentally achieved, and sequencing revealed that ≈18% of clones [number of clones sequenced (n) = 50, 17.3% expected by the binomial distribution using the 1/32 probability of finding TAG at any NNK randomized site] contained the amber codon before phage production. After phage production in SY-X-E. coli, Keto-X-E. coli, Bpy-X-E. coli, or Boro-X-E. coli., the resulting phage population had between 7 and 16% of clones containing the amber codon as revealed by sequencing (n = 100). On a population level, this represents a 1.1- to 2.5-fold expression bias in favor of sequences containing only the canonical amino acids (Fig. S1). χ2 tests suggest that these values are consistent with the bias typified by individual clones (Table S3). This mild expression bias should be easily overcome by functional performance.
A Known Sulfotyrosine-Containing Anti-gp120 Antibody Can Be Selected out from a Doped Randomized Library Using SY-X-E. coli.
It is known that HIV infection requires gp120 binding to the CCR5 coreceptor, that sulfation of CCR5 is obligatory for this interaction to be productive, and that neutralizing anti-gp120 sulfated human antibodies isolated from serum exploit this feature (17, 18). In fact, the recent crystal structure of one such antibody, 412d, bound to gp120 reveals that its two sulfotyrosines contribute to ≈20% of the total buried surface, with one of the two residues accounting for almost 100 Å2 (19). We reasoned, therefore, that high-affinity anti-gp120 antibodies could be evolved in SY-X-E. coli because sulfated tyrosine, otherwise a result of posttranslational modification in complex eukaryotes, could be incorporated into any sequence as an unnatural amino acid in E. coli.
We first conducted an evolution experiment to see whether antibody 412d containing its two sulfotyrosines (residues 100 and 100c) could be selected from a randomized library based on affinity for gp120. To adapt 412d for phage, the scFv 412d-2SY with two amber codons introduced at the two sites of human 412d tyrosine sulfation (residues 104 and 107 of the scFv form) was cloned into the pSEX backbone to create pSEX-412d2TAG. 412d-2SY was then displayed on phage using SY-X-E. coli. To generate a 412d library, the 412d scFv-pIII coding sequence was randomized by saturation mutagenesis with NNK at residues in close proximity to the sulfotyrosines (Pro-101, Asn-105, Ala-108, Pro-109, Gly-112, Met-113) as well as the two locations where sulfation occurs: residues 104 and 107 (see SI Text) (20). The resulting phagemid library (experimental maximal complexity of 2 × 108) was transformed into SY-X-E. coli from which phage were then produced. This phage population was spiked with phage displaying 412d-2SY at a ratio of one 412d-2SY phage to 2,000 library phage and panned against gp120 immobilized on a microtiter plate. After four rounds of selection in which phage amplification was conducted in SY-X-E. coli after each round, the population converged onto one sequence, 412d-2SY, demonstrating that evolution for gp120 binding in SY-X-E. coli can yield a sequence containing unnatural amino acids. In fact, the population after the third round (nearly complete convergence occurred after the fourth) also had sulfotyrosine-containing sequences that originated from library rather than spiked phage (Table S4). This shows that new sequences containing an unnatural amino acid, and not just 412d-2SY, are also evolved to bind gp120. For more information please see Tables S5 and S6.
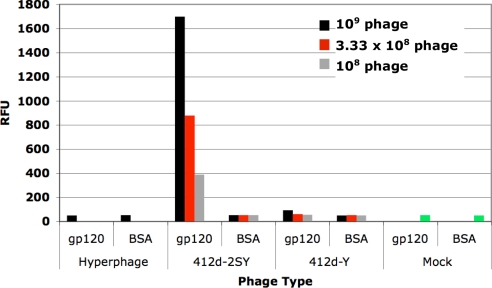
To ensure that 412d-2SY was selected based on its functional merit, we determined the expression level of phage displaying 412d-2SY and compared it to the expression level of the initial library phage and the library phage at round three, both of which contained some-to-many sequences that had no unnatural amino acids. This comparison revealed that phage displaying 412d-2SY were produced in lower yields than the library phage (Fig. S2); therefore, the enrichment for 412d-2SY phage must have been because of a functional advantage rather than any general expression advantage. 412d-2SY phage were also isolated to test for gp120-binding by ELISAs. As a natural comparison, phage displaying 412d from pSEX-412d2TAT, in which the two sulfotyrosines are replaced by tyrosines (412d-Y), were produced and similarly tested. As Fig. 2 shows, 412d-2SY effectively binds gp120 over BSA control. Furthermore, the ELISA signal is ≈10-fold higher for gp120 binding by 412d-2SY than by 412d-Y. Thus, 412d-2SY was selected based on its binding affinity resulting directly from one or both of its sulfotyrosine residues.
Fig. 2.
Phage ELISA for gp120-binding with 412d-2SY selected from a doped 412d library compared with 412d-Y, where sulfotyrosines were replaced by tyrosines. For each sample, 0.33-μg gp120 was coated onto a Maxisorp plate, blocked with 2% milk, and bound with the respective phage. Phage was detected with an anti-M13 antibody. BSA-binding was used as a control to show specific gp120-binding. “Mock” refers to signal generated without any phage added.
Selection for Anti-gp120 Antibodies from a Naïve Germline Library in SY-X-E. coli Yields a Sulfated Antibody.
We next attempted an evolution experiment for gp120-binding in which a completely naïve germline antibody library, instead of one based on any known gp120-binding sequence, was subjected to selection for binding gp120. Because we did not want to bias this library toward any target, a mixture of human germline heavy-chain variable regions were amplified from human genomic DNA and assembled with the A27 germline light chain, using overlap PCR, into scFvs with six consecutive residues in the CDR3-loop region completely randomized using site-saturation mutagenesis by NNK (see SI Text). Two partially randomized residues flanking these six residues were inserted to mimic the natural junctional diversity because VH gene regions do not end in frame. The library was then cloned into a phagemid vector, yielding an experimental maximal complexity of 2 × 109. DNA sequencing revealed that an expected ≈17% of clones contained at least one amber codon.
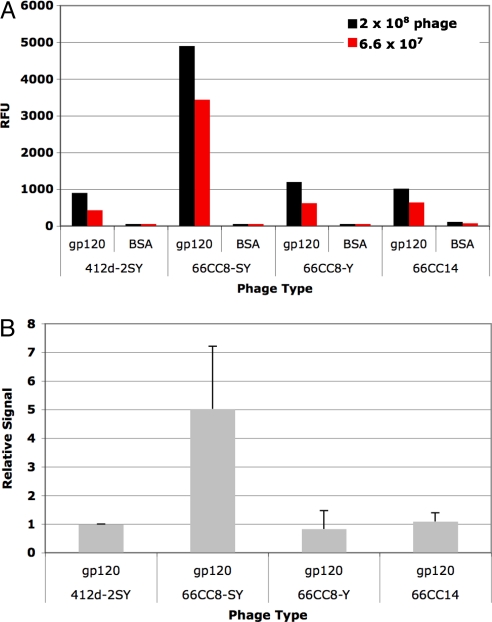
SY-X-E. coli were then used to generate phage from this library, which were subsequently panned against gp120. Bound phage were amplified in SY-X-E. coli and this process was repeated for a total of four rounds, where each additional round showed enrichment for gp120-binding clones over the previous rounds (Fig. S3a and Table S7). Sequencing revealed a concurrent enrichment for sulfotyrosine-containing clones (Fig. S3b). After three rounds, ≈40% of the phage population contained a sulfotyrosine; after the fourth round, the library converged primarily on one of these sulfotyrosine-containing antibodies with the sequence …E sY G S P R G Y… (sY, sulfotyrosine; amino acids corresponding to the locations of partial randomization are italicized; amino acids corresponding to the locations of full randomization are underlined) and heavy-chain V region VH 3–38. This clone represented 60% of the population (n = 20). Yield of phage displaying this scFv (66CC8-SY) in the presence of sulfotyrosine was >100-fold higher than yield of phage in the absence of sulfotyrosine, confirming specific incorporation of sulfotyrosine (5). The yield of phage displaying 66CC8-SY was also compared to the yield of phage displaying the initial library and that from round three, and in both cases phage displaying 66CC8-SY were produced in lower amounts (Fig. S4), as expected from the fact that 66CC8-SY contains an unnatural amino acid. Thus, 66CC8-SY was selected over other sequences, including ones containing only canonical amino acids, based on binding affinity. This result demonstrates that evolution with an unbiased, naïve library in a strain encoding sulfotyrosine yields a sulfated antibody as a solution to gp120-binding.
Gp120-binding by 66CC8-SY was then characterized by ELISA. As Fig. 3 shows, phage-displayed 66CC8-SY binds gp120 specifically over BSA control and also outperforms, by ≈fivefold, the known antibody 412d-2SY. When compared to the most enriched antibody that contained only natural amino acids from the same selection (antibody 66CC14 with the sequence …E R R G R E G H… and which represented 20% of the population after round 4, n = 20), 66CC8-SY had roughly a fourfold higher ELISA signal (see Fig. 3). We also compared 66CC8-SY with its analog where tyrosine replaces sulfotyosine (66CC8-Y). 66CC8-Y was produced using an aaRS encoding tyrosine in response to TAG and the resulting phage were tested for gp120-binding by ELISA in comparison with phage displaying 66CC8-SY. As Fig. 3 shows, phage-displayed 66CC8-SY produces an approximately fivefold higher ELISA signal for gp120-binding than does 66CC8-Y, indicating that sulfotyrosine is properly displayed on phage and demonstrating that the sulfate residue specifically contributes to the affinity of 66CC8-SY. This result is expected because 66CC8-Y and many related sequences should have been present in the starting library population from which 66CC8-SY emerged. We note that in all these cases, the display level of scFv on phage is consistent across clones because of the enforced multivalency of hyperphage, making comparison across samples straightforward.
Fig. 3.
Phage ELISA for gp120-binding with 66CC8-SY, 66CC8-Y, and 66CC14. For each sample, 0.3-μg gp120 was coated onto a microtiter plate well, blocked with 2% milk, and bound with the respective phage. Phage was detected with an anti-M13 antibody. BSA-binding was used as a control to show specific gp120-binding. (A) ELISA for a representative gp120-binding experiment done with two phage concentrations. (B) Average ELISA signals representing four separate experiments done with two separate phage preparations. Each experiment used the same amount of phage across all samples. Averages were calculated from signals normalized to 412d-2SYs binding within the same experiment. Error bars represent plus standard deviations. We note that this consolidated graph exaggerates variation because the separate ELISA experiments use different phage concentrations, and represent different incubation, washing, development, and detection times.
We next attempted to express and characterize free scFvs without fusion to phage. Although expression of the scFv 66CC8-SY using an orthogonal tRNA/aaRS pair specific for sulfotyrosine gave yields comparable to expression of 66CC8-Y (≈7 mg/L), all protein produced was insoluble and resisted multiple attempts at refolding. When converted to the Fab format, 66CC8-SY, 66CC8-Y, and 66CC14 (purified yields of 0.8, 1.0, and 1.0 mg/ml, respectively) lost activity for gp120-binding (Fig. S5). We believe that the germline variable regions corresponding to these selected antibodies are unstable and require the scFv linker, explaining both the loss of activity upon conversion to Fab format and the resistance to refolding in free scFv format. This is in contrast to expression of free 412d-2SY and 412d-Y (purified Fab yields of 0.25 and 0.7 mg/ml, respectively), both of which gave high yields of folded scFvs, and retained all activity for gp120-binding when converted to Fab format. Although there is no literature Kd value for comparison, Fab 412d-2SY bound gp120 effectively and in a sulfate-dependent manner, with >15-fold higher affinity than Fab 412d-Y by ELISA (Fig. S5c). This is analogous to mammalian sulfated 412d, which immunoprecipitates gp120 only when sulfated. Selection for stability or the use of libraries derived from Fabs rather than naïve germline scFvs should prevent such problems of folding and format conversion in future selection experiments.
Discussion
We have developed a system for protein evolution in which unnatural amino acids beyond the canonical 20 are available in the exploration of protein function space. In contrast to previous attempts that include unnatural or noncanonical amino acids in directed evolution experiments (21, 22), unnatural amino acids in our system are genetically encoded and require only the unique codon, TAG, for site-specific incorporation. Therefore, translation with unnatural amino acids utilizes the same fundamental paradigm as translation with natural amino acids, resulting in unrestricted 21 aa protein evolution. There is, however, one qualification: the yield bias favors sequences containing only natural amino acids. Our phage-based system is therefore optimized to mitigate this bias such that diversification strategies resulting in populations with sequences containing unnatural amino acids in conjunction with sequences containing only canonical amino acids can be subject to evolution for function. Because the yield bias is minimized but not completely removed, selections, including all those we have described, require that unnatural amino acids contribute to function for sequences containing them to be maintained in the population.
X-E. coli strains that incorporate four unnatural amino acids were optimized for phagemid-based directed evolution by manipulation of growth conditions, and one strain encoding the unnatural amino acid sulfotyrosine has been used to evolve gp120-binding antibodies from an unbiased, naïve germline antibody library. In this experiment, the phage-displayed antibody population converged on a unique clone that contains a sulfotyrosine, which directly contributes to gp120-binding affinity. The selection of this clone from a completely naïve library provides the first evidence that an expanded genetic code can confer a selective advantage through the functional contribution of an unnatural amino acid.
Optimization of this system to minimize bias was done with sequences that contain only one unnatural amino acid. Phage-yield bias against sequences that contain more than one unnatural amino acid may therefore be greater, thus requiring a more pronounced functional advantage for such sequences to prevail across multiple rounds. Any such bias against selection of sequences containing multiple unnatural amino acids can be overcome by iterative selection experiments. For example, if the fittest sequences in a given selection contain a single unnatural amino acid, a second selection experiment can be done where the unnatural amino acid from the winning sequences is fixed and other surrounding amino acids are randomized. With this constrained library, the incorporation of a second UAA would again experience only a minimized bias, as all members of the population would already contain a first unnatural amino acid. Naturally, this approach is not required if the functional advantage of having multiple unnatural amino acids overcomes the associated expression bias. For example, 412d-2SY, which contains two of the unnatural amino acid sulfotyrosine, is selected directly from a 412d-based library that is initially dominated by sequences containing only natural amino acids using our system.
Although this system was optimized to allow evolution with the four unnatural amino acids sulfotyrosine, bipyridyl-alanine, 4-borono-phenylalanine, and para-acetyl-phenylalanine, chosen on the basis of their unique chemical properties, phage-based evolution with expanded genetic codes should be general to all unnatural amino acids incorporated by orthogonal tRNA/aaRS pairs (≈45 so far). This fact, combined with the versatility of phage display and the array of available library design and diversification strategies (13), should now allow for evolution of unique binding modes, catalytic activities, and structures where unnatural amino acids expand the range and type of function that can be achieved. Moreover, the notion that an artificially expanded genetic code can increase evolutionary fitness can be further explored.
Materials and Methods
X-E. coli Plasmid Design and Construction.
tRNA/aaRS pairs were encoded on pCDF plasmids. The pCDF plasmid carries a replicon derived from CloDF13 (23) and encodes MjtRNATyrCUA and a mutant MjTyrRS that specifically charges MjtRNATyrCUA with an unnatural amino acid. To construct pCDF-Keto that encodes a mutant aaRS that charges MjtRNATyrCUA with para-acetyl-phenylalanine, an insert gene bearing the CloDF13 replicon was amplified from vector pCDF-1b (Novagen) by PCR using primers MT01: 5′-GTGTTGTTGTTGTTGTATACCAAATAGCTAGCTCACTCGGTC-3′ and MT02: 5′-GTTGTTGAGCTCGATAAATTGCACTGAAATCTAGAGCGGTTC-3′ then digested with restriction enzymes AccI and SacI, purified, and ligated into an AccI/SacI cut pSup vector (24). Sequencing revealed a G-to-A mutation in the proK promoter of the second tRNA cassette in the plasmid, which resulted in more viable clones that were functionally active. This mutation was therefore allowed to remain. Once pCDF-Keto was made, other synthetases were swapped in using the restriction sites NdeI and PstI to generate pCDF-Bpy, which incorporates bipyridyl-alanine; pCDF-SY, which incorporates sulfotyrosine; and pCDF-1BG11, which incorporates 4-borono-phenylalanine, all in response to the amber codon. Each of these pCDF plasmids were then transformed into Top10 F' (Invitrogen) to create Keto-X-E. coli, Bpy-X-E. coli, SY-X-E. coli, and Boro-X-E. coli and maintained on the antibiotics chloramphenicol (30 μg/ml) and tetracycline (30 μg/ml).
pSEX-GermTAG and pSEX-GermTAT Plasmid Construction.
A Fab-format antibody containing VH 3–23 and VL A27 with a TAG in the VH CDR3 loop was synthesized by Blue Heron. To construct pSEX-GermTAG, first the light-chain gene was amplified from this synthetic Fab antibody gene with primers LC1: 5′-GCACGCGTAGAAATTGTGTTGACG-3′ and LC2: 5′-CTTTGGATCCAGCGGCCGCCCGTTTGATTTCCACCTTGGTCCCTTGGCC-3′. It was then digested with Mlu and BamHI and subsequently inserted into a similarly digested pSEX81 (Progen) to create pSEX-GermA27. The heavy-chain gene was then amplified from the synthetic Fab antibody gene with primers HC1: 5′-CGGCCATGGCTGAGGTGCAGCTGTTGGAGTCTGG-3′ and HC2: 5′-CTTCAAGCTTTGGGGCGGATGCACTCCCTGAGGAGACGGTGACCAGGGTTCCTTGG-3′ and then digested with NcoI and HindIII. This sequence was inserted into a similarly digested pSEX-GermA27 to create pSEX-GermTAG. Quickchange (Stratagene) site-directed mutagenesis was used to create pSEX-GermTAT where the TAG codon in pSEX-GermTAG is replaced by a TAT codon.
In these pSEX plasmids, a trypsin site is present between the scFv and pIII, which allows for the removal of displayed scFvs to reveal infective pIII. This is important for elution and infection.
pSEX-412d2TAT and pSEX-412d2TAG Construction.
The scFv coding region was amplified from a mammalian 412d expression vector (18) using primers 412dF: 5′-CACGCCATGGCTGAGGTGCAGCTGGTGCAGTCTGGGGCTGAGGTG-3′ and 412dB: 5′-CTTTGGATCCAGCGGCCGCCCGTTTGATTTCCACCTTGGTCCCCTGGCCAAA-3′. The resulting scFv was digested with NcoI and BamHI and inserted into a similarly digested pSEX81 to yield pSEX-412d2TAT. Quickchange (Stratagene) site-directed mutagenesis was used to replace the two locations of mammalian tyrosine sulfation (residues 104 and 107) with amber codons to yield pSEX-412d2TAG.
Phage Production.
In all experiments carried out under optimized conditions, the following phage expression protocol was used. First, an X-E. coli culture transformed with either phagemid library or single phagemid clone DNA was grown overnight in 2YT supplemented with 100-μg/ml ampicillin, 30-μg/ml tetracycline, 30-μg/ml chloramphenicol, and 1% glucose. After saturation was reached, a total volume equal to 12.5% of the final phage expression volume was removed from the culture, spun down, and resuspended in a volume of 2YT equal to 25% of the final phage-expression volume. The 2YT was supplemented with 100-μg/ml ampicillin, 30-μg/ml tetracycline, and 30-μg/ml chloramphenicol. To this culture was added hyperphage (Progen) at 20 MOI (MOI, multiplicity of infection). The resulting culture was then incubated at 37 °C and centrifuged 100 rpm for 1 h, after which the cells are spun down and the media removed. The phage were then resuspended in the desired final phage expression volume of 2YT supplemented with 100-μg/ml ampicillin, 50-μg/ml kanamycin, and 30-μg/ml chloramphenicol. The unnatural amino acid corresponding to the X-E. coli used was added directly to the media and the phage culture was incubated at 280 rpm for 18 h at 30 °C. For Keto-X-E. coli, Bpy-X-E. coli, SY-X-E. coli, and Boro-X-E. coli, unnatural amino acids were added at 8 mM, 15 mM, 1.5 mM, and 6.5 mM (13 mM of the racemic mixture), respectively. For Bpy-X-E. coli, 20 mM FeSO4 was also added to prevent toxicity associated with iron depletion. For Boro-X-E. coli, 25-mM NaOH was added to solubilize the amino acid. Para-acetyl-phenylalanine was synthesized by Synchem based on published procedures (7), sulfotyrosine was purchased from Senn Chemicals or Bachem, and 4-borono-phenylalanine was purchased from Aldrich as a mixture of D and L isomers. For the synthesis of bipyridyl-alanine, see SI Text.
After phage production, the media was collected and cells were discarded. The media was then concentrated down to a convenient volume using 10-kDa cutoff concentrators (Amicon). The concentrated phage was then precipitated by the addition of 5 times phage precipitation buffer (20% PEG 8000, 2.5-mM NaCl) and dissolved in a convenient volume of PBS, pH 7 (PBS). Precipitation was done twice.
Phage Quantification.
Two methods were used to titer phage according to published protocols (16). For applications, such as selection where infective phage are the relevant population, phage was quantified by infection of Top10 F' cells. Specifically, a small volume of phage was digested with 1.75-μg/ml trypsin (Worthington Biochemicals) and used to infect Top10 F' cells. The amount of infected cells was then determined by plating dilutions on selective plates (ampicillin plates were used for pSEX-based phagemids). For applications such as ELISA, where the total particles of phage is the relevant population, phage were coated onto ELISA plates, blocked with 2% milk in PBS, washed several times with PBST (PBS plus 0.025% Tween 20), bound by an anti-M13 polyclonal antibody (NEB) in 2% milk PBST, and detected with a QuantaBlu fluorogenic substrate (Pierce). This sample was compared to a standard curve where known amounts of hyperphage were adsorbed onto the plates and similarly treated. When multiple samples were being compared, samples were also standardized to all give a similar titer signal.
Selection of gp120-Binding Phage.
Soluble ADA gp120 (0.5 μg) was coated onto the surface of a MaxiSorp (Nunc) microtiter plate well in 100-μl PBS for 12 h at 37 °C. After blocking for 2 h with 2% milk (Bio-Rad) in PBS and washing three times with 200-μl PBST, the concentrated phage library was added in 100 μl 2% milk PBST and incubated at 37 °C for 4 h. Washing was done with PBST and PBS after which phage were eluted with 1.75-μg/ml trypsin (Worthington Biochemicals) for 12 min. Eluted phage were used to infect 20 ml of a 0.4 O.D. culture of SY-X-E. coli in 2YT supplemented with 30-μg/ml chloramphenicol and 30-μg/ml tetracycline. Infection was allowed to occur for 1 h at 37 °C, after which a small aliquot of cells were plated to determine the number of eluted phage. The rest of the cells were spun down and resuspended in 25-ml 2YT supplemented with 30-μg/ml chloramphenicol, 30-μg/ml tetracycline, 100-μg/ml ampicillin, and 1% glucose. After overnight growth, the enriched library was used to produce phage for the subsequent round.
Determination of gp120-Binding by ELISA.
Per sample, 0.33 μg of soluble ADA gp120 was coated onto the surface of a MaxiSorp (Nunc) microtiter plate well in 100-μl PBS for 12 h at 37 °C. After blocking for 2 h with 200 μl 2% milk (Bio-Rad) in PBS, equal amounts of concentrated phage previously expressed, precipitated, and quantified by ELISA according to the procedures described above, were loaded in 100 μl 2% milk PBST and incubated at 37 °C for 2 h. After washing five times with PBST, an anti-M13 polyclonal antibody (NEB) was added in 110 μl of 2% milk in PBST and incubated at 37 °C for 2 h. After washing eight times with PBST, QuantaBlu fluorogenic substrate (Pierce) was added and the ELISA signal was determined using a fluorescence plate reader (SpectraMax Gemini).
Supplementary Material
Acknowledgments.
C.C.L. thanks Luke Lairson, Eric Brustad, David Horning, Susan Cellitti, and Bernhard Geierstanger for helpful discussions and technical assistance, and The Fannie and John Hertz Foundation and the National Science Foundation for predoctoral fellowships. This research was supported by the National Institutes of Health Grant GM62159.
Footnotes
Conflict of interest statement: A provisional patent on this work has been filed.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809543105/DCSupplemental.
References
- 1.Vetsigian K, Woese C, Goldenfeld N. Collective evolution and the genetic code. Proc Natl Acad Sci USA. 2006;103(28):10696–10701. doi: 10.1073/pnas.0603780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 3.Xie J, Liu W, Schultz PG. A genetically encoded bidentate, metal-binding amino acid. Angewandte Chemie (International ed) 2007;46(48):9239–9242. doi: 10.1002/anie.200703397. [DOI] [PubMed] [Google Scholar]
- 4.Brustad E, et al. A genetically encoded boronic acid. Angewandte Chemie. 2008;46:9239–9242. International ed. [Google Scholar]
- 5.Liu CC, Schultz PG. Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat Biotechnol. 2006;24(11):1436–1440. doi: 10.1038/nbt1254. [DOI] [PubMed] [Google Scholar]
- 6.Liu CC, Brustad E, Liu W, Schultz PG. Crystal structure of a biosynthetic sulfo-hirudin complexed to thrombin. J Am Chem Soc. 2007;129(35):10648–10649. doi: 10.1021/ja0735002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc Natl Acad Sci USA. 2003;100(1):56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia G, et al. Directed evolution of novel polymerase activities: mutation of a DNA polymerase into an efficient RNA polymerase. Proc Natl Acad Sci USA. 2002;99(10):6597–6602. doi: 10.1073/pnas.102577799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebar EJ, Pabo CO. Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science. 1994;263(5147):671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 10.Rader C, Barbas CF., 3rd Phage display of combinatorial antibody libraries. Curr Opin Biotechnol. 1997;8(4):503–508. doi: 10.1016/s0958-1669(97)80075-4. [DOI] [PubMed] [Google Scholar]
- 11.Martin A, Sieber V, Schmid FX. In-vitro selection of highly stabilized protein variants with optimized surface. J Mol Biol. 2001;309(3):717–726. doi: 10.1006/jmbi.2001.4698. [DOI] [PubMed] [Google Scholar]
- 12.Famm K, Winter G. Engineering aggregation-resistant proteins by directed evolution. Protein Eng Des Sel. 2006;19(10):479–481. doi: 10.1093/protein/gzl032. [DOI] [PubMed] [Google Scholar]
- 13.Brakmann S, Johnsson K. Directed Molecular Evolution of Proteins. Weinheim: Wiley-VCH; 2002. [Google Scholar]
- 14.Tian F, Tsao ML, Schultz PG. A phage display system with unnatural amino acids. J Am Chem Soc. 2004;126(49):15962–15963. doi: 10.1021/ja045673m. [DOI] [PubMed] [Google Scholar]
- 15.Rondot S, Koch J, Breitling F, Dubel S. A helper phage to improve single-chain antibody presentation in phage display. Nat Biotechnol. 2001;19(1):75–78. doi: 10.1038/83567. [DOI] [PubMed] [Google Scholar]
- 16.Broders O, Breitling F, Dubel S. Hyperphage. Improving antibody presentation in phage display. Methods in molecular biology. 2003;205:295–302. doi: 10.1385/1-59259-301-1:295. [DOI] [PubMed] [Google Scholar]
- 17.Farzan M, et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96(5):667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 18.Choe H, et al. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell. 2003;114(2):161–170. doi: 10.1016/s0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- 19.Huang CC, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317(5846):1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang CC, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci USA. 2004;101(9):2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo TH, Link AJ, Tirrell DA. Evolution of a fluorinated green fluorescent protein. Proc Natl Acad Sci USA. 2007;104(35):13887–13890. doi: 10.1073/pnas.0701904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love KR, Swoboda JG, Noren CJ, Walker S. Enabling glycosyltransferase evolution: a facile substrate-attachment strategy for phage-display enzyme evolution. ChemBioChem. 2006;7(5):753–756. doi: 10.1002/cbic.200600018. [DOI] [PubMed] [Google Scholar]
- 23.Nijkamp HJ, et al. The complete nucleotide sequence of the bacteriocinogenic plasmid CloDF13. Plasmid. 1986;16(2):135–160. doi: 10.1016/0147-619x(86)90072-7. [DOI] [PubMed] [Google Scholar]
- 24.Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nature methods. 2006;3(4):263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.