Abstract
The influenza surface glycoprotein hemagglutinin (HA) is a potential target for antiviral drugs because of its key roles in the initial stages of infection: receptor binding and the fusion of virus and cell membranes. The structure of HA in complex with a known inhibitor of membrane fusion and virus infectivity, tert-butyl hydroquinone (TBHQ), shows that the inhibitor binds in a hydrophobic pocket formed at an interface between HA monomers. Occupation of this site by TBHQ stabilizes the neutral pH structure through intersubunit and intrasubunit interactions that presumably inhibit the conformational rearrangements required for membrane fusion. The nature of the binding site suggests routes for the chemical modification of TBHQ that could lead to the development of more potent inhibitors of membrane fusion and potential anti-influenza drugs.
Keywords: crystallography, drug design
Influenza A virus membranes contain 3 proteins: hemagglutinin (HA), neuraminidase (NA), and the proton channel (M2). HA is responsible during the initial stages of infection for sialic acid-receptor binding and, after virus uptake into endosomes, for fusion of virus and cell membranes (1). M2 transfers protons into the infecting virus in endosomes, and at acidic pH the matrix protein, M1, is dissociated from the genome–transcriptase complex, so that the uncoated complex is transported to the nucleus after membrane fusion (2). At the end of infection, NA cleaves sialic acid from virus and cell glycoconjugates to ensure release of newly-made viruses from infected cells (3). Both M2 and NA are targets of current anti-influenza drugs (4–6). The proton channel, M2, is blocked by the drugs amantadine and rimantadine, and NA is inhibited by the drugs zanamivir and oseltamivir. All 4 drugs are effective upon prompt administration after infection or prophylactically, but concern has been raised by the isolation of viable mutant viruses that are resistant to them (7–11). There is therefore a need to develop new antivirals to act on additional virus targets. Their availability would make possible drug combination therapies to avoid the selection of resistant viruses, a strategy that has been successful in highly active antiretroviral therapy against HIV (12) and was recently reported for the combined use of amantadine and oseltamivir against influenza (13). To address the need for new antivirals against influenza a number of studies have been made of inhibitors of the receptor binding or membrane fusion activities of HA, particularly the latter (14–17).
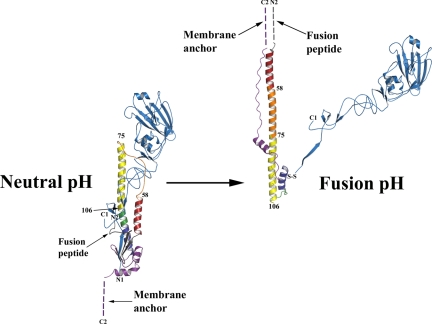
The membrane fusion potential of HA is activated in endosomes at acidic pH by the induction of an irreversible reorganization of HA structure (18, 19). Comparison of the neutral-pH and fusion-pH structures indicates that at fusion pH the membrane-distal domains of HA dissociate, and extensive structural reorganization occurs that involves extrusion of the “fusion peptide” from the interior of the neutral-pH structure, presumably toward the target endosomal membrane with which the virus membrane is to fuse. In its new position in the fusion-pH structure (19), the fusion peptide is at the N terminus of a new 100-Å-long triple-helical coiled-coil, while the C-terminal membrane anchor is repositioned at the same end of the refolded molecule (Fig. 1) (20). Structural similarities between the low-pH form of HA and equivalent regions of the ectodomains of other viral membrane fusion proteins in their postfusion states suggest that this juxtaposition of termini is a common outcome in membrane fusion (1, 21).
Fig. 1.
Diagrams of the structure of a subunit of the HA trimer at neutral pH (18) (Left) and after incubation at fusion pH (pH 5.0, 20°) (19) (Right). The disulfide-linked polypeptide chains, HA1 and HA2, are colored blue and multicolored, respectively. Their N and C termini are labeled N1, C1, and N2, C2, respectively. The fusion pH diagram was constructed by combining the HA1 domain structure (39) and the fusion pH HA2 structure (20). Comparison of the diagrams shows that at fusion pH the HA1 membrane distal domains (blue) detrimerize. The fusion peptide at the N terminus of HA2 (gray) is relocated to become the N terminus of a new 100-Å-long α-helix that forms a triple-helical coiled-coil in the fusion pH trimer. Each monomer of the new coiled-coil is composed of the repositioned (turned through 180°) short α-helix (red) from the neutral pH α-helical hairpin (red-orange-yellow); the interhelical extended chain of the hairpin (orange), refolded into an α-helix; and the N-terminal half (yellow) of the central, neutral pH, long α-helix. Two turns (green) of the central, neutral pH long α-helix refold at fusion pH to form a 180° reverse turn at the C terminus of the new coiled-coil. As a consequence the C-terminal half of the neutral pH long α-helix (dark blue) and the C-terminal region of HA2 (purple) are reoriented and refolded in the fusion pH structure, antiparallel to the new α-helix and packed into the grooves of the coiled-coil, positioning the C-terminal membrane anchor at the same end of the fusion pH HA as the relocated fusion peptide (dashed lines indicate regions of structure that are not determined). The numbered residues in HA2, 58, 75, and 106 are in regions that differ in structure between group 1 and group 2 HAs.
Numerous small molecules have been identified that block virus infectivity by inhibiting the conformational changes required for HA-mediated membrane fusion (14–16), and attempts have been made to “dock” some of them in silico to potential binding sites on HA. However, the development of more effective compounds has been limited by the lack of crystal structures of relevant HA complexes. Another potential limitation or disincentive to development for some of these compounds is that they appear to be effective against only certain subtypes of HA. Because the HAs of current seasonal influenza viruses, H1 and H3, are members of different phylogenetic groups, as are the highly pathogenic viruses of the H5 and H7 subtypes, it will be important to understand the structural basis of drug sensitivity and group specificity.
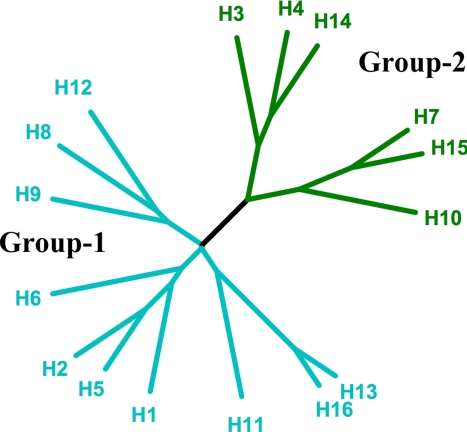
The 16 subtypes of HA form 5 clades and further segregate into 2 groups (Fig. 2) (22, 23). Comparison of the available HA structures indicates that they differ on a group-specific basis in regions that are prominent in the changes required for membrane fusion (24, 25). The fact that some of the fusion inhibitors work only on certain subtypes of HA suggests that they may bind in one of these regions. To explore this possibility we have analyzed by X-ray crystallography the structures of 2 HAs in complex with tert-butyl hydroquinone (TBHQ). This small hydrophobic compound was shown in previous studies to inhibit both the fusion-pH conformational change in H3 subtype HA and viral infectivity at concentrations between 5 and 10 μm (15). Our results show that TBHQ binds at a largely hydrophobic interface between HA monomers. They also show how the binding site is formed in just 1 of the 2 phylogenetic groups of HA and that the interactions between the inhibitor and the HA trimer would be expected to increase the stability of the complex and hence account for the mechanism of action of the inhibitor.
Fig. 2.
Phylogenetic tree of influenza A HAs. The 2 groups are colored cyan (group 1) and green (group 2), each of which can be further subdivided into 3 clades (H8, H9, and H12; H1, H2, H5, and H6; H11, H13, and H16) and 2 clades (H3, H4, and H14; H7, H10, and H15). The tree was drawn by using the program TreeIllustrator (40).
Results
We have determined the crystal structures of TBHQ bound to an H14 [A/mallard/Astrakhan/263/1982(H14N5)] HA and an H3 (X31) HA. H14 and H3 are in the same clade within group 2 HAs (Fig. 2) and share 60% sequence identity overall and 87% within HA2. The native crystal structure of H14 HA was solved by molecular replacement using H3 HA as a search model and, as expected from the sequence similarity, they have very similar structures (rmsd of 0.6 Å for 459 Cα atoms).
Location of TBHQ Binding Site.
Crystals of H14 HA soaked in a 5 mM solution of TBHQ for 30 min diffracted to a Bragg spacing of >2.5 Å. From in silico docking analyses TBHQ was suggested to bind to HA in a cavity near the fusion peptide (15); however, electron density maps (Fig. 3 A and B) clearly showed the compound bound uniquely, adjacent to the C terminus of the short α-helix of the HA2 α-helical hairpin (Fig. 3A). TBHQ was manually positioned into the electron density and then subjected to standard crystallographic refinement. The thermal factors for the atoms of TBHQ refined to similar values to those of the surrounding protein atoms, indicating that high site occupancy had been achieved.
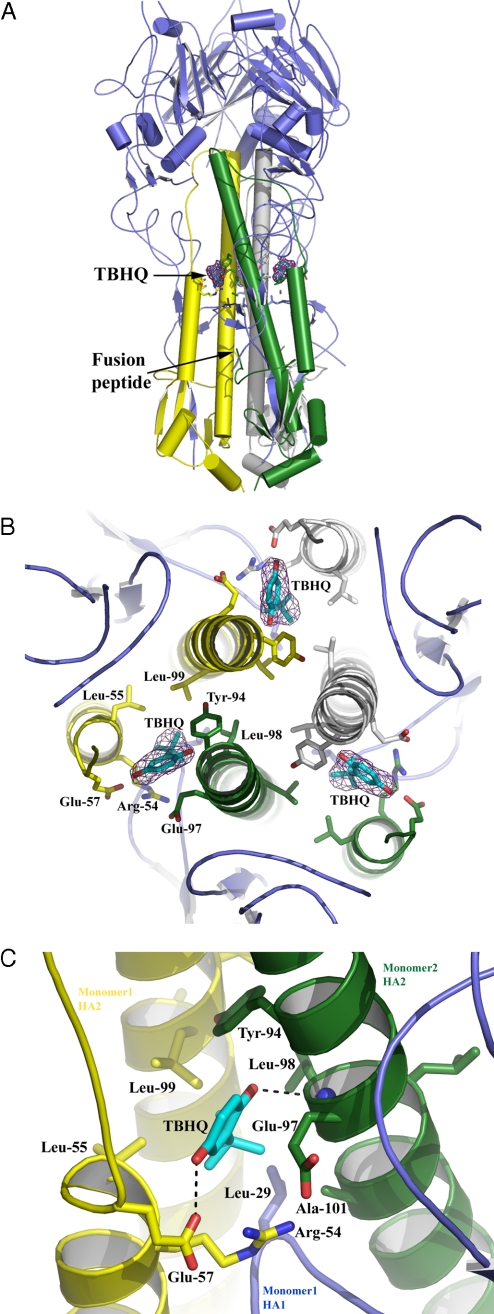
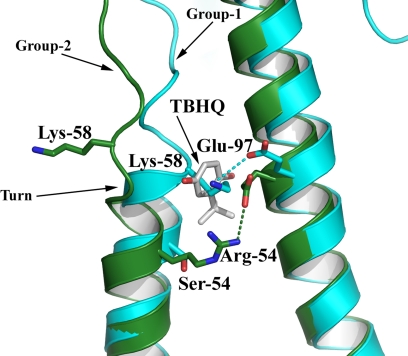
Fig. 3.
The location and composition of the TBHQ binding site. (A) Representation of the H14 HA trimer. The 3 HA1s are colored in blue, and the 3 HA2s are in yellow, green, and gray. The locations of the 3 TBHQ molecules and the fusion peptide are highlighted. (B) View down the 3-fold axis of H14 HA to show the 3 TBHQ binding sites. A 2 Fo − Fc electron density map (contoured at 1σ) is shown for TBHQ. Selected residues are shown as sticks. The same color scheme as in A is used. (C) Close-up view of a single TBHQ binding site. Selected residues are shown as sticks, and potential hydrogen bonds are shown as dotted lines.
The TBHQ binding site is located at the interface between 2 monomers of the HA trimer, and there are therefore 3 TBHQ sites per trimer (Fig. 3B). The site is formed by residues from the long HA2 α-helices of each monomer (monomer 1 is colored in yellow and monomer 2 in green in Fig. 3) and from the short HA2 α-helix of monomer 2. Monomer 1 also fills in the bottom of the site via residue 291 of the highly conserved β-hairpin of HA1 (residues 26–34). The interactions between TBHQ and HA are predominantly (85%) hydrophobic, largely accounted for by the highly hydrophobic base of the site, comprising the group-conserved residues Leu-291, Leu-982, and Ala-1012 of monomer 1, and Leu-552 and Leu-992 of monomer 2, into which the tert-butyl group of TBHQ packs (subscript 1 denotes a residue from HA1 and subscript 2 denotes a residue from HA2). There are 3 ionizable amino acids associated with the site: Arg-542 and Glu-572 from monomer 2 and Glu-972 from monomer 1. The conformations of Arg-542 and Glu-972 are such that the aliphatic parts of their side chains face the binding site, whereas the ionizable groups are oriented away from it. Indeed, the aliphatic side chain of Glu-972 packs across the face of the quinone ring of TBHQ. In contrast, the carboxyl group of Glu-572 is involved in a hydrogen-bond network with both the side chain and the main-chain carbonyl of Arg-542, and with the O1 oxygen of TBHQ. The other oxygen atom of TBHQ, O2, makes a hydrogen bond with the main-chain amide of Leu-982 (Fig. 3C). Presumably, these hydrogen bonds through which the short and long α-helices of the site are bridged, together with the hydrophobic interactions, enhance the stability of the HA trimer. Only O1 and the top of the quinone ring of bound TBHQ are partially accessible to solvent, because 90% of the surface area of TBHQ is solvent inaccessible. The binding site has a normalized complementarity of 0.7 (a value of 1 representing a perfect fit) (26).
To establish whether this binding site is conserved in other group 2 HAs we determined the cocrystal structure of TBHQ in complex with X31 H3 HA. The resolution of the diffraction data obtained was lower by comparison with the H14 data. The resulting electron density maps were of good quality, and strong difference density for TBHQ was apparent in the same site as in H14 [supporting information (SI) Fig. S1]. We are therefore, confident that the detailed description of the molecular interactions of TBHQ from the high-resolution H14 structure will be indicative of TBHQ binding to all group 2 HAs that have highly conserved structures in this region (24).
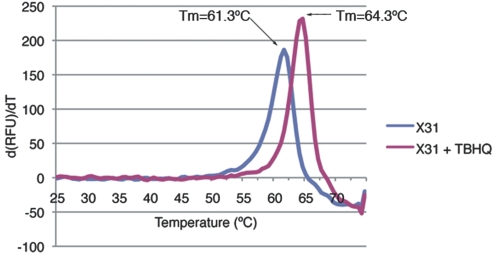
To establish the mechanism of action of TBHQ thermal shift assays were performed in the presence and absence of TBHQ. Fig. 4 shows that there is a 3°C upward shift in melting temperature (Tm) for X31 H3 HA in the presence of 1 mM TBHQ, demonstrating a significant stabilization of X31 H3 HA by TBHQ binding.
Fig. 4.
A representative fluorometric trace of the first derivative of the change in RFU against temperature, demonstrating the thermostabilization of X31 H3 HA by TBHQ. Applying the methodology of Mayhood and Windsor (41), a binding affinity of TBHQ for X13 H3 HA of between 5 and 50 μM at 20 °C can be calculated.
Structural Basis of the Specificity for Group 2 HAs.
As shown in Fig. 2, the 5 phylogenetic clades of HA fall into 2 groups. Previous studies have shown that membrane fusion by H3, but not H1, HAs is inhibited by TBHQ, suggesting that TBHQ may be a group 2-specific inhibitor (27). Comparison of the crystal structures of group 1 and group 2 HAs in the vicinity of the TBHQ binding site reveals important structural differences that could account for the group-specific activity of the compound (Fig. 5).
Fig. 5.
The structural basis for HA group-specific inhibition by TBHQ. Superposition of H14 (green) and H5 (cyan) HAs clearly shows that the additional turn of helix A in the group 1 HAs precludes TBHQ binding. Potential hydrogen bonds are shown as dotted lines.
In group 1 HAs access, to what would be the TBHQ binding site, is blocked because of an extra turn at the C terminus of the short α-helix (residues 56–58). This additional turn of helix in group 1 HAs results from the formation of an intermonomer salt bridge between Lys-582 and Glu-972. In group 2 HAs this interaction is not made (although Lys-582 is conserved in all HAs) because Glu-972 prefers to form a salt bridge with the group 2-conserved residue Arg-542. Consequently, Lys-582 is not well ordered in group 2 HAs, and the TBHQ site is accessible.
Discussion
By X-ray crystallographic analyses of HA-inhibitor complexes we have shown that the inhibitor of influenza virus-mediated membrane fusion, TBHQ, binds to HA at 3 identical sites, cross-linking the trimer through interactions at each site with the long central α-helices of 2 monomers and the short α-helix of one of them (Fig. 3). As a consequence, TBHQ binding stabilizes the neutral pH conformation of HA, rendering it membrane fusion inactive. Our description of the mode of TBHQ binding, together with the report that TBHQ inhibits membrane fusion at a concentration of <20 μm (15), suggest that development of an effective antiviral drug with the necessary potency may be possible. For example, modification of the tert-butyl group to improve hydrophobic interactions and the creation of additional polar interactions may lead to increased efficacy. The structure also shows that space for larger/additional polar substituents on TBHQ exists in the membrane distal region of the site, near residue Thr-592, which is highly polar and thus might be exploited in a structure-based drug design program. It has been shown that drug-like molecules require a balance between polar and hydrophobic properties.
Support for the proposal that the mechanism of inhibition of membrane fusion by TBHQ results from stabilizing the nonactivated conformation of HA is provided by mutation data. These data show that resistance mutations that destabilize the neutral pH conformation of HA counteract the stabilizing effect of TBHQ binding. Unlike other mutations in HA that map directly to the site of action and have been used to identify directly the sialic acid receptor binding site (28) and sites to which infectivity-neutralizing antibodies bind (29), the TBHQ-resistant mutations do not map to the TBHQ-binding site described here. Instead they involve mainly residues that interact with the fusion peptide (15). The phenotypes of these mutants included increases in the pH of membrane fusion. The amino acid substitutions involved (Asp-112 → Gly, Asp-112 → Asn, and Lys-117 → Glu) are the same as reported from studies of lysosomotropic membrane fusion inhibitors, such as amantadine in vivo (30). In those cases the pH of fusion and the thermal stability of the mutant HAs were strongly correlated; the higher the pH of fusion, the lower the Tm, and vice versa, implying that their mode of action was the destabilization of the neutral form of HA.
The TBHQ binding site we have described highlights the importance of the region near residue 582 (Fig. 5) at the C terminus of the short helix of the α-helical hairpin of HA2, in the low pH-mediated structural change in HA required for membrane fusion. It is particularly interesting that group 1 and group 2 HAs adopt different structures in this region and that group 2 HAs are sensitive to TBHQ, whereas group 1 HAs are not. Other group-specific structural features near the C terminus of the long α-helix of the α-helical hairpin (residue 752) and near the C terminus of the fusion peptide (residue 1062) have also been noted before (Fig. 1) (24). However, until now, these group-specific differences in HA structure had not been correlated with any group-specific biological properties. The fact that HAs of the 2 groups occupy 2 discrete conformational states, rather than a distribution of forms, demonstrates that these 2 observed forms represent at least local free-energy minima. In this context it is notable that the region containing Lys-582 has among the highest thermal factors for any part of the group 2 HA structures but has normal values in group 1 HAs. Clearly, sufficient favorable interactions are generated elsewhere to offset the relatively unstable conformation of the turn around Lys-582. The ability of this region of the structure to adopt 2 discrete conformations may be related to the fact that it plays a key part in the molecules' low pH-induced structural changes, perhaps indicating that details of the refolding pathway may differ for HAs of the different groups.
A further indication of the distinct properties of the 2 phylogenetic groups of HAs is the report of another set of antiviral compounds that inhibit membrane fusion by group 1, but not group 2, HAs (16). The binding sites for these inhibitors have not been determined but it seems likely that they may involve one or another of the group-specific regions of structure already identified near residues 752 and 1062 (Fig. 1).
Methods
Preparation of X31 H3 HA and H14 HA from A/mallard/Astrakhan/263/1982 grown in hens' eggs was by bromelain digestion as described (31). H14 HA crystals were grown by vapor diffusion in hanging drops consisting of 2 μL of reservoir solution [0.1 M Tris hydrochloride (pH 8.5), 8% (wt/vol) polyethylene glycol 8,000] and 2 μL of concentrated protein solution (10 mg/mL in 10 mM Tris·HCl, pH 8.0). X31 HA crystals were grown by vapor diffusion in hanging drops consisting of 2 μL of reservoir solution (1.9 M ammonium citrate) and 2 μL of concentrated protein solution (10 mg/mL in 10 mM Tris·HCl, pH 8.0). Native H14 HA data were collected at 100K on an in-house Rigaku-MSC RU200 rotating anode coupled to a RaxisIIc detector. H14 crystals were soaked for ≈30 min in 5 mM TBHQ made up in cryo buffer (reservoir solution augmented with 20% glycerol). Data were collected at 100K on an in-house Rigaku Micromax-007 rotating anode with osmic mirrors coupled to a Rigaku RaxisIV++ detector. X31 H3 HA crystals were soaked in 200 μM TBHQ for ≈30 min, and data were collected on beamline IO3 at the Diamond Light Source (Oxfordshire, U.K.) at 100K with an ADSC Q315 CCD. Diffraction data were integrated by using Denzo and scaled with Scalepack (32). Native H14 HA was solved by molecular replacement with Phaser (33) using H3 HA as a search model. Standard refinement, with Refmac (34) or PHENIX (35), and manual model building, with O (36), was performed for both H14 structures and X31 H3 structure. Crystallographic statistics are given in Table 1.
Table 1.
Data collection and refinement statistics
| Statistic | H14 native | H14–TBHQ complex | X31 H3–TBHQ complex |
|---|---|---|---|
| Data collection | |||
| Space group | P213 | P213 | P41 |
| Cell dimensionsa, b, c, Å | 139.91, 139.91, 139.91 | 139.22, 139.22, 139.22 | 160.21, 160.21, 176.68 |
| Resolution, Å | 2.6 (2.69–2.6) | 2.5 (2.59–2.5) | 2.8 (2.93–2.8) |
| Rsym | 12.6 (53.6) | 13.0 (68.5) | 10.9 (52.5) |
| I /σI | 10.5 (3.9) | 10.2 (2.9) | 19.6 (4.3) |
| Completeness, % | 99.5 (99.7) | 95.8 (91.9) | 99.8 (100) |
| Redundancy | 7.2 | 6.3 | 7.6 |
| Refinement | |||
| Resolution, Å | 30.0–2.6 | 30–2.5 | 30–2.8 |
| No. reflections | 26,079 | 28,412 | 103,381 |
| Rwork/Rfree | 19.6/24.5 | 19.4/24.8 | 19.8/21.8 |
| No. of atoms | |||
| Protein | 3,836 | 3,836 | 12,133 |
| Ligand/ion | 0 | 12 | 36 |
| Water | 368 | 338 | 0 |
| B factors | |||
| Protein | 31.5 | 34.6 | 45.6 |
| Ligand/ion | NA | 52.9 | 79.9 |
| Water | 35.6 | 41.3 | NA |
| rmsd | |||
| Bond lengths, Å | 0.008 | 0.012 | 0.017 |
| Bond angles, ° | 1.15 | 1.51 | 1.78 |
Numbers in parentheses indicate data for the highest resolution bin. NA, not applicable.
Differential scanning fluorimetry was performed as described (37) using a Bio-Rad iCycler iQ. Briefly, 50 μL of reactions containing 5 μg of X31 HA, 5X SYPRO Orange (Invitrogen), 100 mM Hepes (pH 7.0), and 150 mM NaCl were arranged in 96-well plates. In addition, 1 μL of DMSO was included with or without 50 mM TBHQ (to give a final concentration of 1 mM TBHQ). Samples were incubated at 25 °C for 30 s, with temperatures rising by 0.5 °C every 30 s for 50 min, while the relative fluorescence units (RFU) were recorded as the temperature increased. Tm were calculated from the maxima of the first derivative of RFU/temperature by using Bio-Rad iQ5 software. Experiments were repeated 4 times.
Supplementary Material
Acknowledgments.
We thank David Hall at the Diamond Light Source for assistance in the X31 data collection and Lesley Haire for help with the X31 data collection. Work at the National Institute for Medical Research was funded by the Medical Research Council. R.J.R. was supported by the Scottish Funding Council and the Medical Research Council. D.A.S. was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Contract HHSN266200700006C. The University of St. Andrews is a charity registered in Scotland (no. SC013532).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3EYJ, 3EYK, and 3EYM).
This article contains supporting information online at www.pnas.org/cgi/content/full/0807142105/DCSupplemental.
References
- 1.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 2.Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 3.Murphy BR, Webster RG. In: Fields Virology. 3rd Ed. Fields DBN, Knipe M, Howley PM, editors. Philadelphia: Lippincott–Raven; 1996. pp. 1397–1445. [Google Scholar]
- 4.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Von Itzstein M, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 6.Kim CU, et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: Design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J Am Chem Soc. 1997;119:681–690. doi: 10.1021/ja963036t. [DOI] [PubMed] [Google Scholar]
- 7.Ives J, et al. Antiviral drug resistance: An oseltamivir treatment-selected influenza A/N2 virus with a R292K mutation in the neuraminidase gene has reduced infectivity in vivo. J Clin Virol. 2000;18:251–269. [Google Scholar]
- 8.Gubareva LV, Kaiser L, Matrosovich MN, Soo-Hoo Y, Hayden FG. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J Infect Dis. 2001;183:523–531. doi: 10.1086/318537. [DOI] [PubMed] [Google Scholar]
- 9.Carr J, et al. Influenza virus carrying neuraminidase with reduced sensitivity to oseltamivir carboxylate has altered properties in vitro and is compromised for infectivity and replicative ability in vivo. Antiviral Res. 2002;54:79–88. doi: 10.1016/s0166-3542(01)00215-7. [DOI] [PubMed] [Google Scholar]
- 10.Ward P, Small I, Smith J, Suter P, Dutkowski R. Oseltamivir (Tamiflu) and its potential for use in the event of an influenza pandemic. J Antimicrob Chemother. 2005;55(Suppl 1):i5–i21. doi: 10.1093/jac/dki018. [DOI] [PubMed] [Google Scholar]
- 11.Hayden FG, Hay AJ. Emergence and transmission of influenza A viruses resistant to amantadine and rimantadine. Curr Top Microbiol Immunol. 1992;176:119–130. doi: 10.1007/978-3-642-77011-1_8. [DOI] [PubMed] [Google Scholar]
- 12.Centre for Disease Control and Prevention. Epidemiology of HIV/AIDS–United States, 1981–2005. Morbid Mortal Wkly Rep. 2006;55:589–592. [PubMed] [Google Scholar]
- 13.Ilyushina NA, Bovin NV, Webster RG, Govorkova EA. Combination chemotherapy, a potential strategy for reducing the emergence of drug-resistant influenza A variants. Antiviral Res. 2006;70:121–131. doi: 10.1016/j.antiviral.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman LR, Kuntz ID, White JM. Structure-based identification of an inducer of the low-pH conformational change in the influenza virus hemagglutinin: Irreversible inhibition of infectivity. J Virol. 1997;71:8808–8820. doi: 10.1128/jvi.71.11.8808-8820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodian DL, et al. Inhibition of the fusion-inducing conformational change of influenza hemagglutinin by benzoquinones and hydroquinones. Biochemistry. 1993;32:2967–2978. doi: 10.1021/bi00063a007. [DOI] [PubMed] [Google Scholar]
- 16.Yu KL, et al. Structure-activity relationships for a series of thiobenzamide influenza fusion inhibitors derived from 1,3,3-trimethyl-5-hydroxy-cyclohexylmethylamine. Bioorg Med Chem Lett. 2002;12:3379–3382. doi: 10.1016/s0960-894x(02)00761-8. [DOI] [PubMed] [Google Scholar]
- 17.De Clercq E. Antiviral agents active against influenza A viruses. Nat Rev Drug Discov. 2006;5:1015–1025. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson IA, Skehel JJ, Wiley DC. Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3-Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 19.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Skehel JJ, Wiley DC. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc Natl Acad Sci USA. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison SJ. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO. A revision of the system of nomenclature for influenza viruses: A WHO memorandum. Bull WHO. 1980;58:585–591. [PMC free article] [PubMed] [Google Scholar]
- 23.Fouchier RA, et al. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell RJ, et al. H1 and H7 influenza hemagglutinin structures extend a structural classification of hemagglutinin subtypes. Virology. 2004;325:287–296. doi: 10.1016/j.virol.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 25.Thoennes S, et al. Analysis of residues near the fusion peptide in the influenza hemagglutinin structure for roles in triggering membrane fusion. Virology. 2008;370:403–414. doi: 10.1016/j.virol.2007.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 27.Shangguan T, Alford D, Bentz J. Influenza virus-liposome lipid mixing is leaky and largely insensitive to the material properties of the target membrane. Biochemistry. 1996;35:4956–4965. doi: 10.1021/bi9526903. [DOI] [PubMed] [Google Scholar]
- 28.Rogers GN, et al. Single amino acid substitutions in influenza hemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 29.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 30.Steinhauer DA, Wharton SA, Skehel JJ, Wiley DC, Hay AJ. Amantadine selection of a mutant influenza virus containing an acid-stable hemagglutinin glycoprotein: Evidence for virus-specific regulation of the pH of glycoprotein transport vesicles. Proc Natl Acad Sci USA. 1991;88:11525–11529. doi: 10.1073/pnas.88.24.11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci USA. 2001;98:11181–11186. doi: 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. In: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S, editors. Warrington, U.K.: SERC Daresbury Laboratory; 1993. pp. 556–562. [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 36.Jones TA, Zhou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 37.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protocols. 2007;9:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 39.Bizebard T, et al. Structure of influenza virus hemagglutinin complexed with a neutralizing antibody. Nature. 1995;37:92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- 40.Trooskens G, De Beule D, Decouttere F, Van Criekinge W. Phylogenetic trees: Visualizing, customizing and detecting incongruence. Bioinformatics. 2005;21:3801–3802. doi: 10.1093/bioinformatics/bti590. [DOI] [PubMed] [Google Scholar]
- 41.Mayhood TM, Windsor WT. Ligand binding affinity determined by temperature-dependent circular dichroism: Cyclin-dependent kinase 2 inhibitors. Anal Biochem. 2005;345:187–197. doi: 10.1016/j.ab.2005.07.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.