Abstract
Newly translated tubulin molecules undergo a series of complex interactions with nascent chain-binding chaperones, including prefoldin (PFD) and chaperonin-containing TCP-1 (CCT). By screening for oryzalin hypersensitivity, we identified several mutants of Arabidopsis that have lesions in PFD subunits. The pfd6–1 mutant exhibits a range of microtubule defects, including hypersensitivity to oryzalin, defects in cell division, cortical array organization, and microtubule dynamicity. Consistent with phenotypic analysis, proteomic analysis indicates several isoforms of tubulins were reduced in pfd6–1. These results support the concept that the function of microtubules is critically dependent on the absolute amount of tubulins.
Keywords: chaperonin, cytoskeleton, morphology, TCP ring complex, TRiC
The biogenesis of actin and tubulin is of fundamental interest because these highly conserved proteins are involved in various processes, including cellular motion, morphogenesis, polarity, intracellular transport, and cell division. Actin and tubulin appear to have coevolved with 2 molecular chaperones, TCP-1 (CCT, also called TCP-ring complex or TriC) and prefoldin (PFD). CCT contains 8 orthologous subunits that form a cylindrical cavity that interacts in a subunit-specific manner with actins and tubulins (1). PFDs, also known as “genes involved in microtubule biogenesis complex,” bind to the nascent protein and transfer it to CCT for correct folding (2, 3). Archaeal PFD is an ≈90-kDa complex that consists of 2 α- and 4 β-subunits. The eukaryotic PFD also consists of 6 subunits of 2 paralogous α-type (Pfd3 and Pfd5) and 4 β-type (Pfd1, Pfd2, Pfd4, and Pfd6).
It has been suggested that a few cytoskeletal proteins, mainly actin and tubulins, are the major target proteins for CCT (4, 5). CCT appears to bind specific regions within actins and tubulins, as determined by the capacity of truncated target proteins to bind to CCT (6). Actin has 3 CCT recognition regions situated between residues 125–179, 244–285, and 340–375, whereas α- and β-tubulin contain 1 hydrophobic region (residues 244–285) that interacts with CCT. Both actin and α-/β-tubulin have been shown to interact with PFD in mammals (7). Although α-tubulin and β-actin are nonhomologous, each contains 2 regions important for binding to PFD (5). Disruption of genes encoding subunits of PFDs results in cytoskeletal defects that are similar to those of mutants in CCT. The Saccharomyces cerevisiae mutant pfd5Δ is viable but displays stunted growth and cold sensitivity. The deletion of PFD5 is synthetic lethal with a-tubulin (tub1–1), c-cpn (tcp1–1 and tcp1–2), and several actin mutants (3). PFD accelerates actin folding at least 5-fold and prevents the premature release of nonnative protein from CCT as demonstrated by pulse-chase labeling experiments followed by immunoprecipitation of CCT (8). In S. cerevisiae, deletion of the gene encoding the PFD component Pac10p leads to lower levels of both α- and β-tubulin (9). Single or double deletions of PFD subunits in S. cerevisiae results in phenotypes similar to those of mutants affecting the microtubule cytoskeleton, including cold-sensitive growth defects and an increased sensitivity toward the microtubule-depolymerizing drug benomyl (4).
In contrast to actin, facilitated folding of α- and β-tubulin requires cofactors in addition to CCT and PFD complex. Both α- and β-tubulin are released from the complex after binding to tubulin folding cofactors (TFC) A, B, C, D, and E (10, 11). Consistent with its function in microtubule folding, deletion of TFC in S. cerevisiae results in a cold-sensitive microtubule defect and increased sensitivity to benomyl (12, 13). Map-based cloning of several genes required for early embryo development identified Arabidopsis homologues of TFC-A, TFC-C, TFC-D, and TFC-E (14–17). Identification of 29 predicted genes encoding members of the chaperonin family of chaperones (CPN60 and CCT), cochaperonins, and cofactor PFD in Arabidopsis prompts questions about the complexities of plant CPN systems, most of which have not been studied experimentally (18). In particular, the Arabidopsis genome encodes 1 copy of each PFD subunit, none of which has been characterized.
We describe here the characterization of mutations in several Arabidopsis PFD genes. Our results suggest tubulin abundance may be important in controlling the dynamic properties of microtubules.
Results
Mutant Isolation and Characterization.
A mutant line designated AP90–5, carrying a mutation designated pfd6–1, was recovered from a screen of approximately 50,000 ethyl-methane sulfonate-mutagenized Columbia-0 seedlings for oryzalin hypersensitivity (19). A transgenic derivative line of AP90–5, called AP90–5M1, expressing a GFP-MAP4 microtubule binding domain marker was produced so that the microtubules could be examined in live tissue (20).
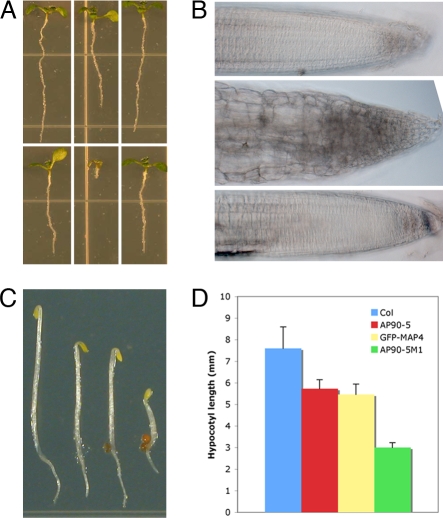
The pfd6–1 mutant exhibited root swelling on 100 nM oryzalin (Fig. 1 A and B) and showed a 25% reduction in hypocotyl length compared with that of dark-grown wild type (Fig. 1 C and D). Line AP90–5M1 showed a 45% reduction in hypocotyl length compared with the control (Fig. 1 C and D), indicating potential involvement of microtubules in the phenotype. Cell division in cortical or epidermal cells, as measured by cell number, was not affected significantly by the lesion in pfd6–1 (data not shown). However, the cell length in pfd6–1 was reduced 30%, especially in the elongation zone (results not shown).
Fig. 1.
Phenotypes of pfd6–1. (A) The effect of oryzalin on seedling growth in wild type and pfd6–1. From left to right are shown wild type, line AP90–5 (pfd6–1), and line AP90–5C1 (pfd6–1 transformed with the PFD6 genomic fragment). Shown are MS medium (Upper) and MS medium with 150 nM oryzalin (Lower). (B) The effect of oryzalin on root morphology in wild type and pfd6–1. Images are of root tips from A Lower. From top to bottom are shown wild type, AP90–5, AP90–5C1. (C) Three-day-old dark grown seedlings. From left to right are shown wild type, AP90–5, GFP-MAP4 control, and GFP-MAP4 in pdf6–1 background (line AP90–5M1). (D) Measurement of hypocotyl length of 3-day-old dark-grown seedlings. Data were collected from 50 seedlings. Error bars indicate standard deviation.
Positional Cloning of pfd6–1.
The pfd6–1mutation was mapped by bulk segregant mapping (21) on chromosome I between marker CIW12 and F27G20. Fine mapping limited the mutation to 4 genes between T1P2_RsaI-3 and T1P2_FnuDII. DNA sequencing of the 4 genes revealed a nucleotide substitution (G to A) in At1g29990 that resulted in an amino acid change R (83) Q [supporting information (SI) Fig. S1A]. At1g29990 encodes a 14-kDa polypeptide with 32% sequence identity to PFD subunit 6 from S. cerevisiae but weaker similarity to the other 5 S. cerevisiae genes. Alignment of putative PFD6 orthologs from Caenorhabditis elegans, Homo sapiens, S. cerevisiae, Oryza sativa, and Arabidopsis thaliana indicated that R83 is conserved in all of these proteins. Thus, the R-to-Q amino acid substitution may have a detrimental effect on protein structure or function (22).
To confirm that At1g29990 corresponded to PFD6, a 3.5-kb genomic fragment containing the entire gene and upstream promoter region was cloned into the binary vector pCambia3300 to produce pYG101. The resulting construct was introduced into pfd6–1 by Agrobacterium-mediated transformation. Twelve independent transformant lines rescued the oryzalin hypersensitive phenotype (Fig. 1B), indicating that At1g29990 was responsible for the pfd6–1 mutant phenotype. We also identified 2 T-DNA insertion lines of PFD6 (named pfd6–2 and pfd6–3) from the public SIGNAL collection (23). pfd6–2 carries a T-DNA insertion in the third exon, and full-length PFD6 RNA could not be detected (Fig. S2 A and B). pfd6–2 shows a similar level of hypersensitivity to oryzalin to the pfd6–1 mutant, whereas pfd6–3 is slightly less sensitive to oryzalin. The pfd6–3 mutant is not a null mutant because its T-DNA insertion at the 5′UTR did not fully disrupt the expression of PFD6 (results not presented). The residual PFD6 in pfd6-3 may explain the reduced oryzalin sensitivity (Fig. S2C).
pfd6–1 Has Defects in Organization of Cortical Arrays and Cell Division.
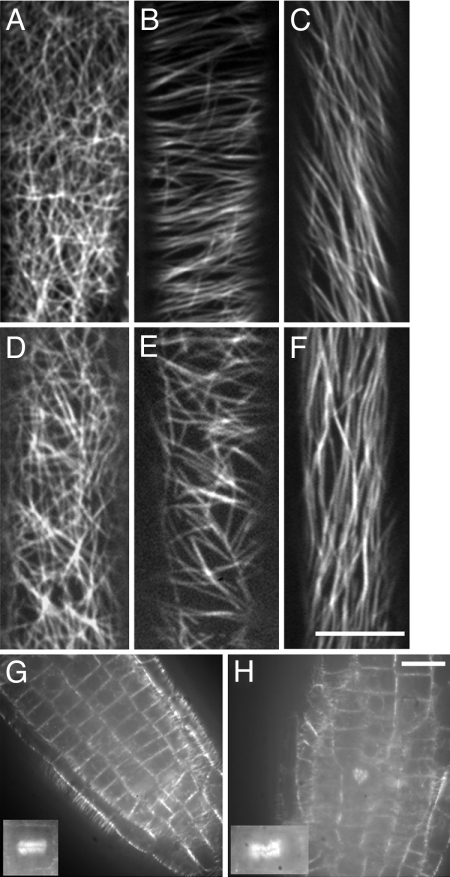
Elongation of diffusely expanding cells is thought to be mainly under the control of cortical microtubules (24). We sought to determine whether the cell size reduction in pfd6–1 has a corresponding microtubule-related defect by examining the microtubule organization of epidermal hypocotyl cells. The epidermal cells at the apical hook of etiolated 3-day-old hypocotyls had random cortical microtubule organization in both control and pfd6–1 tissues (Fig. 2 A and D). After a short transition zone that lies between the apical hook and the elongation zone, the cortical arrays become increasingly organized as cells prepare for expansion. The cortical arrays in the elongation zone are generally transverse, which is perpendicular to the axis of growth. Microtubules in the elongation zone of pfd6–1 appeared less organized than those in wild-type cells (Fig. 2 B and E). Only 12% of the pfd6–1 cells displayed transverse microtubule orientations compared with 79% for the control (Table S1). No difference was observed in fully elongated cells between the control line and pfd6–1 (Fig. 2 C and F). The increased randomness of microtubule organization may explain the reduction of hypocotyl cell expansion displayed in pfd6–1 cells. pfd6–1 also showed increased randomness of microtubule organization in light grown seedlings (data not shown), suggesting that altered microtubule organization in pfd6–1 was not because of a difference in growth conditions.
Fig. 2.
pfd6–1 affects microtubule organization in hypocotyl cells. GFP-MAP4-labeled microtubules in wild type (A–C) and pfd6–1 (D–F) hypocotyl cells. Images were collected at different positions of hypocotyl cells. Microtubule organization has distinct patterns for cells at the apical hook (A and D), 2–3 mm below apical hook (B and E), and 2–3 mm away from root (C and F). (Scale bar, 10 μm.) (G and H) Abnormal orientation of cell division planes in pfd6–1. Cells were visualized with GFP-MAP4 in roots of 3-day-old de-etiolated wild type (G) and pfd6–1 (H). An abnormal phragmoplast may lead to the cell plate defects observed in pfd6–1. (Scale bar, 10 μm.)
Epidermal cells of wild-type roots are rectangular with their division planes perpendicular to the long axis, and 99% of the new division planes in wild type were perpendicular to the long axis of the mother cell (Fig. 2). Epidermal cells in pfd6–1 were more variable in shape, and their division planes were frequently skewed in relation to the root axis (Fig. 2H). Aberrant divisions were more frequently observed in pfd6–1 (>30%), resulting in the reduction of transverse divisions from 99% (control) to 76% (pfd6–1). To explore whether aberrant divisions may be because of abnormal cell plate formation, we examined the appearance of the GFP-MAP4 marker on cytokinetic microtubule arrays. Abnormal phragmoplasts were observed in line AP90–5M1 grown on MS plates without oryzalin (Fig. 2 G and H). In control root tips, 99% of phragmoplasts were oriented perpendicular to the root axis and microtubules were uniformly distributed across phragmoplasts. By contrast, 11 of 32 phragmoplasts were misoriented and showed skewed microtubule distributions in pfd6–1. The degree of disorganization of root cell files was exacerbated by growth on oryzalin at 100 nM (data not shown), a concentration that has no effect on cell file organization in wild-type seedlings.
Microtubule Dynamics Are Altered in the pfd6–1 Mutant.
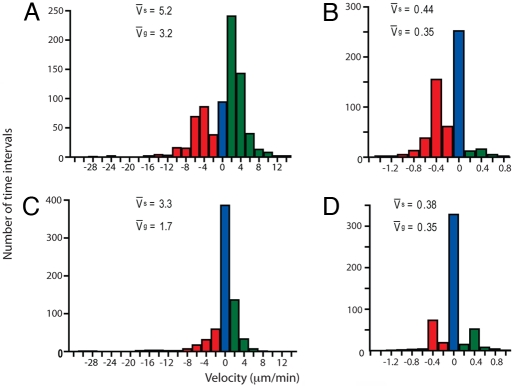
Microtubule dynamics are one of the key factors that contribute to the organization of cortical arrays (25). To investigate whether microtubule dynamics were altered in pfd6–1, we visualized microtubule dynamics by confocal microscopy of AP90–5T1, a transgenic derivative of AP90–5 carrying a YFP-tubulin fusion, YFP-TUA5 (26). Seedlings were dark-grown for 3 days and, to minimize the disturbance of microtubule orientation because of blue-light-induced reorientation (24), all measurements were taken in the first 5 min after exposure to the confocal laser. Using both the GFP-MAP4 marker line (AP90–5M1) and YFP-TUA marker line (AP90–5T1), we found that the microtubule density in pfd6–1 was not obviously different from that in wild type. Differences in microtubule array organization were apparent in upper hypocotyl (Fig. 2) and root cells (data not shown). Measurement of microtubule dynamics indicated that pfd6–1 had a significant reduction (−47%) in plus end growth with a rate of 1.7 μm/min as compared with 3.2 μm/min for control cells (Table 1). The shrinkage rate at the plus end in pfd6–1 had a similar but less pronounced reduction (−37%). A change in the proportion of time spent in growth or shortening phases for the plus end in pfd6–1 correlates well with microtubule behavior as shown in Fig. 3. There was a 27% increase of individual microtubules maintained at the pause state in pfd6–1 as compared with control, consistent with the observation that microtubule dynamics appeared to be more static in pfd6–1 than those of control (Movie S1). The reduction of time spent in the growth phase (≈25%) at the plus end also contributes to the static appearance of microtubule dynamics.
Table 1.
In vivo measurements of single microtubule growth phases, dynamicity, and net polymer gain/loss in the wild type (Col-0) and mutant background (pfd6–1)
| Time in phase | Col-0 |
pfd6–1 |
||
|---|---|---|---|---|
| (+) | (−) | (+) | (−) | |
| Growth, % | 63.5 | 8.7 | 38.7 | 18.4 |
| Pause, % | 3.3 | 41.9 | 30.6 | 60.2 |
| Shorten, % | 33.2 | 49.4 | 30.7 | 21.4 |
| Dynamicity, μm/min | 3.75 ± 2.9 | 0.3 ± 0.2 | 1.53 ± 2.8 | 0.15 ± 0.2 |
| Net polymer gain, μm/min | 0.31 | −0.19 | 0.29 | −0.01 |
Measurements consist of 1393 and 1216 velocities (2-s time intervals) for control and pfd6–1, respectively.
Fig. 3.
pfd6–1 reduces microtubule dynamicity. Growth and shrinkage velocities for the leading end (A and C) and lagging end (B and D) of single microtubules in 3-day de-etiolated hypocotyls of wild type (A and B) and pfd6–1 (C and D). Velocities were calculated from 57 (leading end) and 53 (lagging end) polymer ends for control cells and 42 per each end for pfd6–1. Total measurements consist of 1393 and 1216 velocities (2-s time intervals) for control and pfd6–1, respectively. The histogram is color-coded for growth (green), pause (blue), and shrinkage (red) velocities. Mean velocities for both growth (vg) and shrinkage (vs) are indicated within each panel.
Tubulins Are Targeted by the PFD Complex for Proper Folding.
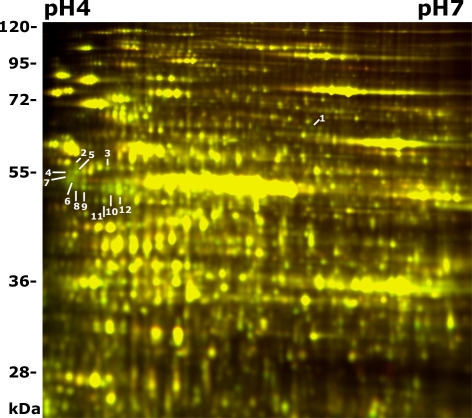
It has been suggested that the PFD complex binds to nascent tubulins and transports them to CCT for folding (3, 4). The altered microtubule organization and dynamicity of cortical microtubules in pfd6–1 suggests that microtubule organization has been affected because of the lesion in PFD6. To test whether the plant PFD complex plays a role in tubulin folding, we studied the proteomic changes caused by the lesion in PFD6 using two-dimensional difference gel electrophoresis (2D DIGE) coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS). Both pfd6–1 and wild-type (Col-0) seedlings were grown hydroponically in continuous light for 5 days before 2D DIGE analysis. Triplicate pairs of pfd6–1 and control (Col-0) were analyzed by using 2D DIGE. Representative spots that were down-regulated in pfd6–1 were detected and analyzed (Fig. 4). Spots of interest were then picked with a robotic spot picker, in-gel trypsin digested, and analyzed with LC-MS/MS. A total of 12 PFD-targeted protein spots were identified as products of 12 unique genes (Table 2). Several different isoforms of tubulins were reduced in pfd6–1, including β-1, β-4, β-7, and β-9. The reduction level (≈30%) was roughly similar among them. In addition, several other non-cytoskeletal putative substrates appeared to be reduced in pfd6–1 (Table 2).
Fig. 4.
2-D DIGE analysis of pfd6–1. Proteins from pfd6–1 and wild-type seedlings grown in liquid growth medium were extracted and labeled with DIGE Cy5 (red) or Cy3 (green), respectively. Proteins were then separated on pH 3–10 nonlinear IPG strips, followed by SDS/PAGE (12.5%) and scanning for Cy5 or Cy3 fluorescence. Color overlay of 2D-DIGE images (pH 4–7 range) of the control (Col-0) and pfd6–1 was shown. Proteins decreased in pfd6–1 display green, and those increased display red, whereas those unaffected display yellow. Protein spots of interest are marked with spot numbers and their identities are shown in Table 2.
Table 2.
Comparative proteomic analysis between the pfd6–1 mutant and wild type
| Spot no. | Peptide matched | Gene ocus | Protein identity | Ratio | P value |
|---|---|---|---|---|---|
| 1 | 13 | At2 g17980 | Sec1-like protein (SLY1) | −1.6 | 5.9E-5 |
| 2 | 11 | At1 g79340 | Metacaspase 4 (ATMC4) | −1.48 | 7.6E-3 |
| 3 | 74 | At1 g48600 | Phosphoethanolamine methyltransferase 2 | −1.31 | 2.4E-4 |
| 4 | 3 | At3 g02540 | Ubiquitin family protein | −1.36 | 1.2E-2 |
| 5 | 4 | At4 g26110 | Nucleosome assembly protein1 (NAP1) | −1.77 | 1.2E-5 |
| 6 | 18 | At1 g75780 | TUB1 | −1.54 | 2.0E-4 |
| 7 | 15 | At4 g20890 | TUB9 | −1.41 | 3.9E-5 |
| 8 | 13 | At2 g29550 | TUB7 | −1.41 | 3.9E-5 |
| 9 | 67 | At5 g44340 | TUB4 | −1.4 | 2.4E-3 |
| 10 | 23 | At4 g32720 | LA protein 1 (ATLA1) | −1.62 | 2.7E-5 |
| 11 | 8 | At1 g76030 | V-ATPase B subunit | −1.28 | 1.0E-3 |
| 12 | 51 | At3 g55800 | Sedoheptulose-1,7-bisphosphatase | −1.78 | 1.2E-4 |
Representative proteins down-regulated in pfd6–1 were identified. Number of peptides obtained from tandem MS analysis of each spot is listed. Average protein level ratios of pfd6–1/wild type and the t test P values are calculated from 3 replicates. Negative ratios indicate that spots were down-regulated in pfd6–1. Spots are numbered as in Fig. 4.
PFD6 Does Not Associate with Microtubules.
The defect in pdf6–1 significantly reduces microtubule dynamicity in vivo. Microtubule dynamics are influenced by interactions of microtubules with cellular factors that are associated with microtubules. To test the possibility that the effect of reduced microtubule dynamicity was because of direct association of PFD6 with microtubules, we examined intracellular localization of PFD6 in vivo. To visualize PFD6, we produced transgenic Arabidopsis plants, designated AP90–5F, with a C-terminal fusion of citrine yellow fluorescent protein (YFP) to the 2.7-kb genomic fragment containing the native PFD6 promoter and the entire gene except 3′UTR. The construct fully complemented mutant phenotypes of reduced hypocotyls and root elongation upon exposure to oryzalin, indicating the fusion protein was functional (Fig. S2C). PFD-YFP was observed in epidermal cells of 3-day-old dark-grown hypocotyls and roots. As expected, PFD6-YFP fluorescence was not associated with microtubule-like structures. As shown in Fig. S2D, PFD6-YFP was evenly distributed throughout the cytoplasm in epidermal cells of roots and hypocotyls.
PFD4 and PFD6 Share Nonredundant Function in the Same Complex.
To gain further insight about the plant PFD complex, we sought to identify interaction partners for PFD6 subunits using the Arabidopsis Interaction Viewer (http://bar.utoronto.ca/interactions/). Of 93 interactions identified for PFD6, a high cut off Interlog confidence value (IC >10) was used to retrieve the 6 top interactors (Table S2). Among them, 4 encode putative subunits of a hypothetical PFD complex in Arabidopsis. Interestingly, tubulin folding factor A (TFC-A) is also predicted to be directly associated with the PFD complex. In a model of the tubulin folding pathway based on biochemical data, nascent β-tubulin first associates with the chaperonin complex, then is released from the complex after binding to TFC-A. Consistent with the model, TFC-A is predicted to associate with the PDF complex (Table S2). Interestingly, γ-tubulin was predicted to be in the PFD complex rather than α- or β-tubulin. This prediction echoes previous results indicating physical and genetic interaction between PFD and Tub4p (4). Nevertheless, these data provide indications that Arabidopsis may encode a fully functional PFD complex involved in microtubule biogenesis.
The PFD complex consists of 6 sequence-related subunits, all of which are represented in the Arabidopsis genome. For a better understanding of the functional relevance of PFD subunits in the same complex in plants, we attempted to identify additional T-DNA insertion mutants in PFD subunits. A fully viable homozygous T-DNA insertion line of PFD4 (named pfd4–1) was isolated. pfd4–1 carries a T-DNA insertion at the second exon-intron junction resulting in loss of full-length RNA (data not shown). A quantification of hypersensitivity to oryzalin was carried out for pfd4–1 and pfd6–1 by measuring the primary root length of 5-day-old light-grown seedlings on increasing concentrations of oryzalin as indicated in Fig. 5 and Table 3. Both pfd4–1 and pfd6–1 were 4-fold more sensitive to oryzalin as compared with wild type. The existence of 2 nonredundant oryzalin-hypersensitive loci suggested that PFD4 and PFD6 are part of the same complex.
Fig. 5.
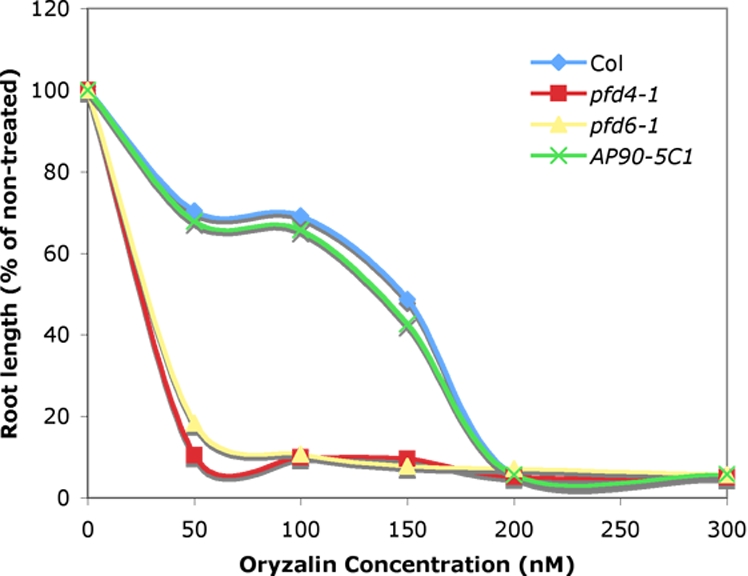
PFD4 and PFD6 are in the same PFD complex. Dose-response curve of the effect of oryzalin on primary root length. Seedlings were grown vertically on agar for 5 days on MS plates supplemented with indicated concentrations of oryzalin. Root length is expressed in percentage of the non-treated samples.
Table 3.
Average root length (as % of control) of ≈30 individuals at 0 nM and 100 nM concentrations of oryzalin
| Oryzalin, nM | Col-0 |
pfd4–1 |
pfd6–1 |
AP90–5C1 |
||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| 0 | 100 | 6.2 | 100 | 10.9 | 100 | 9.3 | 100 | 7.8 |
| 100 | 69.2 | 7.0 | 9.9 | 1.8 | 10.7 | 1.2 | 65.7 | 5.3 |
Discussion
Microtubules are important structural components involved in many cellular processes in plants, including mitosis, cytokinesis, morphogenesis, and vesicular transport. Normal microtubule function is thought to require coordination among various components of the cytoskeleton including nucleation processes, the microtubule organization center, motor proteins, and various microtubule associated proteins (MAPs). Microtubule function is also critically dependent on factors involved in microtubule biogenesis. Tubulin folding factors (TFCs) are involved in regulation of dimer formation and microtubule stability. Four (TFC-A, TFC-C, TFC-D, and TFC-E) of six TFCs have been identified and functionally characterized so far in higher plants (14–16). Mutations in PORCINO (POR, TFC-C ortholog), CHAMPIGNON (CHO, TFC-D ortholog), and PEIFFERLING (PFI, TFC-E ortholog) are embryo lethals and fail to form microtubules during cell division in mutant embryos (14). KIESEL (KIS, TFC-A ortholog) mutants display weaker and more variable defects associated with impaired microtubule function (15). Little is known about the early steps of microtubule biogenesis, but it appears that the hexameric PFD complex is involved in the first step of binding nascent tubulins and transferring them to CCT for proper folding.
The characterization of pfd6–1 reported here supports the idea that Arabidopsis depends upon the PFD and CCT complexes for stability of tubulin and a small number of other proteins. Lesions in PFD6 resulted in impaired microtubule function, which was associated with reduced plant size, abnormal orientation of cell division planes, altered microtubule organization, reduced microtubule dynamicity, and reduced trichome branching (data not shown). Rapidly expanding cells in the etiolated hypocotyls have transverse microtubule arrays that are perpendicular to the long axis. The reduction in coherence of microtubule orientation in those rapidly expanding cells in pfd6–1 is associated with a restriction of expansion in longitudinal direction. Therefore, overall hypocotyl length is reduced in pfd6–1 compared with wild type.
As in S. cerevisiae, tubulins are among the major target proteins for CCT in Arabidopsis. 2D-DIGE and MS/MS analysis indicated that different isoforms of tubulins were reduced in pfd6–1. Presumably, loss of the chaperonin caused abnormal folding resulting in accelerated turnover of the incorrectly folded tubulin. Consistent with this finding, the pfd6–1 mutant had reduced rates of both dimer addition and loss at the plus end. Polymerization events at the minus end were too small for meaningful comparisons. Microtubules assemble by adding α/β-tubulin dimers at both ends of growing microtubules. The addition occurs preferentially at the leading end (+ end) when the concentration of dimers is above critical concentrations (Cc). When the concentration of α/β-tubulin dimers is below Cc, the microtubules disassemble twice as rapidly at the (+) end as at the (−) end (27). Thus, the observation that pfd6–1 reduces both shrinkage and growth rate more significantly at the (+) end is consistent with microtubule behaviors below the Ccs. It is also formally possible that misfolded tubulin in the mutant may inhibit microtubule assembly to some extent.
The cortical arrays of elongating pfd6–1 are significantly disordered. Several factors could potentially affect the organization of microtubule arrays in this way. It is thought that microtubule dynamics is one of the key factors that contribute to the organization of arrays (28). Recently identified tubulin point mutations caused directional changes in handedness of cortical microtubule arrays. A right-handed mutant (tua5D251N) bears a mutant residue that affects GTP hydrolysis in β-tubulin. Consequently, microtubule depolymerization is inhibited and overall dynamics is decreased. A mutation at the intradimer interface (tua4S178Δ), on the other hand, results in highly dynamic microtubules and left-handed cortical arrays. We propose that the reduced growth and shrinkage rate of cortical microtubule in pfd6–1 is because of reduced concentration of tubulin pools that affects microtubule dynamics. Exactly how microtubule dynamics contributes to final organization of cortical microtubule arrays is yet to be understood. Polymerization-biased microtubule instability at the + ends and slow depolymerization at the − ends make cortically attached microtubules migrate across the cell (26). The resulting sustained microtubule treadmilling causes widespread microtubule repositioning and contributes to reorientation of microtubules and formation of bundles in rapidly expanding hypocotyls. The properties of the pfd6–1 mutant suggest that organization of microtubule arrays is critically dependent on the amount of tubulin.
Other eukaryotic PFD complexes bind to nascent chains and assist folding of actin and tubulin (4, 5, 7). In our 2D-DIGE analysis using pfd6–1, actins were not identified among the down-regulated proteins and actin organization was not obviously changed in pfd6–1 as compared with wild type (data not shown). Thus, either actin is not a substrate for PFD6 or the mutated pfd6–1 may have conformational changes that do not affect binding of PFD complex to all of its substrates. Although it cannot be excluded that PFD could also play a role in actin folding, PFD function is more critical to support the assembly of microtubule arrays than the formation of the actin cytoskeleton. The viability of pfd6–1 mutant raises the possibility that either the PFD complex is not necessary under the growth conditions used, or that a paralog may substitute for PFD6. Alternatively there may be a PFD independent folding mechanism that can compensate for loss of pfd6–1. Recently, a novel CCT cofactor phosducin-like protein 1 (PLP1) was identified to interact with CCT and to modulate the efficiency of β-tubulin and actin folding, at an early step distinct from the PFD complex (29–31). Temperature-sensitive alleles of S. cerevisiae PLP2 exhibit cytoskeletal and cell cycle defects (29). Consistent with the model that PLP1 participates in an early step of β-tubulin folding and in parallel with PFD, plp1Δ interacted synergistically with mutants in any of 5 PFD subunits (31). More recently, PLP3 homologs (PLP3a and PLP3b) from Arabidopsis were identified and impairing PLP3a and PLP3b expression resulted in disrupted microtubule arrays, defective cytokinesis, and disoriented cell growth (32). Further investigation is required to know whether PLP3 modulates CCT-mediated tubulin folding in parallel with PFD. Although few other putative non-cytoskeletal substrates appeared to be transferred to CCT by the PFD complex in Arabidopsis, it is apparent that the PFD complex has similar roles in plants as in other eukaryotes.
Methods
Plant Materials, Growth Conditions, and Genetic Methods.
A. thaliana Columbia (Col-0) seedlings were grown on vertical MS plates (1/2 × MS salts, 0.8% agar, and 0.05% Mes, pH 5.7) at 22 °C under continuous light. Soil-grown plants were grown at 22 °C under 16-hr light and 8-hr dark.
For mapping of the pdf6–1 mutation, AP90–5 (pfd6–1) was crossed to Ler and F2 seedlings showing hypersensitivity to 150 nM oryzalin were selected. pfd6–1 was mapped on chromosome I between marker CIW12 and F27G20 (21) and fine mapped to the region between TIP2_RasI-3 and TIP2_FnuDII (Table S3). Genetic complementation tests were performed by introducing a 3.5-kb genomic fragment containing the entire gene and upstream promoter region of At1g29990. The genomic fragment was amplified by using primer 29990_5′EcoRI and 29990_3′PstI (Table S3) and was cloned into pGEM-Teasy vector (Promega). The resulting construct (pYG100) was sequenced and cloned into pCambia3300 to produce pYG101, which was then introduced into pfd6–1 by Agrobacterium-mediated transformation, resulting in 54 transgenic lines.
Seeds of PFD6 and PFD4 knockout lines from the SIGNAL collection were obtained from the Arabidopsis Biological Resource Center (http://www.biosci.ohio-state.edu/∼plantbio/Facilities/abrc/abrchome.htm). Primers used for T-DNA genotyping of pfd6 alleles were 5′29990_TDNA and 3′29990_TDNA (Table S3). Primers used for T-DNA genotyping of the pfd4 allele were 5′08780 and 3′08780 (Table S3)
Construction of Transgenic Lines.
A 2.7-kb fragment containing the entire gene except 3′UTR and upstream promoter region of At1g29990 was amplified by using forward primer 5′PFD6_311 and reverse primer 3′PFD6_101 (Table S3) from Columbia-0 genomic DNA. The purified PCR fragment was introduced into pDONR/Zeo through attB×attP reaction mediated by Gateway BP Clonase II (Invitrogen) to produce pYG102, then the insert was introduced into a modified version of pEarleyGate 311 (a gift from D. Ehrhardt, Carnegie Institution, Stanford, CA) through an attL×attR reaction mediated by Gateway LR Clonase II. The construct (pYG103) was then introduced into pfd6–1 by Agrobacterium-mediated transformation. Line AP90–5 was crossed to a GFP-MAP4 marker line that facilitates visualization of MTs in live root cells (20). A resulting F3 line (AP90–5M1), homozygous for both pfd6–1 and GFP-MAP4, was chosen for further analysis. For generating AP90–5T1, crosses were made between AP90–5 and YFP-TUA5 and lines homozygous for both mutant and YFP-TUA5 were identified.
Confocal Microscopy and Image Analysis.
For microtubule dynamics analysis, seeds were germinated on MS agar plates and grown vertically in the darkness for 3 days at 22 °C. Seedlings were mounted between 2 cover slips in water. Imaging was performed on a spinning-disk confocal microscope (24) by using a Leica x63-oil objective. EGFP and YFP were excited at 488-nm by using an Argon laser. A band-pass filter (520/50 nm) was used for emission filtering. Image analysis was performed by using Metamorph (Molecular Devices) and ImageJ (W. Rasband, National Institutes of Mental Health, Bethesda, MD) software.
2D-DIGE and LC-MS/MS.
Proteomic analysis of pfd6–1 was performed as previously described by using 24-cm, pH 3 to 10, isoelectric-focusing strips and 12.5% SDS/PAGE gels (33). Proteins were extracted by a phenol-methanol method (33) from Arabidopsis seedlings of pfd6–1 and Columbia-0 grown hydroponically in MS media with 1.5% sucrose for 5 days. The identities of proteins were identified by tandem mass spectrometry (LC-MS/MS) by Stanford University Mass Spectrometry (http://mass-spec.stanford.edu).
Database Search and Sequence Alignment.
Predicted amino acid sequences of PFD6 were retrieved from the TAIR database (www.arabidopsis.org) and used to identify other PFD subunits in Arabidopsis and rice. Aligned sequences were processed by using Boxshade (www.ch.embnet.org/software/BOX_form.html) in a fraction of 0.5 and output as new RTF format. Predicted interaction partners with PFD6 were retrieved from the Bio-Array Resource for Arabidopsis Functional Genomics (http://bbc.botany.utoronto.ca/) (34).
Supplementary Material
Acknowledgments.
We thank K. Hematy, S. Li, and J. Estévez for reading the manuscript and providing suggestions. This work was supported in part by an award from The Balzan Foundation, U.S. Department of Energy Grant DOE-FG02-03ER20133), and a grant from Energy Biosciences Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808652105/DCSupplemental.
References
- 1.Leroux MR, Hartl FU. Protein folding: Versatility of the cytosolic chaperonin TRiC/CCT. Curr Bio. 2000;10:R260–264. doi: 10.1016/s0960-9822(00)00432-2. [DOI] [PubMed] [Google Scholar]
- 2.Deuerling E, Bukau B. Chaperone-assisted folding of newly synthesized proteins in the cytosol. Critic Rev Biochem Mol Biol. 2004;39:261–277. doi: 10.1080/10409230490892496. [DOI] [PubMed] [Google Scholar]
- 3.Vainberg IE, et al. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- 4.Geissler S, Siegers K, Schiebel E. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J. 1998;17:952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rommelaere H, et al. Prefoldin recognition motifs in the nonhomologous proteins of the actin and tubulin families. J Biol Chem. 2001;276:41023–41028. doi: 10.1074/jbc.M106591200. [DOI] [PubMed] [Google Scholar]
- 6.Rommelaere H, De Neve M, Melki R, Vandekerckhove J, Ampe C. The cytosolic class II chaperonin CCT recognizes delineated hydrophobic sequences in its target proteins. Biochemistry. 1999;38:3246–3257. doi: 10.1021/bi9815905. [DOI] [PubMed] [Google Scholar]
- 7.Hansen WJ, Cowan NJ, Welch WJ. Prefoldin-nascent chain complexes in the folding of cytoskeletal proteins. J Cell Biol. 1999;145:265–277. doi: 10.1083/jcb.145.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegers K, et al. Compartmentation of protein folding in vivo: Sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO J. 1999;18:75–84. doi: 10.1093/emboj/18.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacefield S, Magendantz M, Solomon F. Consequences of defective tubulin folding on heterodimer levels, mitosis and spindle morphology in Saccharomyces cerevisiae. Genetics. 2006;173:635–646. doi: 10.1534/genetics.105.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian G, et al. Pathway leading to correctly folded β-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- 11.Tian G, et al. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J Cell Biol. 1997;138:821–832. doi: 10.1083/jcb.138.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoyt MA, Macke JP, Roberts BT, Geiser JR. Saccharomyces cerevisiae PAC2 functions with CIN1, 2 and 4 in a pathway leading to normal microtubule stability. Genetics. 1997;146:849–857. doi: 10.1093/genetics/146.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Archer JE, Vega LR, Solomon F. Rbl2p, a yeast protein that binds to β-tubulin and participates in microtubule function in vivo. Cell. 1995;82:425–434. doi: 10.1016/0092-8674(95)90431-x. [DOI] [PubMed] [Google Scholar]
- 14.Steinborn K, et al. The Arabidopsis PILZ group genes encode tubulin-folding cofactor orthologs required for cell division but not cell growth. Genes Dev. 2002;16:959–971. doi: 10.1101/gad.221702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirik V, et al. Functional analysis of the tubulin-folding cofactor C in Arabidopsis thaliana. Curr Biol. 2002;12:1519–1523. doi: 10.1016/s0960-9822(02)01109-0. [DOI] [PubMed] [Google Scholar]
- 16.Kirik V, et al. The Arabidopsis TUBULIN-FOLDING COFACTOR A gene is involved in the control of the α/β-tubulin monomer balance. Plant Cell. 2002;14:2265–2276. doi: 10.1105/tpc.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutsche I, Essen LO, Baumeister W. Group II chaperonins: New TRiC (k) s and turns of a protein folding machine. J Mol Biol. 1999;293:295–312. doi: 10.1006/jmbi.1999.3008. [DOI] [PubMed] [Google Scholar]
- 18.Hill JE, Hemmingsen SM. Arabidopsis thaliana type I and II chaperonins. Cell Stress Chaperones. 2001;6:190–200. doi: 10.1379/1466-1268(2001)006<0190:attiai>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paredez A, Persson S, Ehrhardt DW, Somerville C. Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiol. 2008;147:1723–1734. doi: 10.1104/pp.108.120196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marc J, et al. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell. 1998;10:1927–1939. doi: 10.1105/tpc.10.11.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukowitz W, Gillmor CS, Scheible WR. Positional cloning in arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 2000;123:795–805. doi: 10.1104/pp.123.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegert R, Leroux MR, Scheufler C, Hartl FU, Moarefi I. Structure of the molecular chaperone prefoldin: Unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell. 2000;103:621–632. doi: 10.1016/s0092-8674(00)00165-3. [DOI] [PubMed] [Google Scholar]
- 23.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 24.Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 25.Paradez A, Wright A, Ehrhardt DW. Microtubule cortical array organization and plant cell morphogenesis. Curr Opin Plant Biol. 2006;9:571–578. doi: 10.1016/j.pbi.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Shaw SL, Kamyar R, Ehrhardt DW. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 2003;300:1715–1718. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- 27.Lodish H, et al. Molecular Cell Biology. New York: W. H. Freeman and Company; 2000. p. 973. [Google Scholar]
- 28.Ehrhardt DW, Shaw SL. Microtubule dynamics and organization in the plant cortical array. Annu Rev Plant Biol. 2006;57:859–875. doi: 10.1146/annurev.arplant.57.032905.105329. [DOI] [PubMed] [Google Scholar]
- 29.Stirling PC, et al. PhLP3 modulates CCT-mediated actin and tubulin folding via ternary complexes with substrates. J Biol Chem. 2006;281:7012–7021. doi: 10.1074/jbc.M513235200. [DOI] [PubMed] [Google Scholar]
- 30.Stirling PC, et al. Functional interaction between phosducin-like protein 2 and cytosolic chaperonin is essential for cytoskeletal protein function and cell cycle progression. Mol Biol Cell. 2007;18:2336–2345. doi: 10.1091/mbc.E07-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 32.Castellano MM, Sablowski R. Phosducin-like protein 3 is required for microtubule-dependent steps of cell division but not for meristem growth in Arabidopsis. Plant Cell. 2008;20:969–981. doi: 10.1105/tpc.107.057737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng Z, et al. A proteomic study of brassinosteroid response in Arabidopsis. Mol Cell Proteomics. 2007;6:2058–2071. doi: 10.1074/mcp.M700123-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geisler-Lee J, et al. A predicted interactome for Arabidopsis. Plant Physiol. 2007;145:317–329. doi: 10.1104/pp.107.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.