Abstract
Treatment of yeast and human cells with DNA-damaging agents elicits Rad6–Rad18-mediated monoubiquitination of proliferating cell nuclear antigen (PCNA) at its Lys-164 residue [ubiquitin (Ub)-PCNA], and this PCNA modification is indispensable for promoting the access of translesion synthesis (TLS) polymerases (Pols) to PCNA. However, the means by which K164-linked Ub modulates the proficiency of TLS Pols to bind PCNA and take over synthesis from the replicative Pol has remained unclear. One model that has gained considerable credence is that the TLS Pols bind PCNA at 2 sites, to the interdomain connector loop via their PCNA-interacting protein (PIP) domain and to the K164-linked Ub moiety via their Ub-binding domain (UBD). Specifically, this model postulates that the UBD-mediated binding of TLS Pols to the Ub moiety on PCNA is necessary for TLS. To test the validity of this model, we examine the contributions that the PIP and Ub-binding zinc finger (UBZ) domains of human Polη make to its functional interaction with PCNA, its colocalization with PCNA in replication foci, and its role in TLS in vivo. We conclude from these studies that the binding to PCNA via its PIP domain is a prerequisite for Polη's ability to function in TLS in human cells and that the direct binding of the Ub moiety on PCNA via its UBZ domain is not required. We discuss the possible role of the Ub moiety on PCNA in TLS.
Keywords: PIP domain, UBZ domain, PCNA ubiquitination, exchange
Translesion synthesis (TLS) promotes replication through DNA lesions. In humans, TLS is carried out by a number of DNA polymerases (Pols) that include Pols η, ι, κ, and Rev1, which are all members of the Y family, and Polζ, a B family member. Biochemical and structural studies with the Y family Pols have indicated a high degree of specialization in their structure and function, which enables them to synthesize DNA opposite a diverse array of DNA lesions (1). For example, Polη is highly efficient in replicating through UV-induced cyclobutane pyrimidine dimers (CPDs) because of its ability to accommodate the CPD in its active site, and inactivation of Polη in humans causes the cancer-prone syndrome, the variant form of xeroderma pigmentosum (XPV) (2–6).
One of the important questions regarding TLS relates to the means by which TLS Pols gain access to the replication fork and take over synthesis from the replicative Pol at the site of a DNA lesion. Genetic and biochemical studies in both yeast and humans have indicated that proliferating cell nuclear antigen (PCNA) plays a critical role in the Pol exchange process. The TLS Pols, such as Polη from yeast, and Pols η, ι, and κ from humans, have been shown to interact physically and functionally with PCNA, and PCNA binding greatly enhances their DNA synthetic activity on both undamaged and damaged DNAs (7–10). The TLS Pols bind PCNA at its interdomain connector loop via their PCNA-interacting protein (PIP) domain, and genetic studies with yeast Polη (yPolη) have shown that mutational inactivation of its PIP domain abolishes its ability to function in TLS in vivo (7).
The various lesion bypass processes, including TLS, are regulated by the Rad6–Rad18 ubiquitin (Ub)-conjugating (UBC) enzyme complex (11, 12). In yeast and human cells treated with DNA-damaging agents, PCNA is monoubiquitinated at its Lys-164 residue by the Rad6–Rad18 enzyme complex (13), and genetic studies in yeast have shown that PCNA monoubiquitination is essential for TLS (14, 15). However, despite a number of studies that have been done to elucidate the role of PCNA K164 ubiquitination (Ub-PCNA) in TLS, it has remained unclear how this PCNA modification regulates the Pol exchange process. One view that has received considerable attention is that in addition to their binding of PCNA at the interdomain connector loop (IDCL) via their PIP domain, the TLS Pols bind to the Ub moiety on PCNA, and that both of these PCNA-binding modes are necessary for TLS (16). The identification of Ub-binding domains (UBDs) in TLS Pols and the finding that mutations in UBDs inactivate their function in lesion bypass have added support to this idea (16–18).
Similar to yPolη, human Polη (hPolη) harbors a PIP domain, Q T L ES F F, from residues 702–708, which is characterized by the conserved hydrophobic residues (underlined). Mutational inactivation of this PIP domain by changing the 2 F residues to As adversely affects the physical binding of hPolη to PCNA in vitro and impairs the enhanced DNA synthetic activity observed in the presence of PCNA, replication factor C (RFC), and replication protein A (RPA) (8). However, even though the hPolη PIP mutation conferred a considerable reduction in its ability to physically and functionally interact with PCNA, the mutant protein still retained residual PCNA binding ability (8). By contrast, the mutational inactivation of the yPolη PIP domain causes a complete loss in its ability to physically and functionally interact with PCNA (7). Also, whereas the PIP mutation in yPolη abolishes its ability to function in TLS in vivo (7), the inactivation of the hPolη PIP domain does not confer the same high degree of impairment in Polη function in human cells (16, 19). Rather, the UV survival of XP-V cells is enhanced upon transfection with the PIP mutant of hPolη, albeit not to WT levels (16).
The Ub-binding zinc finger (UBZ) domain of hPolη binds Ub (16). Because mutational inactivation of the Ub-binding ability of this domain results in defects in Polη's function in TLS in vivo, the inference has been drawn that the binding of the Ub moiety on PCNA via the UBZ domain is a prerequisite for hPolη's ability to access PCNA (16). In particular, the observation that the C638A mutation in the hPolη UBZ domain confers a much higher degree of UV sensitivity than the PIP mutation has even suggested that the binding of the Ub moiety on PCNA via its UBZ domain is more important for the targeting of hPolη to PCNA than is its binding to the PCNA IDCL domain via the PIP domain (16).
In the studies reported here, we analyze the contributions that the PIP and UBZ domains of hPolη make to its ability to bind PCNA and carry out TLS. Here, we identify an additional PIP motif in hPolη and show that in the absence of the previously identified C-terminal PIP domain, which we now designate as PIP2, the newly identified PIP1 domain can promote the PCNA binding of hPolη. Further, we provide evidence that the binding of PCNA via its PIP domain is essential for hPolη's ability to function in TLS in human cells, whereas the direct binding of the K164-linked Ub moiety on PCNA is not required for Polη function. We discuss the implications of various observations we report here and suggest that the Ub-binding ability of the hPolη UBZ domain has no direct bearing on Pol exchange and TLS.
Results
Identification of an Additional PCNA-Binding PIP Motif in hPolη.
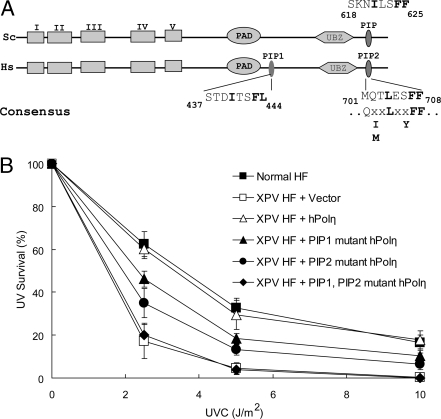
Although mutational inactivation of the PIP domain in yPolη abrogates its ability to physically and functionally interact with PCNA and carry out TLS in yeast cells (7), mutational inactivation of the hPolη PIP domain does not confer the same high degree of defectiveness in its ability to interact with PCNA and perform TLS in human cells (8). The observation that residual PCNA-binding ability is still retained in the hPolη PIP mutant protein raised the possibility that hPolη harbors an additional PIP domain that can promote PCNA binding. In support of such a possibility, we noted the presence of an additional PIP-like motif in hPolη that lies between residues 437 and 444, just C-terminal to the polymerase-associated domain (PAD) region (Fig. 1A), and in the studies described below we show that this motif does, in fact, function in PCNA binding. We have named this motif PIP1 and the C-terminal motif that lies between residues 701–708 PIP2 (Fig. 1A).
Fig. 1.
Requirement of PCNA-binding PIP domains of hPolη for its in vivo function in TLS. (A) Alignment of PIP domains of yeast (Sc) and human (Hs) Polη. In addition to the C-terminal PIP domain present in both yPolη and hPolη, hPolη harbors a PIP domain that is located just after the PAD domain. The highly conserved hydrophobic residues of the PIP domain are shown in bold. The positions of the 5 conserved Pol domains (I–V), the PAD domain, and the UBZ domain are also indicated. (B) Effects of PIP1 and PIP2 mutations on the UV sensitivity of hPolη. The survival of XPV cells transfected with different Polη constructs was examined after exposure to different doses of UV irradiation and incubation on growth media in the presence of 1 mM caffeine.
To test the significance of the PIP1 motif in Polη function, we changed the F443, L444 residues to alanines (F443A, L444A) and examined the UV sensitivity of XPV cells transfected with the plasmid carrying this mutant Polη. As shown in Fig. 1B, the ability of the PIP1 mutation to confer WT levels of UV resistance to XPV cells was significantly impaired. To determine whether the PIP1 and PIP2 domains could substitute for one another in hPolη function in TLS in vivo, we compared the effects of the PIP1 (F443A, L444A) and PIP2 (F707A, F708A) mutations alone with that of the PIP1 PIP2 double mutation where both domains were inactivated. Interestingly, we find that whereas the PIP1 or PIP2 mutation confers an intermediate level of UV resistance to XPV cells, the PIP1, PIP2 double mutant is completely defective in imparting UV resistance to XPV cells. These observations suggested that the PIP1 and PIP2 domains can substitute for one another and that the mutational alteration of both of these domains causes a complete loss in hPolη's ability to function in TLS in human cells.
Binding of PCNA by hPolη Via Its PIP Domain is Necessary for the Enhancement of its DNA Synthetic Activity.
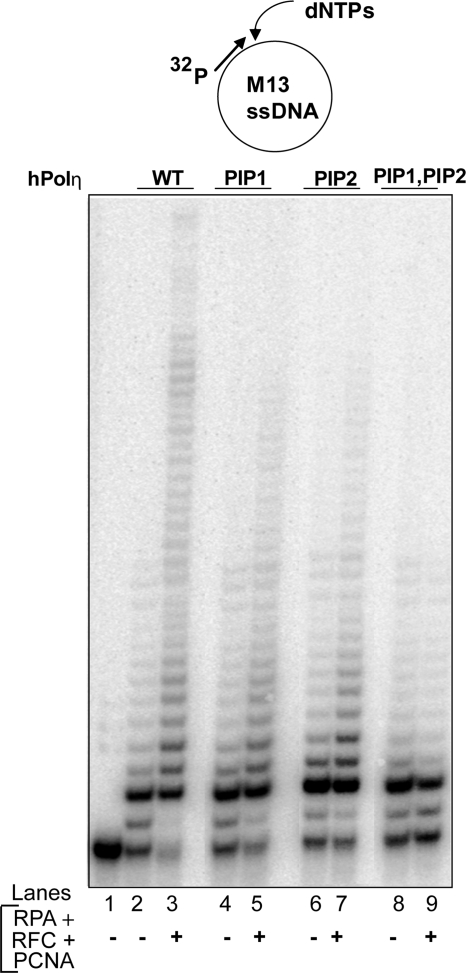
Next, we examined whether the F443A, L444A mutations in the PIP1 domain adversely affect the PCNA-dependent stimulation of hPolη DNA synthetic activity. For this purpose, we examined DNA synthesis by hPolη in the presence of PCNA, RFC, and RPA by using single-stranded M13 circular DNA primed with a 5′ 32P-labeled oligomer at a unique site. As we have shown before (8), the DNA synthetic activity of hPolη is enhanced markedly upon the addition of PCNA, RFC, and RPA (Fig. 2, lanes 2 and 3). An enhancement of activity also occurs with the PIP1 and PIP2 mutant proteins but the degree of enhancement is somewhat reduced compared with that for WT hPolη (Fig. 2, lanes 4–7). Importantly, hPolη bearing mutations in both the PIP1 and PIP2 domains shows no evidence of enhancement of DNA synthetic activity in the presence of PCNA, RFC, and RPA (Fig. 2, lanes 8 and 9). We conclude from these observations that hPolη has 2 PCNA binding PIP domains that can functionally substitute for one another and that in the absence of both of these domains the ability of hPolη to bind PCNA on DNA and the consequent stimulation of its DNA synthetic activity is abolished.
Fig. 2.
Requirement of PIP domains in hPolη for stimulation of its DNA synthetic activity by PCNA. DNA synthesis by WT hPolη (lanes 2 and 3), PIP1 mutant hPolη (lanes 4 and 5), PIP2 mutant hPolη (lanes 6 and 7), or PIP1, PIP2 mutant hPolη (lanes 8 and 9) protein (1 nM each) was examined on M13 circular DNA annealed to a radiolabeled primer (5 nM) in the absence or presence of PCNA (50 ng), RFC (15 ng), and RPA (200 ng). Lane 1, DNA substrate with no hPolη added.
Requirement of PCNA-Binding PIP Domains for Accumulation of hPolη in Replication Foci.
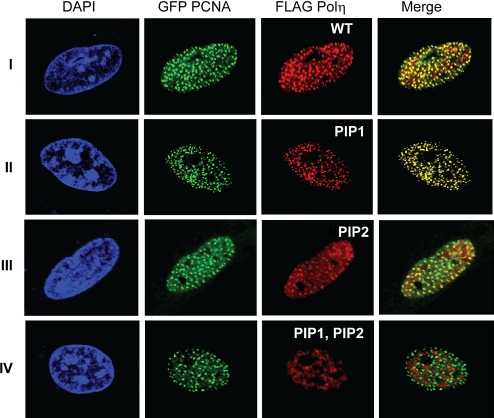
hPolη colocalizes with PCNA in replication foci, and the level of hPolη accumulation in foci is greatly increased in UV-irradiated human cells (20). To evaluate the contribution of the PIP1 and PIP2 domains to the accumulation of hPolη in foci, we cotransfected MRC5 fibroblasts with WT GFP PCNA and with WT or PIP1 and/or PIP2 FLAG - Polη. As shown in Fig. 3I, WT Polη (stained red) and PCNA (stained green) both accumulate in foci in UV-irradiated human cells, and a great majority of hPolη foci colocalize with PCNA as indicated from the appearance of yellow foci. Interestingly, hPolη foci formation or its colocalization with PCNA are not affected by the F443A, L444A PIP1 (Fig. 3II) or the F707A, F708A PIP2 (Fig. 3III) mutations. Importantly, hPolη protein bearing mutations in both the PIP1 and PIP2 domains did not accumulate in replication foci, and there was no evidence for the colocalization of this mutant hPolη with PCNA as indicated from the absence of any yellow foci (Fig. 3IV). We conclude from these observations that the coincident accumulation of hPolη with PCNA in replication foci requires that the ability to bind PCNA via one of its PIP domains be retained.
Fig. 3.
PCNA binding by PIP domains is essential for accumulation of hPolη in replication foci. MRC5 cells were cotransfected with WT GFP PCNA and WT FLAG Polη (I) or PIP1 (F443A, L444A) (II), PIP2 (F707A, F708A) (III), or PIP1, PIP2 (F443A, L444A, F707A, F708A) (IV) mutant FLAG Polη. Twenty-four hours after transfection, cells were UV-irradiated with 40 J/m2 followed by incubation for 6 h before fixation. FLAG-tagged hPolη proteins were imunostained with anti-FLAG mAb. (Magnification: 600×.)
Mutational Analyses of the hPolη UBZ Domain.
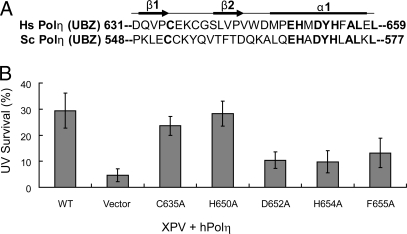
Solution structure of the hPolη UBZ domain (21) has indicated that it adopts a classical C2H2 zinc finger structure characterized by a ββα fold in which residues 632–634 and 641–643 form 2 short antiparallel β-strands and residues 647–658 form an α-helix (Fig. 4A). In the structure, the α-helix packs against the β-strands with a zinc ion coordinated by the 2 cysteines located on the fingertip made by the 2 β-strands and the 2 histidines located on the α-helix. The UBZ domain binds Ub via the exposed surface of the α-helix, and the conserved residues such as H650, D652, Y653, H654, F655, and others are directly involved in Ub binding (21).
Fig. 4.
Role of UBZ domain in hPolη function in vivo. (A) Sequence alignment of human (Hs) and yeast (Sc) Polη UBZ domains. Highly conserved residues are shown in bold. The positions of 2 short β-sheets and the α-helix as determined from the solution structure of hPolη UBZ are indicated above the sequences. (B) Complementation of the UV sensitivity of XPV cells by hPolη UBZ mutations. The survival of XPV cells transfected with WT or UBZ mutant Polη constructs was examined after exposure to 5 J/m2 of UV irradiation followed by incubation on growth media in the presence of 1 mM caffeine.
In a previous study, the C638A UBZ mutation of hPoln was shown to be highly defective in its ability to confer UV resistance to XPV cells, and the defect imparted to UV survival by this mutation was greater than that seen for the PIP mutation that lies in the C-terminal PIP2 domain (Fig. 1A) (16). In addition, the D652A mutation of the UBZ domain was shown to inactivate the formation of hPolη foci in UV-irradiated cells (16). From such observations and the fact that hPolη can be preferentially immunoprecipitated from human cells in association with Ub-PCNA (22), the inference has been drawn that the direct binding of Ub moiety on PCNA via its UBZ domain is important for hPolη's ability to bind PCNA and carry out its function in TLS (16).
Because a role for the hPolη UBZ domain in the direct binding of K164-linked Ub on PCNA has been inferred from analysis of only a few UBZ mutations, it becomes important to carry out a more extensive mutational analyses of the UBZ domain of hPolη to determine whether the loss of Ub binding by the UBZ domain is strictly associated with a defect in hPolη's ability to function in TLS or whether the inability to bind Ub and the ability to function in TLS are mutationally separable. For this reason, we constructed the C635A, H650A, and H654A mutations that lie in the C2H2 zinc finger domain and are completely conserved in all of the UBZ domains. In addition, we constructed 2 mutations, D652A and F655A, in the conserved residues in the α-helix. These residues in the zinc finger and the α-helix all are required for Ub binding by the UBZ.
Effects of UBZ Mutations on hPolη Function in Vivo.
To evaluate the importance of UBZ domain in hPolη function in TLS in human cells, we transfected the various UBZ mutant Polη in XPV cells and determined their ability to confer UV resistance. Of the 3 mutations in the C2H2 zinc finger domain examined, we found that they differ in the level of UV resistance they impart to XPV cells. Whereas the H650A mutant Polη restored WT levels of UV resistance and the C635A mutant also conferred near to WT level of UV resistance, the H654A mutation was greatly impaired in its ability to restore UV resistance to XPV cells (Fig. 4B). The D652A and F655A mutations that lie in the α-helix were also impaired in their ability to confer WT levels of UV resistance to XPV cells (Fig. 4B). From these observations we conclude that a loss in the Ub-binding ability of the UBZ domain is not concomitantly associated with a defect in hPolη's ability to function in TLS in human cells.
Mutations in the UBZ Domain Have No Adverse Effect on the Stimulation of the DNA Synthetic Activity of hPolη by PCNA or Ub-PCNA.
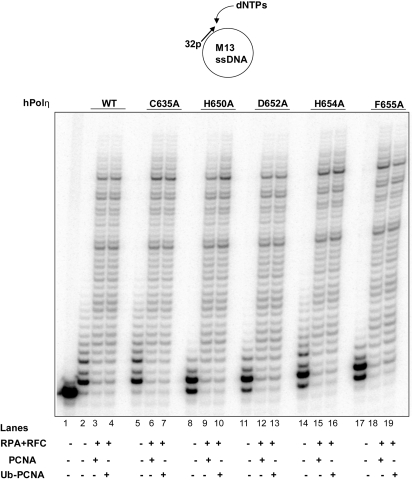
The proposal that TLS Pols bind PCNA at the IDCL via their PIP domain and bind the K164-linked Ub moiety via their UBD implies that Ub-PCNA would be much more effective than unmodified PCNA in binding to TLS Pols and that mutational inactivation of the UBZ domain would impair their preferential binding to Ub-PCNA. In studies with yPolη, we have found no evidence for greater stimulation of activity by Ub-PCNA over unmodified PCNA (23), and in another study, only a small additional increase in the activation of yPolη was reported with Ub-PCNA than with PCNA (24). Even though no further stimulation of yPolη activity could be observed with Ub-PCNA, we could not exclude the possibility that the binding of the Ub moiety on PCNA via its UBZ domain plays a more important role in effecting the PCNA binding of hPolη than yPolη. To verify that Ub-PCNA confers no further stimulation of hPolη activity over PCNA, we examined its activity in the presence of unmodified PCNA and K164-linked monoubiquitinated PCNA that we have reconstituted by using the purified human Rad6–Rad18 enzyme (25). As shown in Fig. 5, we observed no further stimulation of hPolη activity with Ub-PCNA over that with PCNA (compare lanes 3 and 4). And, importantly, none of the UBZ mutations, C635A, H650A, D652A, H654A, or F655A, were found to have any adverse effect on hPolη DNA synthetic activity with PCNA or Ub-PCNA (Fig. 5, lanes 5–19).
Fig. 5.
hPolη UBZ mutant proteins are not defective in functional interaction with PCNA. DNA synthesis by WT hPolη (lanes 2–4) or the UBZ mutant hPolη proteins C635A (lanes 5–7), H650A (lanes 8–10), D652A (lanes 11–13), H654A (lanes 14–16), and F655A (lanes 17–19) (1 nM each) was examined by using M13 circular DNA annealed to a radiolabeled primer (5 nM) in the presence of each of the 4 dNTPs under standard reaction conditions. As indicated, the reactions were carried out in the presence or absence of PCNA or Ub-PCNA (50 ng), RFC (15 ng), and RPA (200 ng). Lane 1, DNA substrate with no hPolη added.
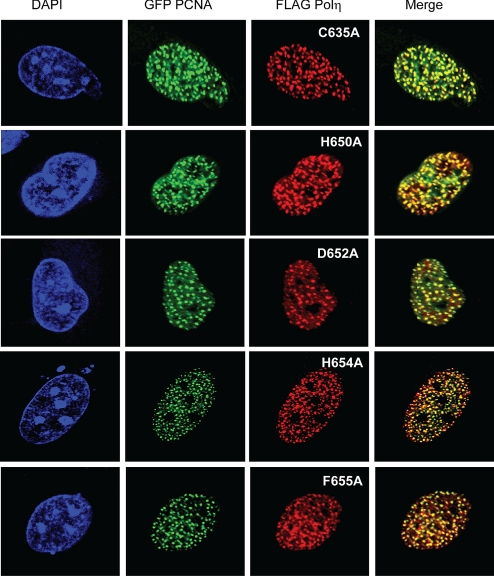
Accumulation of hPolη UBZ Mutant Proteins in Replication Foci.
Our findings that the inactivation of PCNA binding by the PIP1, PIP2 mutation results in a complete loss in hPolη's ability to accumulate in replication foci with PCNA and that this PIP mutation abolishes hPolη function in TLS in human cells have indicated that binding to PCNA via its PIP domain is critically important for Polη's function in vivo. For the UBZ mutations, however, we find that the stimulation of hPolη activity by PCNA or Ub-PCNA is not affected by any of them, indicating that they have no adverse effect on the binding of hPolη to PCNA or Ub-PCNA. Because the inability to bind PCNA in the PIP1, PIP2 hPolη mutant protein results in a complete loss of its capacity to accumulate in foci, one would have expected that the retention of PCNA-binding ability in the UBZ mutant proteins would enable them to accumulate in foci. The reported lack of accumulation of D652A UBZ mutant hPolη in replication foci, however, runs counter to this proposal (16). For this reason, we decided to examine the effects of different UBZ mutations, including that of the D652A mutation, on the accumulation of hPolη in replication foci in UV-irradiated human cells. We found that the abilities to accumulate in replication foci and to colocalize with PCNA are retained by the various UBZ mutations that we studied (Fig. 6). Our observations that the C635A, H650A, and H654A mutations in the C2H2 zinc finger do not affect foci formation in UV-irradiated human cells, and the previously reported observation that the H654A mutation in this zinc finger affects foci formation in UV-irradiated cells only somewhat (≈2-fold) (18), imply that the mutational inactivation of the zinc finger of the UBZ domain and the consequent inactivation of its ability to bind Ub have no significant impact on the ability of hPolη to accumulate in foci and colocalize with PCNA. Our analyses with the D652A and F655A mutations, which also inactivate the Ub-binding ability of hPolη but not the colocalization with PCNA, add further support to our conclusion that foci formation and colocalization with PCNA can still occur in hPolη UBZ mutant proteins that have lost the ability to bind Ub.
Fig. 6.
UBZ mutant hPolη proteins accumulate in replication foci. MRC5 cells were cotransfected with WT GFP PCNA and UBZ mutant FLAG Polη as indicated. Twenty-four hours after transfection, cells were UV-irradiated with 40 J/m2 followed by incubation for 6 h before fixation. FLAG-tagged proteins were immunostained with anti-FLAG mAb. For the accumulation of WT hPolη, see Fig. 3. (Magnification: 600×.)
Discussion
Based on studies with hPolη, the proposal has been made that hPolη and other Y family TLS Pols bind PCNA at 2 sites, the IDCL, which they bind by their PIP domain, and K164-linked Ub, which they bind via their UBZ domain, and that the binding of the Ub moiety on PCNA by the UBZ domain is essential for Polη's localization in replication foci and its function in TLS in vivo (16, 22). The key point of this model is that it assigns a function to the Ub moiety on PCNA in the direct binding of hPolη and other TLS Pols and postulates that this binding is mediated by the UBD of the TLS Pol. An alternative possibility, however, is that hPolη and other TLS Pols bind PCNA only at the IDCL and the Ub moiety on PCNA plays no significant role in binding directly to the TLS Pol; rather, when the progressively moving replicative Pol stalls at a DNA lesion, the K164-linked Ub moiety effects a conformational change in PCNA, which enables the TLS Pol to bind PCNA at the IDCL and take over synthesis from the replicative Pol. Hence, the indispensable role of the Ub moiety on PCNA would then be in promoting the Pol exchange process. We discuss the implications of our observations with the hPolη PIP and UBZ domains for these alternate models.
Although genetic studies with yPolη have indicated an indispensable role of the PIP domain in PCNA binding (7), genetic studies with the hPolη PIP domain, which resembles the yPolη PIP domain and is similarly localized in the C terminus, have shown that the hPolη PIP mutant still retains a considerable ability to function in TLS in human cells (16, 19)., Here, we provide evidence that the reason for the lack of an absolute requirement of the hPolη PIP domain is the presence of an additional PIP domain in this protein. In addition to the previously identified C-terminal PIP domain, hPolη harbors another PIP domain situated just after the PAD domain; we refer to these domains as PIP2 and PIP1, respectively. The mutational inactivation of either domain results in a partial deficiency in hPolη's proficiency for PCNA binding and carrying out its function in TLS, and inactivation of both PIP domains completely abrogates PCNA-mediated stimulation of hPolη activity and its ability to function in TLS in vivo. Also, although the PIP1 or PIP2 mutant protein shows evidence of colocalization with PCNA in replication foci in UV-irradiated cells, the accumulation of hPolη in replication foci and its coincident localization with PCNA do not occur for the PIP1, PIP2 double mutant. From these observations we conclude that the PIP1 and PIP2 domains can functionally substitute for one another. An important implication of these results is that hPolη's binding to PCNA via its PIP domain is essential for its ability to access PCNA and carry out TLS in human cells.
To evaluate the role of the UBZ domain in hPolη's ability to access PCNA and function in TLS, we examined the effects of mutations in 5 different residues that are highly conserved in the UBZ domain. We find that not all of the UBZ mutations affect the proficiency of hPolη function in TLS in vivo, as indicated from the ability of C635A and H654A to restore an almost WT level of UV resistance to XPV cells. Importantly, we show that all of the UBZ mutant proteins can function with PCNA or Ub-PCNA and that both forms of PCNA stimulate their DNA synthetic activity to the same degree as seen for WT hPolη. In agreement with this observation, we find that in UV-irradiated human cells, all of the UBZ mutant proteins show evidence of accumulation in replication foci and colocalization with PCNA. We conclude from these observations that the ability of hPolη to bind PCNA is not affected by the K164-linked Ub moiety on PCNA or by mutational inactivation of the UBZ domain, and consequently, the ability to colocalize with PCNA in foci is retained in the UBZ mutant proteins.
Our proposal that the UBZ domain is not required for hPolη's ability to access PCNA on the replication fork raises the question of why mutations such as D652A, H654A, and F655A adversely affect hPolη's ability to complement the UV sensitivity of XPV cells. We suggest that these mutations affect hPolη function in ways unrelated to PCNA binding; for example, they may directly or indirectly affect the proficiency of hPolη to physically interact with proteins bound at the replication fork, which are important for the coordination of hPolη function in TLS with the replicative Pol.
We draw the following conclusions from our study. (i) hPolη binds PCNA via its PIP domain and this PCNA binding mode is essential for hPolη's ability to function in TLS in vivo. (ii) The K164-linked Ub moiety on PCNA does not increase the proficiency of hPolη for PCNA binding. (iii) Because mutational inactivation of the UBZ domain has no adverse effect on the stimulation of the DNA synthetic activity of hPolη with PCNA or Ub-PCNA and does not impair the colocalization of hPolη with PCNA in replication foci in UV-irradiated cells, the UBZ domain makes no significant contribution to hPolη's PCNA binding. (iv) And, we surmise that the adverse effect of some UBZ mutations on hPolη function in TLS in vivo as indicated from the lack of their ability of complement the UV sensitivity of XPV cells results from effects unrelated to PCNA binding.
Our conclusion that the binding of hPolη to PCNA requires only the PIP domain and that the Ub moiety on PCNA is not directly involved in the binding to hPolη implies that the indispensable role of K164-linked Ub on PCNA concerns some other aspect of TLS. We suggest that the Ub moiety effects a conformational change on PCNA that enables Polη to access the IDCL in PCNA and take over synthesis when the replicative Pol stalls at a DNA lesion. Thus, in this proposal Rad6–Rad18-mediated PCNA ubiquitination effects the Pol exchange process at the lesion site. Support for this idea is indicated from recent biochemical experiments in which processive synthesis by yPolδ was carried out on primed single-stranded M13 circular DNA in the presence of PCNA or K164-linked monoubiquitinated PCNA together with RFC and RPA (26). When processively moving yPolδ was stalled by nucleotide omission, yPolη could take over synthesis from yPolδ only in the presence of Ub-PCNA and not when unmodified PCNA was used, and the PIP domain of yPolη was necessary for this Pol exchange to occur. Importantly, the exchange with yPolη could occur only if processively moving yPolδ was stalled, whereas in the absence of stalling, yPolη was unable to take over synthesis from yPolδ even in the presence of Ub-PCNA (26). From these biochemical studies we can also infer that Ub-PCNA has no adverse effect on processive synthesis by Polδ and that only when the processively moving Polδ is stalled does Polδ-bound Ub-PCNA undergo a conformational change that is conducive for PCNA binding by Polη and its takeover of DNA synthesis from the replicative Pol.
Methods
Generation of hPolη Mutations.
The generation of the hPolη F707A, F708A mutations in the PIP2 domain has been described (8). Site-directed mutagenesis was performed on WT hPolη to generate mutations in the PIP1 domain (F443A, L444A) and on PIP2 mutant hPolη to generate mutations in the PIP1 domain (F443A, L444A, F707A, F708A), yielding PIP1, PIP2 mutant hPolη. UBZ site-specific mutations (C635A, H650A, D652A, H654A, and F655A) in the UBZ domain were generated in WT hPolη.
Proteins.
Human PCNA, RFC, and RPA were purified as described. WT and mutant hPolη proteins fused to GST were purified as described, and the GST portion was removed by treatment with PreScission protease.
DNA Pol Assays.
The circular DNA substrate used for the DNA synthesis studies was a 7.2-kb M13mp18 ssDNA primed with a radiolabeled 38-nt oligomer (LP273, 5′-GGG TTT TCC CAG TCA CGA CGT TGT AAA ACG ACG GCC AG-3′). The standard DNA Pol reaction mixture contained 10 nM DNA substrate, 40 mM Tris·HCl (pH 7.5), 8 mM MgCl2, 150 mM NaCl, 1 mM DTT, 100 μg of BSA/ml, 500 μM ATP, and 100 μM each dGTP, dATP, dTTP, and dCTP. The reaction was carried out in the absence or presence of PCNA or Ub-PCNA (50 ng), RFC (15 ng), and RPA (200 ng) at 37 °C for 10 min after the addition of WT or mutant hPolη (1 nM) protein to the reaction mixture. The reaction was stopped by the addition of loading buffer (40 μL) containing EDTA (20 nM), 95% formamide, 0.3% bromophenol blue, and 0.3% cyanol blue. The reaction products were resolved on 10% polyacrylamide gels containing 8 M urea. The reaction products were visualized with a Molecular Dynamics STORM PhosphorImager.
Stable Expression of WT and Mutant hPolη in XPV Cells.
WT and mutant hPolη genes were subcloned into pSilencer 4.1-CMV puro vector (Ambion). The vectors were transfected into XPV (XP30R0) human fibroblasts (HF) by Lipofectamine 2000 reagent (Invitrogen), and stably expressing cell lines were isolated by using puromycin selection.
UV Cytotoxicity Assays.
Normal and transfected XPV (XP30R0) HF cells were plated in 6-well plates with 50% confluence. After 24 h of incubation, cells were exposed to UV. For UV irradiation, cells were washed with PBS buffer and irradiated with various doses (0–10J/m2) of UVC light in the presence of PBS buffer. After irradiation, fresh growth medium containing 1 mM caffeine was added to cells. Cells were incubated for an additional 48 h after UV irradiation. The UV cytotoxicity was determined by the MTT assay (Promega) following the manufacturers' manual. Briefly, 200 μL of MTT assay solutions were added to each well and incubated for 30 min. Cell viability was determined by measuring the OD at 490 nm. Four independent experiments were carried out for WT and each mutant hPolη.
Immunocytochemistry and Fluorescence Microscopy.
WT and mutant hPolη genes were cloned into p3X Flag CMV7.1 vector (Sigma) and the construct, GFP-PCNAL2, for expressing GFP-PCNA has been described (27). SV40-transformed MRC5 HF (AG10076) cells grown on coverslips were transfected with different plasmid constructs, and 24 h after transfection, cells were UV-irradiated at 40 J/m2 followed by incubation for 6 h. Cells were then rinsed in PBS and fixed in cold methanol for 10 min. Cells were subsequently washed twice with PBS and blocked with PBS + 1% BSA for 20 min followed by incubation with primary antibody diluted in 1% BSA containing PBS for 1 h. Coverslips were washed 5 times with PBS and incubated with Cy3-conjugated anti-mouse antibody again diluted in PBS containing 1% BSA. After washing 5 times with PBS, coverslips were mounted with mounting media containing glycerol and DAPI. Stained cells were analyzed and photographed with an Olympus confocal laser scanning microscope. Antibodies used were mouse anti-FLAG mAb 1:400, M2 (Sigma F3165) and anti-mouse Cy3 1:1,000 (Sigma C2181).
Acknowledgments.
This work was supported by National Institutes of Health Grant ES012411 (to L.P.), National Cancer Institute Training Grant T32CA117834 (to J.-H.Y.), a Wellcome Trust International Senior Research Fellowship (to I.U.), and Howard Hughes Medical Institute Grant 55005612 (to L.H.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 2.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Polη. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 3.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 4.Lone S, et al. Human DNA polymerase κ encircles DNA: Implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 5.Masutani C, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 6.Trincao J, et al. Structure of the catalytic core of S. cerevisiae DNA polymerase η: Implications for translesion DNA synthesis. Mol Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 7.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 8.Haracska L, et al. Physical and functional interactions of human DNA polymerase η with PCNA. Mol Cell Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haracska L, et al. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc Natl Acad Sci USA. 2001;98:14256–14261. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haracska L, et al. Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol Cell Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: A potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 12.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin-conjugating, DNA-binding, and ATP hydrolytic activities. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 13.Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 14.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 16.Bienko M, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 17.Guo C, et al. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plosky BS, et al. Controlling the subcellular localization of DNA polymerases ι and η via interactions with ubiquitin. EMBO J. 2006;25:2847–2855. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gueranger Q, et al. Role of DNA polymerases η, ι, and ζ in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair. 2008;7:1551–1562. doi: 10.1016/j.dnarep.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Kannouche P, et al. Domain structure, localization, and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bomar MG, Pai M-T, Tzeng S-R, Li SS-C, Zhou P. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase η. EMBO Rep. 2007;8:247–251. doi: 10.1038/sj.embor.7400901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 23.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc Natl Acad Sci USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garg P, Burgers PM. Ubiquitinated proliferatinag cell nuclear antigen activates translesion DNA polymerase η and REV1. Proc Natl Acad Sci USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Unk I, et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci USA. 2006;103:18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuang Z, et al. Regulation of polymerase exchange between Polη and Polδ by monoubiquitinatiion of PCNA and the movement of DNA polymerase holoenzyme. Proc Natl Acad Sci USA. 2008;105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leonhardt H, et al. Dynamics of DNA replication factories in living cells. J Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]