Abstract
A fundamental biological question is what forces shape the guanine plus cytosine (GC) content of genomes. We studied the specificity and rate of different mutational biases in real time in the bacterium Salmonella typhimurium under conditions of strongly reduced selection and in the absence of the major DNA repair systems involved in repairing common spontaneous mutations caused by oxidized and deaminated DNA bases. The mutational spectrum was determined by whole-genome sequencing of two S. typhimurium mutants that were serially passaged for 5,000 generations. Analysis of 943 identified base pair substitutions showed that 91% were GC-to-TA transversions and 7% were GC-to-AT transitions, commonly associated with 8-oxoG- and deamination-induced damages, respectively. Other types of base pair substitutions constituted the remaining 2% of the mutations. With regard to mutational biases, there was a significant increase in C-to-T transitions on the nontranscribed strand, and for highly expressed genes, C/G-to-T mutations were more common than expected; however, no significant mutational bias with regard to leading and lagging strands of replication or chromosome position were found. These results suggest that, based on the experimentally determined mutational rates and specificities, a bacterial genome lacking the relevant DNA repair systems could, as a consequence of these underlying mutational biases, very rapidly reduce its GC content.
Keywords: dna repair, experimental evolution, gc bias, mutation spectra, Salmonella typhimurium
A central question in evolutionary genomics is what mechanisms cause the variation observed in DNA base composition between and within genomes and how rapidly and by what mechanisms these biases might change in response to, for example, altered ecology and genetic constitution of the organism. The large range in guanine plus cytosine (GC) content among bacterial species is well established, varying between at least 17% and 75% GC, with an even larger variation in the third codon position (1). Within a genome, GC content usually is quite homogeneous and has a strong phylogenetic signal, but despite this overall homogeneity, there frequently exist strand-specific biases between the two strands of DNA such that the average nucleotide composition deviates from the theoretically expected A = T and G = C within each strand. Thus, most bacterial chromosomes are relatively strongly enriched in G over C and in T over A and are slightly depleted in G+C in weakly selected positions in the leading strand compared with in the lagging strand (2). In addition, highly transcribed genes appear to show a G and T skew on the nontranscribed strand compared with poorly transcribed genes (3).
Although the causes of these biases remain unclear, the biases can be expected to arise at least at three different levels. First, there might exist an underlying bias in the mutation pressure caused by unavoidable spontaneous DNA damage, such as deamination of C→U and 5-meC→T (4, 5) or oxidation of G to form 7,8-dihydro-8-oxoG (8-oxoG) (6). An example of such a bias is the deamination of C and 5-meC, which occurs more rapidly in single-stranded DNA than in double-stranded DNA in vitro (7). During replication and transcription, the leading and nontranscribed strands are in a single-strand state for a longer time than the lagging and transcribed strands, making them more prone to deamination (8, 9). These underlying spontaneous mutational pressures in turn might be modulated by the specificity and efficiency of the different repair systems that remove deaminated and oxidated DNA damages. With regard to deamination damages, two uracil glycosylases, encoded by the ung and mug genes, remove uracil from DNA, leaving an abasic site that can be restored by repair DNA synthesis (10–13), and the Vsr endonuclease encoded by the vsr gene initiates the very short patch repair system that removes thymine in a G-T mispair (14). The common oxidated base 8-oxo-G inserted into DNA is removed by two highly conserved enzymes, MutM and MutY. MutM is a glycosylase that excises 8-oxoG paired with C, thereby initiating base excision repair that restores the GC base pair. Failure to do so before replication allows 8-oxoG to mispair with A. This mispair is a substrate for another glycosylase, MutY, that removes adenine and allows subsequent DNA repair synthesis (15). Mutants defective in mutM or mutY have an elevated rate of GC-to-TA transversion mutations (15). Finally, a GC bias could be introduced by selection. Several theories emphasizing the role of selection in affecting genomic GC content have been suggested (16–21). Not surprisingly, given the complexities of bacterial ecology, none of these theories provides a universal explanation regarding the nature of those putative selective forces to explain the advantage of a high or low GC content.
A central and unanswered question in this context is how genomic GC content might change in response to alterations in the genetic capacity of the cell to repair different types of mutations. If an organism loses its ability to repair deamination and oxidative damages, how rapidly can the genomic GC content change, and what are the dominant mutational biases? These questions previously have been addressed primarily by analysis of sequenced genomes, making it difficult to distinguish between selection and neutral processes. Furthermore, experimental studies typically have been limited by the use of single genes instead of genomes or by experimental conditions that do not fully exclude the influence of selection. To avoid these limitations, we chose an experimental evolution approach in which mutational pressures were assessed in real time under conditions of strongly reduced selection and in the absence of the major DNA repair systems for deaminated and oxidized bases. Furthermore, to avoid any local biases in mutational pattern, we analyzed the mutational spectrum at the genome level by high-coverage DNA sequencing of two complete Salmonella typhimurium genomes. This approach allowed us to examine the presence of local and global mutational biases and to experimentally determine both the rate and the nature of the underlying mutational biases.
Results
Experimental Rationale.
To address the question of mutational biases, random point mutations were allowed to accumulate in the genomes of a wild-type and four different S. typhimurium LT2 mutant strains constructed using linear transformation and phage P22 transduction (see Materials and Methods). Mutation accumulation was achieved experimentally by serially passaging different mutant bacteria with DNA repair system defects by repeated streaking on rich agar plates to generate new colonies initiated by a single cell. For each of the five strains, 12 lineages were serially passaged each day for 200 days. During each serial passage, the bacterial population expanded from 1 cell to 108 cells in a colony, representing ≈5,000 generations of growth for the entire experiment (200 serial passages × 25 generations/passage). The repeated one-cell bottleneck increased genetic drift and allowed all types of mutations to come to fixation with high and similar probabilities. The genotypes of the four mutants tested were ung−, ung− vsr−, mutM− mutY−, and ung− vsr− mug− mutM− mutY−, with the isogenic wild-type S. typhimurium LT2 used as a control. Because inactivation of ung, vsr, and mug results in increased GC-to-AT transitions (4, 5), and inactivation of mutM and mutY causes increased GC-to-TA transversions (15), it was possible to separate the effects of these repair genes in a single strain with all five genes inactivated. To confirm that the mutation rate did not change during the experiment, mutation rates to rifampicin resistance were measured for the ancestral strains and lineages after 200 cycles of serial passage (Fig. 1). No significant change in mutation rate was observed between the ancestral and evolved lineages, but one lineage of the quintuple mutant was excluded for further analysis because of its inability (for unknown reasons) to grow to high cell density in Luria-Bertani (LB) broth.
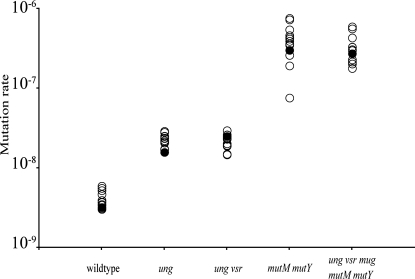
Fig. 1.
Mutation rates to rifampicin resistance for ancestral strains and lineages after 200 growth cycles. Closed circles represent the ancestral strains before mutation accumulation; open circles represent the evolved strains. Mutation rates were increased 6-fold in the ung- and ung- vsr- mutants and ≈100-fold in the mutM- mutY- and ung- vsr- mug- mutM- mutY- mutants compared with the wild type (3 × 10−9), with no significant differences between ancestral and evolved strains.
DNA Sequence Analysis.
To identify any mutations found in the 4.95-Mbp S. typhimurium genome, independent lineages of serially passaged wild-type and mutant bacteria were analyzed by whole-genome DNA sequencing using the 454 technology. The genomes of two random lineages of the ung- vsr- mug- mutM- mutY- quintuple mutant evolved for 200 cycles were sequenced to 8.7× and 7.8× depths. Sequences were assembled into contigs of about 4.8 Mbp of the 4.95-Mbp S. typhimurium genome, including pSLT (97%). The genome of an evolved wild-type strain was sequenced at lower coverage (4.9× depth), to confirm that deletion of repair systems was the cause of the observed mutation accumulation. The contigs were compared with the published S. typhimurium LT2 genome sequence using genomic BLASTn, allowing identification of the position and nature of all mutations. Single base pair deletions and insertions were not counted, because sequence technology−dependent uncertainties in base calling associated with mononucleotide runs prevented exclusion of false positives. Before further analyses, the pSLT plasmid, repeats, and duplicated regions were removed, leaving 4.68 Mbp of the chromosome. Differences between the published genome sequence and the sequenced lineages were excluded from further analysis if identical mutations were found in at least two of the evolved lineages, including the evolved wild-type lineage. A total of 68 differences were removed [see supporting information (SI) Text for details]. This decreased the likelihood that differences between our ancestral strain of S. typhimurium and the sequenced reference strain were counted as mutations and also removed any possible mutations introduced during strain construction. BLASTx analyses of regions surrounding the mutations were performed to identify the mutated genes and the coding context of all mutations.
Mutational Spectra and Biases.
In the two sequenced ung, vsr, mug, mutM, and mutY genomes, a total of 943 base pair substitutions (BPSs) were found, of which 856 (91%) were GC-to-TA transversions and 65 (7%) were GC-to-AT transitions, likely caused by 8-oxoG and deamination, respectively. Other types of BPSs represented the remaining 22 (2%) of the mutations (Table 1). Among the mutations found, 96% had an error rate of <99.99%, which was only slightly lower than the fraction for the entire sequence, suggesting that sequencing errors had only a very limited influence on our results (see Materials and Methods). No apparent differences in the number and types of mutations between the two sequenced strains were found (Table S1). In the serially passaged wild-type control strain, 15 BPSs were found in the 4.31-Mb analyzed sequence, and no mutational bias toward GC-to-TA transversions or GC-to-AT transitions was noted (Table S1), suggesting that ≈98% of the mutations that accumulated during serial passage of the quintuple mutant strain resulted from deletion of the repair systems. To investigate the presence of various mutational biases, GC-to-TA transversions and GC-to-AT transitions were analyzed (Table 2). Surprisingly, no significant bias in terms of leading and lagging strands of replication with regard to either G-to-T or C-to-T mutations were found (Table 1). To determine any potential bias in chromosome position and potential presence of mutational hot spots, Kolmogorov-Smirnov tests were performed against a uniform distribution; no significant deviations were found (P = .45 for G-to-T, P = .14 for C-to-T). The distribution of mutations with regard to chromosomal location is shown in Fig. 2. Data for all of the mutations found, including type of mutation, genomic position, strand of replication and transcription, gene, protein, amino acid substitution, codon change and strain, are given in Table S2.
Table 1.
Mutational spectrum of base substitutions and calculated fixation rates
| Substitution | Mutations | Fixation rate per base pair per generation |
|---|---|---|
| A → C, T → G | 3 | 6.4 × 10−11 |
| A → G, T → C | 9 | 1.9 × 10−10 |
| A → T, T → A | 3 | 6.4 × 10−11 |
| C → A, G → T | 856 | 1.8 × 10−8 |
| G → A, C → T | 65 | 1.4 × 10−9 |
| C → G, G → C | 7 | 1.5 × 10−10 |
| All substitutions | 943 | 2.0 × 10−8 |
Table 2.
Substitution patterns of G → T and C → T mutations
| Substitution | Observed | Expected |
|---|---|---|
| Leading strand G → T | 445 | 447 |
| Lagging strand G → T | 411 | 409 |
| Leading strand C → T | 36 | 31 |
| Lagging strand C → T | 29 | 34 |
| Nontranscribed strand G → T | 424 | 421 |
| Transcribed strand G → T | 359 | 362 |
| Nontranscribed strand C → T | 39 | 28 |
| Transcribed strand C → T | 21 | 32 |
| Synonymous | 230 | 202 |
| Nonsynonymous | 557 | 583 |
| Nonsense | 49 | 52 |
| Intergenic | 85 | 83 |
The observed and expected (if random) numbers of the different substitutions types are shown. Data for the two strains separately are available in Table S1.
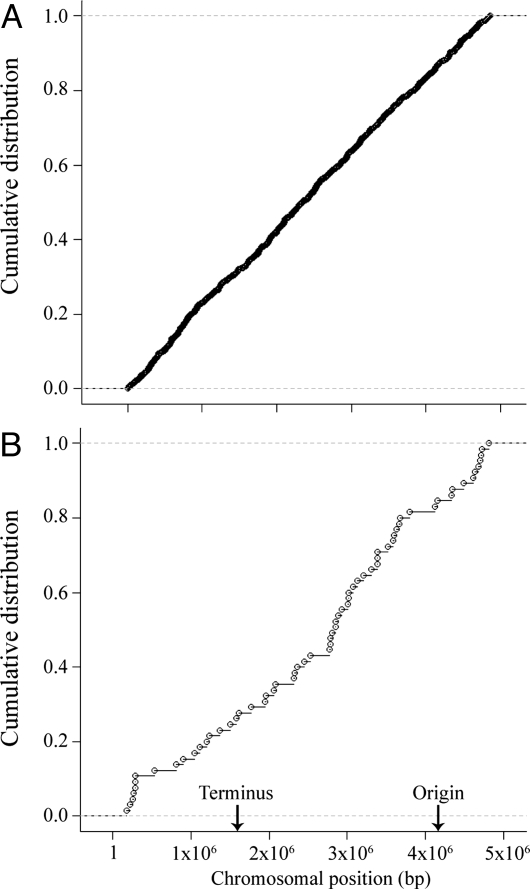
Fig. 2.
Empirical cumulative distribution functions of (A) GC-to-TA transversions and (B) GC-to-AT transitions. If mutations are distributed randomly with regard to chromosomal location, then a linear relationship is expected, with no deviations at the origin of replication (4.08 Mbp) or terminus (1.61 Mbp).
To identify any other potential biases in the mutational spectrum, the expected number of each codon change under a random distribution of mutations was calculated, considering the frequency of the native codon in the S. typhimurium genome. This gave 96 possible codon changes each for both transitions and transversions. This distribution was then compared with the observed codon changes to assess potential biases. The numbers of G-to-T mutations in the nontranscribed and transcribed strands were not significantly different from those that would be expected to occur by chance; however, of the 60 C-to-T transitions, either in coding regions or less than 40 base pairs upstream or downstream of coding regions, 39 were found on the nontranscribed strand and 21 were found on the transcribed strand. If the overrepresentation of G in the nontranscribed strand were taken into account, then 28 mutations in the nontranscribed strand and 32 mutations in the transcribed strand would be expected. Our data indicate a significant increase in C-to-T transitions on the nontranscribed strand (P < .05, Fisher's exact test).
A set of highly expressed genes with a codon adaptation index higher than 0.66 (Table S3) comprising 78 kilobase pairs (kbp) was extracted from the HEG database and used to investigate the influence of expression on mutation rates (22). A comparison of the 60 C-to-T transitions with the list of highly expressed genes revealed 7 mutations in those genes, representing 12% of the total mutations. The mutagenic target of 78 kbp represents only 1.7% of the analyzed sequence, and there were seven times as many C-to-T transitions as would be expected to occur by chance in the highly expressed genes, suggesting a significant increase (P < .05, Fisher's exact test). For G-to-T transversions, 42 of the 856 mutations in the coding regions were found in the highly expressed genes, compared with the expected 14 mutations, which also makes the increase in G-to-T transversions in these genes highly significant (P < .0001, Fisher's exact test).
No significant biases in terms of nonsynonymous, synonymous, or nonsense mutations were found for either transversions or transitions (Table 2). Mutations were no less likely to be found in essential genes than in the rest of the chromosome, based on 301 genes classified as essential in Escherichia coli (Table S4; P = .19, Fisher's exact test). This suggests that purifying selection was significantly reduced during the experiment and that deleterious mutations accumulated at random with limited constraints on the fitness of the mutant, causing the fixation rate to be close to the mutation rate.
Base Pair Substitution Mutation Rates.
During every growth cycle, there were ≈25 cell divisions, giving a total of 5,000 generations during the mutation accumulation experiment. The BPS rate of the quintuple mutant was calculated by taking the average of the mutations per genome and dividing by the number of generations. The mutational spectrum of the quintuple mutant (ung, vsr, mug, mutM, and mutY) was biased heavily toward BSPs, which allowed estimation of the total mutation rate as the BSP rate. The mutation rate was 0.094 mutations per genome and generation, or 2.0 × 10−8 per base pair per generation. Reducing the GC content of the S. typhimurium chromosome by 1% would require ≈48,500 GC-to-AT mutations. Given the mutation rate calculated above, this could occur in about 500,000 generations, provided that the relevant repair systems were absent and that selection was reduced. Assuming a generation time of one generation per day in nature, this would take 1,400 years, a very short time span from an evolutionary perspective. The BSP rate of the wild-type S. typhimurium also can be approximately calculated from the sequencing, but this will provide only an upper limit, because sequencing errors will influence the result to a greater degree (see Materials and Methods). The mutation rate for the wild-type strain, calculated as described above, is then 3.4 × 10−3 BPSs per genome and generation, or 7.0 × 10−10 per base pair per generation. Results from the rifampicin resistance fluctuation test showed an ≈100-fold increase in the mutation rate for the quintuple mutant (Fig. 1), which is close to the 28-fold increase for the quintuple mutant calculated here considering the quality of the data for the wild-type sequence.
Fitness Loss During Serial Passage.
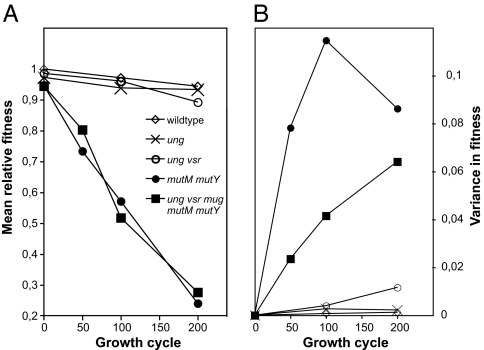
As expected, the average fitness decreased and the fitness variance increased with time for all five strains examined (Fig. 3 A and B). The rate of fitness loss was similar to that for the wild-type strain for ung-, was increased 2-fold for ung- vsr-, and was increased 12-fold for mutM- mutY- and ung- vsr- mug- mutM- mutY-. The decreased fitness of the quintuple mutant during the experiment (1.37 × 10−4 per generation) allowed the average fitness loss per mutation to be estimated as 0.00145.
Fig. 3.
(A) Mean fitness of the evolved strains, measured as the generation time during exponential growth in rich growth medium normalized by the generation time of the ancestral wild type included in each experiment. (B) Variance in fitness between lineages of the evolved strains.
Discussion
This study demonstrates for the first time how, in the absence of the repair systems that normally counteract the intrinsic forces of deamination and oxidation, an underlying mutational bias could rapidly change the global genomic base composition. The experimental setup with population bottlenecks and a resulting strong reduction of selection allowed us to study the presence of several important mutational biases that have been used to explain the characteristics of base composition in bacterial genomes. In addition, the use of complete genome sequences makes the results more general, because it reduces the risk of false conclusions associated with experimental systems based on a single gene. The majority of the mutations found in this study were those associated with oxidation of guanine (91%); mutations associated with deamination of cytosine (7%) contributed much less to the change in GC. By determining the number of accumulated mutations and the associated fitness reduction, we can estimate both the genomic mutation rate and the average fitness loss per mutation. The estimate of the total genomic mutation rate to 3.4 × 10−3 per genome per generation for wild-type S. typhimurium is very close to that of Drake (23) based on lacI and the his operon in E. coli, which was determined to be about 3 × 10−3 per genome per generation. Our estimate of the average fitness loss of 1.5 × 10−3 per BPS is an order of magnitude lower than the upper-bound estimates of the average deleterious mutational effect in E. coli reported by Kibota and Lynch (24) and in S. typhimurium reported by Maisnier-Patin et al. (25). The difference between our present estimate and the estimates from those previous studies can be explained in part by the fact that we consider only BPS, disregarding other types of mutations.
With regard to the occurrence of various mutational biases, we report several significant observations. Highly expressed genes had a higher mutation rate for both transitions and transversions compared with the rest of the genome, suggesting that transcription can influence compositional biases. The phylogenetic observation that highly expressed genes have lower divergence seems to argue against this theory, but it appears that these genes have much fewer neutral or nearly neutral sites, which limits the fixation of mutations in these genes (26–28). Furthermore, C-to-T transitions likely caused by deamination of cytosine were more common in the nontranscribed strand of coding regions, whereas no such bias was found in the case of G-to-T transversions. These results support previous studies on single genes in which transcription was reported to increase mutation rates and a bias between the transcribed and nontranscribed strands for C-to-T transitions was observed (29, 30). The rate constants for cytosine deamination have been determined in vitro to be 10−10 per second in single-stranded DNA and 7 × 10−13 per second in double-stranded DNA under similar conditions as used in our experiments (pH 7.4; 37 °C) (7). If we assume that half of the deaminations will be fixed after replication, which is reasonable because one strand will still carry the correct base, then we can use the rate constants to estimate the number of mutations expected in our experiment. After 200 days, we would then expect about 15 mutations per genome, assuming a constant double-stranded state, and 2,100 mutations for a fully single-stranded state. The 32.5 mutations per genome observed in this study were higher than expected from the double-stranded state, suggesting that the time spent in the single-stranded state can influence strand bias. Assuming that the excess C-to-T transition mutations result from deamination only, the time spent in the single-stranded state can be estimated to be 0.8% of the total time (32.5–15/2,100–15).
Previously observed compositional biases between the leading and lagging strands of replication have been explained by the presence of mutational biases associated with the asymmetry of replication (31). Somewhat unexpectedly, under our experimental setup we found no asymmetry between the two strands for either C-to-T mutations or G-to-T mutations. Potential explanations for this finding include the possibility that the bias was too weak to detect using our experimental approach, that the asymmetry was not linked to the formation of these types of damage, and that the bias was related to an asymmetry conferred by the inactivated DNA repair systems. Regardless of the explanation for the lack of an observable leading-lagging bias, these data suggest that under our experimental setup, the transcription-associated biases were more pronounced than the replication-associated biases. Furthermore, we detected no effect of chromosomal location on mutation rate, as was suggested by Sharp et al. (27), who reported that synonymous mutations were more frequent in the terminus region. Thus, for DNA damages likely caused by deamination and oxidation of bases, no obvious hot spot regions were found, and the mutations appeared to be random in relation to chromosome location.
Finally, these results are of relevance for understanding the AT richness of the size-reduced genomes belonging to intracellular parasites and endosymbionts. This AT richness has been explained by relaxed selective constraints, caused by passage through small population bottlenecks combined with the loss of DNA repair systems, including ung, mutM, and mutY, during reductive evolution (32, 33). Our experiments mimic this evolutionary process and provide support for the importance of these repair systems in maintaining genomic GC content. Furthermore, as discussed above, the loss of these repair systems potentially could result in very rapid reduction in GC content. As shown in Table 3, all of the AT-rich small genomes lack all or some of the relevant repair genes, and it is conceivable that the reduced GC content is, at least in part, a consequence of the resulting stronger GC-to-AT mutational bias conferred by ubiquitous oxidation and deamination damages. A similar reasoning also might apply to protozoans (e.g., Plasmodium) that have AT-rich genomes and also lack many of the relevant repair genes (34).
Table 3.
Absence and presence of genes associated with DNA repair in bacteria with small genomes and low GC content
| Species | ung | mutM | mutY | Genome size, Mbp | GC content, % |
|---|---|---|---|---|---|
| Carsonella ruddii | — | — | — | 0.16 | 17 |
| Wigglesworthia glossinidia brevipalpis | + | — | — | 0.69 | 22 |
| Buchnera aphidicola Sg | —* | — | + | 0.64 | 25 |
| Blochmannia floridanus | + | — | + | 0.70 | 27 |
| Fusobacterium nucleatum ATCC 25586 | + | — | — | 2.17 | 27 |
| Borrelia burgdorfheri B31 | + | — | — | 1.52 | 28 |
| Ehrlichia ruminantium Gardel | — | + | — | 1.50 | 28 |
| Rickettsia prowazekii Madrid E | — | — | — | 1.11 | 29 |
| Mycoplasma genitalium G-37 | + | + | — | 0.58 | 32 |
| Baumannia cicadellinicola | + | + | — | 0.68 | 33 |
| Wolbachia pipientis wBM | — | + | — | 1.08 | 34 |
The ung, mutM, and mutY genes are widespread among members of the major bacterial phyla, whereas mug and vsr genes are present only in a smaller number of species. A gene was considered absent if no annotated gene with the same function was found and no homologues were found in BLAST searches with a cutoff of P < 10−6 using the S. typhimurium sequence and excluding the mutY homologue nth.
*Pseudogene.
Materials and Methods
Strains and Media.
S. enterica var. Typhimurium LT2 (designated S. typhimurium here) and derivatives thereof were used in all experiments. All liquid media used was LB broth, and all solid media was LB agar supplemented with kanamycin, 30 or 50 mg/L, and ampicillin, 100 mg/L, for selection and plasmid maintenance.
Construction of Repair-Deficient Mutants.
Gene deletions in the S. typhimurium chromosome were made using the Lambda Red system as described previously (35). A kanamycin-resistance cassette flanked by FLP recombinase target sequences (FRTs) was amplified from template plasmid pKD4 (GenBank accession AY048743) with primers containing 40 base pair extensions homologous to regions in or adjacent to the ung, vsr, mug, mutM, and mutY genes and used for transformation of the Lambda Red strain. Deleted genes were sequentially moved to S. typhimurium LT2 by phage P22 transduction, and the FRT-flanked kanamycin cassette was removed with a helper plasmid (pCP20) expressing the site-specific FLP recombinase (35). All constructs were verified by colony polymerase chain reaction with primers outside the deleted regions. A detailed description of strain construction is given in the SI, and all primers used are listed in Table S5.
Mutation Accumulation Experiment.
Twelve independent lineages each of wild-type LT2, ung::kan, Δung vsr::kan, ΔmutM mutY::kan, and ΔungΔmugΔmutMΔmutY vsr::kan, were used for the mutation accumulation experiment. Each lineage was passaged through random single-cell bottlenecks on LB agar plates supplemented with 0.2% glucose by always choosing the last visible colony appearing in the streak irrespective of size or appearance every 24 h for 200 cycles at 37 °C.
Growth Rate Measurements.
Exponential growth rates were measured at 37 °C in LB broth. One μL of an overnight culture was used to inoculate 2 ml of LB broth; 350 μl of this was loaded into each well, and absorbance at 600 nm was recorded every 4 min by a BioscreenC reader (Labsystems). All growth rates were normalized to the growth rate of the ancestral S. typhimurium LT2 included in each experiment. Growth rates were measured in two separate experiments in quadruplicate.
Mutation Rate Determination.
Mutation rates were determined for the ancestral strains and for the evolved lineages after 200 cycles. Approximately 103 cells were used to inoculate 10 replicates with 220 μl of LB broth and grown for 24 h at 37 °C with shaking (200 rpm) in a 10-ml tube. Then 200 μl of the overnight culture was spread on LB agar plates with 100 mg/L of rifampicin, and suitable dilutions were spread on LB agar plates without rifampicin for determination of viable cells. The number of colonies was counted after incubation at 37 °C for 30 h, and mutation rates were calculated using the Lea-Coulson method of the median (for low/moderate number of mutations) or the Drake formula (for high mutation number of mutations) as described previously (36).
DNA Sequencing and Analysis.
Genomic DNA was prepared from two lineages of the Δung Δmug ΔmutM ΔmutY vsr::kan strain and one lineage of the wild-type strain evolved for 200 cycles using a Genomic Tip 100G (Qiagen) according to the manufacturer's instructions. The lineages were chosen using a random number generator among the strains with similar mutation rates as its ancestor. (Three lineages of the quintuple mutant were excluded.) Genome sequencing was performed with a Genome Sequencer FLX (Roche) at the KTH Sequencing Facility, Royal Institute of Technology, KTH, Stockholm, Sweden. Contigs longer than 500 bp were used for BLAST searches against the reference S. typhimurium LT2 genome to find mutations. For the two sequenced quintuple mutants, the fraction of bases with quality scores higher than Q40 were 97%, where Q40 represents an error rate of 99.99%. Among the mutations found, 96% were +Q40 bases, only slightly lower than the fraction for the entire sequence, suggesting that sequencing errors had only a limited influence on our results. In the sequenced evolved wild-type strain, 95% were Q40 bases, but only 47% of the mutations were found, suggesting a larger fraction of false positives. Statistical analyses were performed using the R statistics package (R Project for Statistical Computing; http://www.r-project.org).
Supplementary Material
Acknowledgments.
This work was supported by grants from the Swedish Research Council and Uppsala University (to D.I.A.). We thank Otto Berg, Linus Sandegren, and Staffan Svärd for comments and a critical reading of the manuscript.
Footnotes
The authors declare no conflicts of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804445105/DCSupplemental.
References
- 1.Hallin PF, Ussery DW. CBS Genome Atlas Database: A dynamic storage for bioinformatic results and sequence data. Bioinformatics. 2004;20:3682–3686. doi: 10.1093/bioinformatics/bth423. [DOI] [PubMed] [Google Scholar]
- 2.Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 3.Francino MP, Chao L, Riley MA, Ochman H. Asymmetries generated by transcription-coupled repair in enterobacterial genes. Science. 1996;272:107–109. doi: 10.1126/science.272.5258.107. [DOI] [PubMed] [Google Scholar]
- 4.Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- 5.Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- 6.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: Determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 8.Marians KJ. Prokaryotic DNA replication. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 9.Frank AC, Lobry JR. Asymmetric substitution patterns: A review of possible underlying mutational or selective mechanisms. Gene. 1999;238:65–77. doi: 10.1016/s0378-1119(99)00297-8. [DOI] [PubMed] [Google Scholar]
- 10.Lutsenko E, Bhagwat AS. The role of the Escherichia coli mug protein in the removal of uracil and 3,N(4)-ethenocytosine from DNA. J Biol Chem. 1999;274:31034–31038. doi: 10.1074/jbc.274.43.31034. [DOI] [PubMed] [Google Scholar]
- 11.Mokkapati SK, Fernandez de Henestrosa AR, Bhagwat AS. Escherichia coli DNA glycosylase Mug: A growth-regulated enzyme required for mutation avoidance in stationary-phase cells. Mol Microbiol. 2001;41:1101–1111. doi: 10.1046/j.1365-2958.2001.02559.x. [DOI] [PubMed] [Google Scholar]
- 12.Hayakawa H, Kumura K, Sekiguchi M. Role of uracil-DNA glycosylase in the repair of deaminated cytosine residues of DNA in Escherichia coli. J Biochem. 1978;84:1155–1164. doi: 10.1093/oxfordjournals.jbchem.a132231. [DOI] [PubMed] [Google Scholar]
- 13.Duncan BK, Rockstroh PA, Warner HR. Escherichia coli K-12 mutants deficient in uracil-DNA glycosylase. J Bacteriol. 1978;134:1039–1045. doi: 10.1128/jb.134.3.1039-1045.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieb M. Spontaneous mutation at a 5-methylcytosine hotspot is prevented by very short patch (VSP) mismatch repair. Genetics. 1991;128:23–27. doi: 10.1093/genetics/128.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci U S A. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rocha EP, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- 17.Suyama A, Wada A. Correlation between thermal stability maps and genetic maps of double-stranded DNAs. Nucleic Acids Symp Ser. 1982;11:165–168. [PubMed] [Google Scholar]
- 18.Musto H, et al. Genomic GC level, optimal growth temperature, and genome size in prokaryotes. Biochem Biophys Res Commun. 2006;347:1–3. doi: 10.1016/j.bbrc.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 19.Singer CE, Ames BN. Sunlight ultraviolet and bacterial DNA base ratios. Science. 1970;170:822–825. doi: 10.1126/science.170.3960.822. [DOI] [PubMed] [Google Scholar]
- 20.Naya H, Romero H, Zavala A, Alvarez B, Musto H. Aerobiosis increases the genomic guanine plus cytosine content (GC%) in prokaryotes. J Mol Evol. 2002;55:260–264. doi: 10.1007/s00239-002-2323-3. [DOI] [PubMed] [Google Scholar]
- 21.McEwan CE, Gatherer D, McEwan NR. Nitrogen-fixing aerobic bacteria have higher genomic GC content than non-fixing species within the same genus. Hereditas. 1998;128:173–178. doi: 10.1111/j.1601-5223.1998.00173.x. [DOI] [PubMed] [Google Scholar]
- 22.Puigbo P, Romeu A, Garcia-Vallve S. HEG-DB: A database of predicted highly expressed genes in prokaryotic complete genomes under translational selection. Nucleic Acids Res. 2008;36:D524–D527. doi: 10.1093/nar/gkm831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kibota TT, Lynch M. Estimate of the genomic mutation rate deleterious to overall fitness in E. coli. Nature. 1996;381:694–696. doi: 10.1038/381694a0. [DOI] [PubMed] [Google Scholar]
- 25.Maisnier-Patin S, et al. Genomic buffering mitigates the effects of deleterious mutations in bacteria. Nat Genet. 2005;37:1376–1379. doi: 10.1038/ng1676. [DOI] [PubMed] [Google Scholar]
- 26.Sharp PM, Li WH. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 1987;4:222–230. doi: 10.1093/oxfordjournals.molbev.a040443. [DOI] [PubMed] [Google Scholar]
- 27.Sharp PM, Shields DC, Wolfe KH, Li WH. Chromosomal location and evolutionary rate variation in enterobacterial genes. Science. 1989;246:808–810. doi: 10.1126/science.2683084. [DOI] [PubMed] [Google Scholar]
- 28.Berg OG, Martelius M. Synonymous substitution-rate constants in Escherichia coli and Salmonella typhimurium and their relationship to gene expression and selection pressure. J Mol Evol. 1995;41:449–456. doi: 10.1007/BF00160316. [DOI] [PubMed] [Google Scholar]
- 29.Klapacz J, Bhagwat AS. Transcription-dependent increase in multiple classes of base substitution mutations in Escherichia coli. J Bacteriol. 2002;184:6866–6872. doi: 10.1128/JB.184.24.6866-6872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudson RE, Bergthorsson U, Ochman H. Transcription increases multiple spontaneous point mutations in Salmonella enterica. Nucleic Acids Res. 2003;31:4517–4522. doi: 10.1093/nar/gkg651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu CI, Maeda N. Inequality in mutation rates of the two strands of DNA. Nature. 1987;327:169–170. doi: 10.1038/327169a0. [DOI] [PubMed] [Google Scholar]
- 32.Moran NA. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersson SG, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- 34.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosche WA, Foster PL. Determining mutation rates in bacterial populations. Methods. 2000;20:4–17. doi: 10.1006/meth.1999.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.