Abstract
Internal ionizable groups are quite rare in water-soluble globular proteins. Presumably, this reflects the incompatibility between charges and the hydrophobic environment in the protein interior. Here we show that proteins can have an inherently high tolerance for internal ionizable groups. The 25 internal positions in staphylococcal nuclease were substituted one at a time with Lys, Glu, or Asp without abolishing enzymatic activity and without detectable changes in the conformation of the protein. Similar results with substitutions of 6 randomly chosen internal positions in ribonuclease H with Lys and Glu suggest that the ability of proteins to tolerate internal ionizable groups might be a property common to many proteins. Eighty-six of the 87 substitutions made were destabilizing, but in all but one case the proteins remained in the native state at neutral pH. By comparing the stability of each variant protein at two different pH values it was established that the pKa values of most of the internal ionizable groups are shifted; many of the internal ionizable groups are probably neutral at physiological pH values. These studies demonstrate that special structural adaptations are not needed for ionizable groups to exist stably in the hydrophobic interior of proteins. The studies suggest that enzymes and other proteins that use internal ionizable groups for functional purposes could have evolved through the random accumulation of mutations that introduced ionizable groups at internal positions, followed by evolutionary adaptation and optimization to modulate stability, dynamics, and other factors necessary for function.
Keywords: electrostatics, internal ionizable groups, pKa values, dehydration
In his seminal review on the hydrophobic effect and other noncovalent forces responsible for the stability of proteins, Kauzmann (1) suggested that charges would avoid the hydrophobic interior of proteins and favor the protein–water interface. This idea was confirmed as an important organizing principle of globular proteins 2 years later when, in his analysis of the first crystal structure where individual side chains could be observed, Kendrew and coworkers (2, 3) noted “a tendency, perhaps less marked than might have been expected, for polar side chains to project outwards from the molecule and for nonpolar ones to be buried within it.” The wealth of protein structures obtained in the last 2 decades has confirmed that in water-soluble proteins, ionizable groups are usually excluded from the hydrophobic interior and found mostly at the surface, where they can interact with water (4–6). Perutz (7, 8) summarized it eloquently: “Most globular proteins are waxy inside and soapy outside.”
Continuum electrostatics representations of proteins offer a plausible hypothesis for the selection against internal ionizable groups. In these models, the protein is usually treated with the dielectric constants of dry protein powders, which range from 2 to 4 (9, 10). If these dielectric constants described accurately the effective polarizability of the protein interior, the ionization of an internal group (i.e., the free energy for transferring a charge from water into the protein interior) would be a highly destabilizing process involving a free energy far greater than the net stability of the average protein. Were this the case, the relative paucity of internal ionizable groups could be explained in terms of evolutionary pressure to enhance the stability of proteins by eliminating internal ionizable groups that were not needed for specific structural or functional purposes. However, it is necessary to consider the possibility that the internal ionizable groups are actually not in the charged state at physiological pH (11–14) and are therefore not as destabilizing as suggested by continuum electrostatics models.
Internal ionizable groups are actually essential for the fundamental energy transduction processes common to all living organisms. Most of these processes involve the transient presence of charge in hydrophobic environments where the presence of water is tightly regulated. For example, internal ionizable groups are necessary for catalysis (7), redox reactions (15, 16), H+ transport (17, 18), ion homeostasis (19), and light-activated processes (20). Internal ionizable groups are poised to play essential roles in bioenergetics precisely because of the relatively low polarity and polarizability of the protein interior: The presence of charge in a hydrophobic environment can trigger conformational reorganization, which is the basis of many forms of energy transduction (20, 21). The apparent dichotomy between the incompatibility of charge with the hydrophobic interior of proteins and the requirement for internal ionizable groups for energy transduction suggests that specialized polar microenvironments evolved around internal ionizable groups to mitigate the energetic penalty for their burial. Here we show that for proteins with high intrinsic stability, special structural adaptations are not necessary for ionizable groups to reside stably in the hydrophobic interior of proteins without affecting their conformation or biological activity.
Results and Discussion
Thermodynamic Signatures of Internal Ionizable Groups.
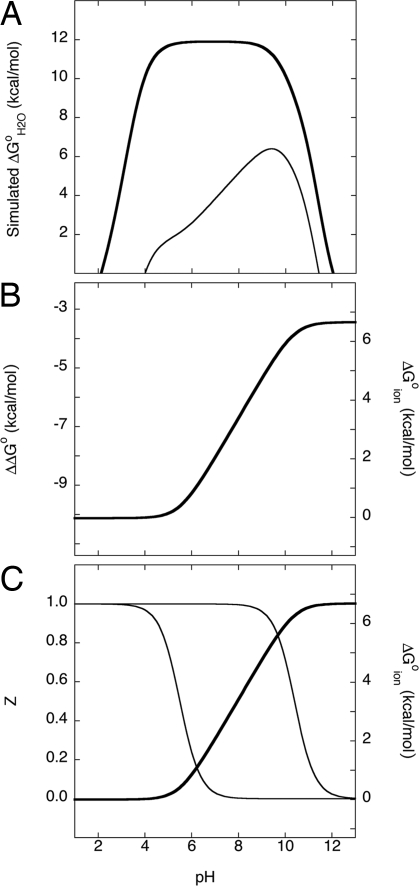
When an ionizable group is removed from water and immersed in a less polar and polarizable material, such as the interior of a protein, its pKa value shifts in the direction that promotes the neutral state unless other factors (e.g., hydrogen bonds, pairing with a charge of the opposite sign, and interactions with internal water molecules) compensate for the loss of hydration (11–14). Ionizable groups with highly perturbed pKa values have significant effects on the pH dependence of the thermodynamic stability (ΔG°H2O) of proteins (11–13). This effect is illustrated graphically in Fig. 1A for a protein with an internal Lys residue with a pKa shifted from its normal value of 10.4 to a value of 6.0. ΔΔG°H2O refers to the difference in ΔG°H2O between a variant with the internal Lys residue with a depressed pKa and the ΔG°H2O of the background protein where this Lys residue is not present. The shape of the pH dependence of ΔΔG°H2O shown in Fig. 1B is characteristic of proteins with basic groups with highly depressed pKa values; proteins with acidic groups with elevated pKa values exhibit the same pattern except that the ΔΔG°H2O values decrease between low and high pH values. At pH > 10.4, Lys in water is neutral. Under these conditions, ΔΔG°H2O primarily reflects the destabilizing consequences of substituting an internal residue with Lys in its neutral state. At pH < 10.4, the stability of the protein decreases by 1.36 kcal/mol per unit of pH owing to the shift in the pKa value of the Lys toward values lower than the normal value of 10.4. The ΔΔG°H2O decreases at this rate until the internal Lys becomes charged.
Fig. 1.
Thermodynamic linkage between pH dependence of stability and shifts in pKa values. (A) Simulated pH dependence of stability (ΔG°H2O) of a protein (thick line) and of one of its variants with an internal Lys with a pKa value of 6 (thin line). (B) Difference in the thermodynamic stability curves (ΔΔG°H2O) in A. (C) H+ titration curves (thin lines) of a Lys with a normal pKa value of 10.4 and with a pKa value of 6.0. The area between these two titration curves (dark line) is equivalent to ΔΔG°H2O in B.
Shifts in the pKa of an internal group can be detected in 2 ways: first, by measuring unfolding free energy as a function of pH (e.g., Fig. 1B) or, second, by direct measurement of the H+ titration curves of the internal Lys residue (Fig. 1C) (11–13, 22). These 2 methods are equivalent, as shown by the simulations in Fig. 1. These simulations illustrate how the measurement of the pH dependence of the stability of 2 proteins that differ only in the presence or absence of an internal ionizable group can reveal whether the pKa value of the internal ionizable group is shifted relative to its normal pKa in water. In practice, the shifts have to be substantial to be detectable with this approach.
One significant consequence of the shifts in pKa values of internal Lys, Asp, and Glu residues is that they can be neutral under physiological conditions of pH, where they are usually fully charged. The magnitude of the net shift in pKa has complex origins; it reflects the polarity and effective polarizability of the microenvironment around the buried ionizable moiety, which can include structural reorganization coupled with the ionization of the internal group (12, 14).
Eighty-Seven Proteins with Internal Lys, Glu, or Asp Residues.
X-ray crystallography and equilibrium thermodynamic studies were used previously to characterize in detail the properties of internal Glu and Lys residues at positions 66 and 92 in staphylococcal nuclease (SNase) (11–13, 22–24). The crystal structures show that these internal groups are buried deeply in the hydrophobic core of the protein. To test the hypothesis that ionizable groups are much more compatible with the hydrophobic interior of proteins than is currently recognized, all of the other internal positions in a hyperstable form of SNase were replaced one at a time with Lys, Asp, or Glu. Internal positions were identified as those in which the Cα–Cβ vector of a side chain points toward the protein interior. The effects of the internal ionizable groups on thermodynamic stability (ΔG°H2O) were measured by chemical denaturation monitored by optical spectroscopy (11, 12). To test the general validity of the results obtained with SNase, Lys and Glu were also introduced into 6 other internal sites chosen randomly in a second unrelated protein, ribonuclease H (RNaseH) from Thermus thermophilus. The thermodynamic consequences of substitutions with Arg were also studied in SNase. The trends observed are similar to those of Lys, Asp, and Glu, but Arg is quite different from the other ionizable groups, and the properties of internal Arg side chains are sufficiently different to warrant detailed discussion elsewhere.
The thermodynamic stability of the parent proteins and of all of the variants was measured by chemical denaturation with guanindinium chloride (GdmCl) monitored by intrinsic fluorescence. Measurements were made at pH 7 and also under conditions of pH where the ionizable groups in water would normally be nearly neutral (i.e., pH 10 for Lys and pH 5 for Asp and Glu). The 2 proteins used in this study are very stable (11.9 kcal/mol for SNase and 12.5 kcal/mol for RNase at pH 7). This high stability was necessary to compensate for the relatively high destabilization incurred by the insertion of ionizable groups in the protein interior with mutagenesis. The complete list of residues that were substituted and all ΔG°H2O values are provided in the SI.
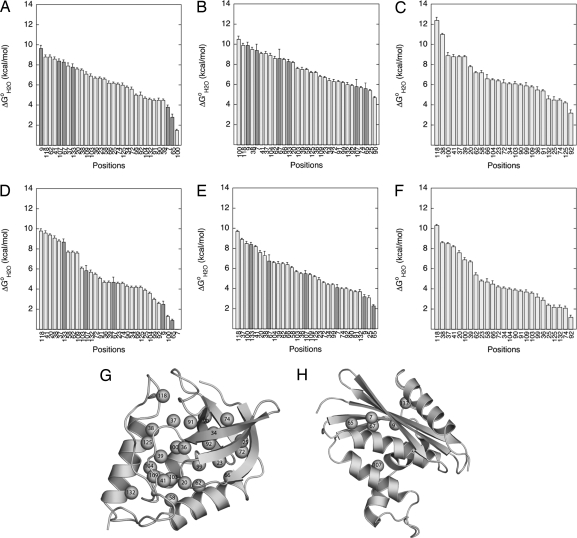
The consequences of replacement of internal positions with ionizable groups on the thermodynamic stability of the proteins are illustrated in Fig. 2. All of the 87 variant proteins but one (L7K variant of RNaseH) were fully folded at physiological pH and unfolded cooperatively and in an apparent 2-state manner. There is at least one naturally occurring, internal ionizable group that is stabilizing (25). In contrast, with the exception of the N118D variant, which is slightly more stable than the background protein, the substitutions of internal positions with ionizable groups were destabilizing at all pH values studied. This result was expected for substitutions of hydrophobic side chains in hydrophobic environments with polar side chains. The median loss of stability at pH 7 [i.e., ΔG°H2O (variant) − ΔG°H2O (background)] was −7.2, −7.8, and −6.4 kcal/mol for substitutions with Lys, Asp, and Glu, respectively. The destabilizing effects of the substitutions were comparable in RNaseH and SNase, suggesting that the results of our study reflect general properties of globular proteins. This ability of proteins to tolerate internal ionizable groups is not widely recognized.
Fig. 2.
Effects of internal ionizable groups on thermodynamic stability. (A–F) Thermodynamic stability (ΔG°H2O) of variants of SNase (light gray) or RNaseH (dark gray) with internal Lys at pH 10 (A), internal Glu at pH 5 (B), internal Asp at pH 5 (C), internal Lys at pH 7 (D), internal Glu at pH 7 (E), and internal Asp at pH 7 (F). (G and H) Ribbon diagram of SNase (G) (32) and RNaseH (H) (33) showing the internal positions that were substituted. All data were measured at 298 K in 100 mM KCl.
The pKa Values of Most of the 87 Internal Ionizable Groups Are Perturbed.
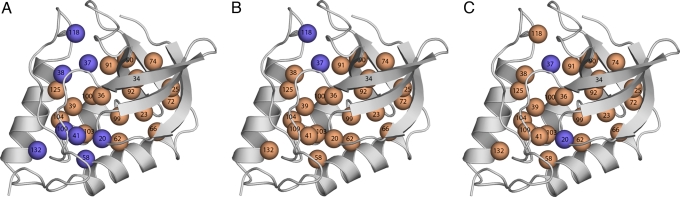
The stability of the Lys-, Asp-, and Glu-substituted proteins measured at pH 7 was compared with the stability measured at a second pH value (pH 5 for Asp and Glu, and pH 10 for Lys) that was close to the normal pKa value of the ionizable group in water. Cases where the stability of the variant relative to the stability of the background protein was pH-sensitive correspond to cases where the pKa values of the internal Lys, Asp, and Glu were shifted relative to their normal pKa values of 10.4, 4.0, and 4.4, respectively. The Lys, Asp, or Glu residues that titrate with shifted pKa values are identified as orange spheres in Fig. 3. The pKa values of the majority of the 87 internal ionizable groups studied were shifted in the direction that promotes the neutral state (elevated for acidic residues and depressed for basic ones). This finding suggests that at least in the case of SNase and RNaseH, all internal microenvironments are less polarizable and polar than water. More Lys residues than Asp or Glu residues appear to titrate with normal pKa values. Only at position 37 did none of the 3 types of ionizable groups show any evidence of a shift in pKa value. Besides showing that most of the internal groups titrate with perturbed pKa values, the data also suggest that many of the internal Lys, Glu, and Asp residues are neutral at pH 7. This result remains to be corroborated by measurement of the pKa values of the internal ionizable groups.
Fig. 3.
pH dependence of difference thermodynamic stability [ΔΔG°H2O = ΔG°H2O (variant) − ΔG°H2O (background)], defined as (ΔΔΔG°H2O/ΔpH) = (ΔΔG°H2O, pH1 − ΔΔG°H2O, pH2)/(∣pH1 − pH2∣). pH1 = 7 and pH2 = 5 for Asp and Glu or 10 for Lys. Blue circles identify positions where ΔΔΔG°H2O/ΔpH = 0, indicating that the pKa values are probably normal; orange circles identify positions where ΔΔΔG°H2O/ΔpH > 0.2, indicating that the pKa values are shifted in the direction that favors the neutral state (depressed for basic groups, elevated for acidic ones). Data are for variants with internal Lys (A), Glu (B), and Asp (C).
Internal Ionizable Groups Do Not Affect the Conformation of the Protein.
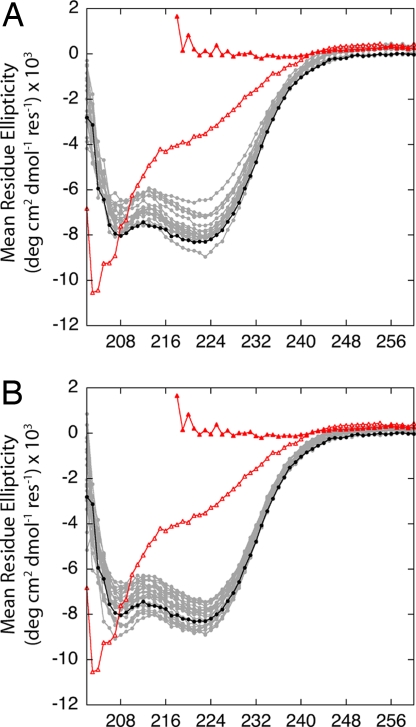
All of the equilibrium unfolding curves of SNase, and most of the curves for RNaseH, were described well in terms of a 2-state unfolding process. The equilibrium thermodynamic data suggested that the substitution of hydrophobic core positions with ionizable groups had no deleterious impact on the conformation of the protein. This suggestion was confirmed by the far-UV CD spectra of the variants. The spectra for SNase variants with internal Lys or Glu at pH 7 are shown in Fig. 4(the data for the Asp-substituted variants are not shown but are comparable). Scans of a permanently unfolded T62P variant of SNase in water and of wild-type SNase denatured in 6 M GdmCl are shown to illustrate by comparison that all variants with internal Lys, Asp, or Glu residues are folded and native-like. The small differences in the spectra of the different proteins are comparable to the differences observed with other types of substitutions in this protein. In all cases studied, the CD spectra show that the substitution of internal positions with ionizable groups did not affect the overall conformation of the native state in a detectable manner.
Fig. 4.
Far-UV CD spectra of SNase variants with internal Glu (A) and internal Lys (B). Also shown are the background protein in water (●), wild-type SNase unfolded in 6 M GdmCl (▲), and an unfolded T62P variant in water (△). The lines are meant only to guide the eye. All measurements at pH 7 in 25 mM Hepes and 100 mM KCl.
Internal Ionizable Groups Do Not Abolish Enzymatic Activity.
Enzymatic activity assays were performed with the SNase variants to examine the impact of internal ionizable groups on catalytic activity (data not shown). The goal was not to quantitate the specific levels of activity in the different variants but to determine whether they were active or not. Although not all of the variants were as active as the background protein, only variants with substitutions at 4 sites (G20, T41, A58, and A109) showed no activity, regardless of pH and of the nature of the ionizable group. The side chains at these positions point toward the active site, which suggests that the loss of activity of these variants might be more related to steric interference of substrate binding to the active site than to disorganization of the protein or direct electrostatic effects. Even the highly destabilized variants in which ionizable groups were introduced in the β-barrel showed activity under some of the conditions of pH that were tested. These data further suggest that the substitution of internal positions with ionizable groups can occur without detectable, deleterious consequences on either structural or functional properties.
Conclusions
The demonstration that a protein can tolerate the substitution of every internal position with either a basic or an acidic residue, without the benefit of special structural adaptation and evolutionary optimization to solvate the internal ionizable group, without loss of enzymatic activity, and without affecting the overall fold and conformation of the protein, suggests that proteins have a much higher intrinsic ability to tolerate internal ionizable groups than is currently recognized. This conclusion is consistent with previous studies with other proteins, which showed that enzymatic activity is not destroyed by substitutions that introduce internal ionizable groups (26). Then why are internal ionizable residues eliminated so efficiently from water-soluble globular proteins when they are not necessary for specific structural or functional purposes? Ionizable groups in the hydrophobic core of a protein are usually destabilizing, and their elimination is certainly advantageous for global stability. However, the fact that under laboratory conditions it was possible to introduce ionizable groups into so many hydrophobic positions in these proteins suggests that folding, solubility, dynamics, and other physical attributes of globular proteins in the cellular milieu contribute to the evolutionary pressure to eliminate unnecessary internal ionizable groups.
The results from this study would appear to contradict the view suggested by continuum-electrostatics representations of the protein interior as a highly hydrophobic environment of low dielectric constant, incompatible with ionizable groups. They do not. Most of the ionizable groups that were introduced by mutagenesis are probably neutral at pH 7. It is therefore incorrect to think about the destabilizing consequences of internal ionizable groups in terms of the presence of charge in the hydrophobic interior of the protein. In the majority of the sites that were studied, the destabilizing consequences of the substitution of internal hydrophobic groups with ionizable ones are governed by the shifts in pKa values and by the cost of replacement of small, internal hydrophobic side chains with larger polar ones (i.e., the ionizable side chains in the neutral state). Further studies are needed to measure the pKa values of these internal groups and the detailed molecular factors that determine them. This information will be useful to describe the structural consequences of the ionization of internal groups in detail, and to test and calibrate computational methods for structure-based calculation of electrostatic energy (14).
These results have interesting implications for the evolution and engineering of enzymes and of proteins for ion and H+ transport and e− transfer. For example, the data suggest that proteins using internal ionizable groups to move H+ across a lipid bilayer could have evolved simply through the random introduction and accumulation of ionizable groups in the protein interior, without the need for the evolution of special microenvironments or structural adaptations to mitigate the destabilizing influence of internal ionizable groups. Many of the proteins that use internal ionizable groups for functional purposes are integral membrane proteins; internal ionizable groups will be less destabilizing to these proteins than to water-soluble globular proteins because the stability of membrane proteins is not defined relative to an unfolded state in water. In contrast, the evolution of active sites with internal ionizable groups in water-soluble enzyme was probably more complex. These internal ionizable groups could have been introduced at a nascent active site randomly, but this would have had to be followed by mutations that increased the stability of the native state to tolerate the internal ionizable groups. Further mutations would also have been necessary to tune the pKa values of the internal groups and to tailor dynamics, internal polarity, and any other factors necessary to achieve catalysis (27–29). Studies are in progress to examine the factors that govern the properties of internal ionizable groups in proteins.
Materials and Methods
Staphylococcal Nuclease.
All studies were performed with the Δ+PHS hyperstable variant of SNase described previously (11, 12). Mutagenesis, protein purification, measurement of unfolding free energies (ΔG°H2O), and measurement of far-UV CD spectra of SNase were performed as described previously (11, 12). The enzymatic activity of SNase and its variants was determined by observing the change in color of a metachromic dye from blue to pink upon hydrolysis of DNA in a DNA-containing agar gel (30). The initial protein concentration in all activity assays was 2.45 mg/mL. Two-microliter volumes of the enzyme solution were spotted onto a Petri plate prepared with 10 mL of agar containing 50 mM Tris·HCl, pH 7.0, 1% NaCl, 1% agar, 300 μg/mL boiled salmon sperm DNA, 20 mM CaCl2, and 0.2 mg/mL toluidine blue O. Mes and 2-(N-cyclohexylamino)ethanesulfonic acid were used as buffers for assays conducted at pH 5.5 and pH 9.5, respectively. Plates were incubated for 24 h at 30 °C and analyzed. The enzyme was determined to be active when spotting of the enzyme on the blue agar led to the formation of a pink halo. Negative controls were performed to demonstrate that direct interference with the active site did abolish activity. For example, the activity of variants with E34Q and D21N, which remove carboxylic groups essential for catalysis, showed low activity and only at very high concentrations and over long periods of time.
Ribonuclease H.
Purification of RNaseH was performed as described previously (31). Fluorescence-monitored GdmCl denaturations of RNase variants were performed with an automated titration fluorometer by Aviv with an excitation wavelength of 298 nm, an emission wavelength of 323 nm, and an excitation bandwidth of 3.2 nm. Experiments at pH 7 were performed in 25 mM Mops. Otherwise, the experimental conditions were identical to those used with SNase.
Supplementary Material
Acknowledgments.
We thank Prof. Susan Marqusee (University of California, Berkeley, CA) for the gift of the RnaseH clone. This work was supported by National Institutes of Health Grant GM-065197 and National Science Foundation Grant MCB-0743422 (to B.G.M.E.). C.A.C. was supported by a predoctoral fellowship from the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805113105/DCSupplemental.
References
- 1.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 2.Perutz MF, Kendrew JC, Watson HC. Structure and function of haemoglobin: II. Some relations betwen polypeptide chain configuration and amio acid sequence. J Mol Biol. 1965;13:669–678. [Google Scholar]
- 3.Kendrew JC, et al. A partial determination by x-ray methods and its correlation with chemical data. Nature. 1961;190:666–670. doi: 10.1038/190666a0. [DOI] [PubMed] [Google Scholar]
- 4.Balaji S, Aruna S, Srinivasan N. Tolerance of the substitution of buried apolar residues by charged residues in the homologous protein structures. Proteins Struct Funct Genet. 2003;53:783–791. doi: 10.1002/prot.10416. [DOI] [PubMed] [Google Scholar]
- 5.Kajander T, et al. Buried charged surface in proteins. Struct Fold Des. 2000;8:1203–1214. doi: 10.1016/s0969-2126(00)00520-7. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Mao J, Gunner MR. Are acidic and basic groups buried in proteins predicted to be ionized? J Mol Biol. 2005;348:1283–1298. doi: 10.1016/j.jmb.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 7.Perutz MF. What are enzyme structures telling us? Faraday Discuss. 1992;93:1–11. doi: 10.1039/fd9929300001. [DOI] [PubMed] [Google Scholar]
- 8.Perutz MF. Electrostatic Effects in proteins. Science. 1978;201:1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- 9.Harvey SC, Hoekstra P. Dielectric relaxation spectra of water adsorbed on lysozyme. J Phys Chem. 1972;76:1987–2994. doi: 10.1021/j100665a011. [DOI] [PubMed] [Google Scholar]
- 10.Bone S, Pethig R. Dielectric studies of protein hydration and hydration-induced flexibility. J Mol Biol. 1985;181:323–326. doi: 10.1016/0022-2836(85)90096-8. [DOI] [PubMed] [Google Scholar]
- 11.Dwyer J, et al. High apparent dielectric constants in the interior of a protein reflect water penetration. Biophys J. 2000;79:1610–1620. doi: 10.1016/S0006-3495(00)76411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karp DA, et al. High apparent dielectric constant inside a protein reflects structural reorganization coupled to the ionization of an internal Asp. Biophys J. 2007;92:2041–2053. doi: 10.1529/biophysj.106.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stites WE, Gittis AG, Lattman EE, Shortle D. In a staphylococcal nuclease mutant the side-chain of a lysine replacing valine 66 is fully buried in the hydrophobic core. J Mol Biol. 1991;221:7–14. doi: 10.1016/0022-2836(91)80195-z. [DOI] [PubMed] [Google Scholar]
- 14.Schutz CN, Warshel A. What are the dielectric “constants” of proteins and how to validate electrostatic models? Proteins Struct Funct Genet. 2001;44:400–417. doi: 10.1002/prot.1106. [DOI] [PubMed] [Google Scholar]
- 15.Varadajaran R, Sewert RT, Gray HB, Boxer SG. Effects of buried ionizable amino acids on the reduction potential of recombinant myoglobin. Science. 1989;243:69–72. doi: 10.1126/science.2563171. [DOI] [PubMed] [Google Scholar]
- 16.Churg AK, Warshel A. Control of the redox potential of cytochrome-C and microscopic dielectric effects in proteins. Biochemistry. 1986;25:1675–1681. doi: 10.1021/bi00355a035. [DOI] [PubMed] [Google Scholar]
- 17.Luecke H, Richter HT, Lanyi JK. Proton transfer pathways in bacteriorhodopsin at 2.3 angstrom resolution. Science. 1989;280:1934–1937. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- 18.Yoshikawa S, et al. Redox-coupled crystal structural changes in bovine heart cytochrome-C oxidase. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 19.Doyle DA, et al. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 20.Luecke H, Lanyi JK. Structural clues to the mechanism of ion pumping in bacteriorhodopsin. Adv Protein Chem. 2003;63:115–130. doi: 10.1016/s0065-3233(03)63005-6. [DOI] [PubMed] [Google Scholar]
- 21.Gennis RB. How does cytochrome oxidase pump protons? Proc Natl Acad Sci USA. 1998;95:12747–12749. doi: 10.1073/pnas.95.22.12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitch CA, et al. Experimental pKa values of buried residues: Analysis with continuum methods and role of water penetration. Biophys J. 2002;82:3289–3304. doi: 10.1016/s0006-3495(02)75670-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Moreno E. B, et al. Experimental measurement of the effective dielectric in the hydrophobic core of a protein. Biophys Chem. 1997;64:211–224. doi: 10.1016/s0301-4622(96)02238-7. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen DM, Reynald RL, Gittis AG, Lattman EE. X-ray and thermodynamic studies of staphylococcal nuclease variants I92E and I92K: Insights into polarity of the protein interior. J Mol Biol. 2004:565–574. doi: 10.1016/j.jmb.2004.05.066. [DOI] [PubMed] [Google Scholar]
- 25.Giletto A, Pace C. Buried, charged, non-ion-paired aspartic acid 76 contributes favorably to the conformational stability of ribonuclease T1. Biochemistry. 1999;38:13379–13384. doi: 10.1021/bi991422s. [DOI] [PubMed] [Google Scholar]
- 26.Bajaj K, Chakrabarti P, Varadajaran R. Mutagenesis-based defitinions and probes of residue burial in proteins. Proc Natl Acad Sci USA. 2005;102:16221–16226. doi: 10.1073/pnas.0505089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warshel A. Energetics of enzyme catalysis. Proc Natl Acad Sci USA. 1978;75:5250–5254. doi: 10.1073/pnas.75.11.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sussman F, Warshel A. Ion-pairs in the interiors of proteins are stable only in polar sites. Biophys J. 1986;49:A291. [Google Scholar]
- 29.Warshel A, Aqvist J, Creighton S. Enzymes work by solvation substitution rather than by desolvation. Proc Natl Acad Sci USA. 1989;86:5820–5824. doi: 10.1073/pnas.86.15.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber DJ, Serpersu EH, Shortle D, Mildvan AS. Diverse interactions between the individual mutations in a double mutant at the active site of staphylococcal nuclease. Biochemistry. 1990;29:8632–8642. doi: 10.1021/bi00489a020. [DOI] [PubMed] [Google Scholar]
- 31.Hollien J, Marqusee S. A thermodynamic comparison of mesophilic and thermophilic ribonucleases H. Biochemistry. 1999;38:3831–3836. doi: 10.1021/bi982684h. [DOI] [PubMed] [Google Scholar]
- 32.Loll PJ, Lattman EE. The crystal structure of the ternary complex of staphylococcal nuclease, Ca2+, and the inhibitor pdTp, refined at 1.65 Å. Proteins Struct Funct Genet. 1989;5:183–201. doi: 10.1002/prot.340050302. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa K, et al. Crystal structure of ribonuclease H from Thermus thermophilus HB8 refined at 2.8 A resolution. J Mol Biol. 1993;230:529–542. doi: 10.1006/jmbi.1993.1169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.