Abstract
Bacteria thrive on and within the human body. One of the largest human-associated microbial habitats is the skin surface, which harbors large numbers of bacteria that can have important effects on health. We examined the palmar surfaces of the dominant and nondominant hands of 51 healthy young adult volunteers to characterize bacterial diversity on hands and to assess its variability within and between individuals. We used a novel pyrosequencing-based method that allowed us to survey hand surface bacterial communities at an unprecedented level of detail. The diversity of skin-associated bacterial communities was surprisingly high; a typical hand surface harbored >150 unique species-level bacterial phylotypes, and we identified a total of 4,742 unique phylotypes across all of the hands examined. Although there was a core set of bacterial taxa commonly found on the palm surface, we observed pronounced intra- and interpersonal variation in bacterial community composition: hands from the same individual shared only 17% of their phylotypes, with different individuals sharing only 13%. Women had significantly higher diversity than men, and community composition was significantly affected by handedness, time since last hand washing, and an individual's sex. The variation within and between individuals in microbial ecology illustrated by this study emphasizes the challenges inherent in defining what constitutes a “healthy” bacterial community; addressing these challenges will be critical for the International Human Microbiome Project.
Keywords: human microbiome, pyrosequencing, skin bacteria
Bacteria thrive on and within the human body, with recent work revealing vast diversity in several human-associated bacterial communities (1, 2). One of the largest human-associated microbial habitats is the skin, a body habitat with complex regional variations in cellular architecture and environmental exposures, where bacterial density may be as high as 107 cells per square centimeter (3). Many of these bacteria are not simply passive or transient colonizers of the skin surface, but rather appear to be adapted to the specific rigors associated with living in different regions of the skin including frequent skin shedding, antimicrobial host defenses, exposure to soaps and detergents during washing, exposure to UV radiation, and low moisture availability (4, 5).
Those bacterial communities that reside on the skin surface appear to be diverse (6, 7), but the full extent of bacterial diversity has not been adequately determined. Likewise, both culture-based and molecular approaches have shown that there may be a core set of bacterial taxa commonly found on skin surfaces (4–6, 8), but there appears to be a significant amount of intra- and interindividual variability in the composition of skin-associated bacterial communities (6, 7). Currently, the factors driving this variability in skin bacterial community composition are not well understood.
Although bacteria are common on all skin surfaces, we focused on bacteria found on the palm because it is likely one of the more dynamic skin microbial habitats given the nearly constant and varied exposure to environmental surfaces and the frequency of perturbations caused by hand washing. In addition, pathogens may inhabit the palmar surface, and efforts to reduce disease transmission by hand washing are a key public health concern (9–11).
We surveyed the bacterial communities found on the palm surfaces of both the dominant and nondominant hands of 51 undergraduate students sampled after taking an examination. Our goal was to assess the intra- and interindividual variability in skin-associated bacterial communities and determine how specific factors (including sex, handedness, and time since last hand washing) may influence the diversity and composition of the bacterial communities. The 16S rRNA genes from the palmar surface bacteria were PCR-amplified by using a universal bacterial primer set with a unique error-correcting barcode for each sample, allowing us to analyze all of the amplified samples in a single pyrosequencing run (12). We extended this technique using Golay codes, which provide a greater degree of error correction than the Hamming codes used in the previous study, allowing us to correct any triple-bit error and detect any quadruple-bit error (versus single-bit correction and double-bit detection in the Hamming codes). Coupling this barcoding technique with the high-throughput capabilities of pyrosequencing, we were able to survey the bacterial communities on each of the swabbed hands at an unprecedented level of detail.
Results and Discussion
After removing sequences of insufficient quality and sequences that could not be adequately classified, nearly 332,000 sequences remained with an average of >3,200 sequences obtained for each of the 102 palm surfaces swabbed (Table 1). For comparison, the total number of sequences included in this study exceeds the total number of sequences obtained from the largest previously published molecular surveys of skin bacterial communities (6, 7) by nearly 2 orders of magnitude. This dataset also provided the most comprehensive survey of bacterial diversity in any human-associated habitat to date.
Table 1.
Summary description of the sampling effort, the number of sequences collected, and the levels of bacterial diversity discovered
| No. of hands sampled | Total no. of sequences | Average length of sequence reads, bp (range) | Total no. of classifiable bacterial sequences | Total no. of phylotypes across all hands sampled | Average no. of sequences per hand (range) | Average no. of phylotypes per hand (range) |
|---|---|---|---|---|---|---|
| 102 (from 27 men and 24 women) | 351,630 | 228 (200–267) | 331,619 | 4,742 | 3,251 (2,410–5,838) | 158 (46–401) |
Phylotypes were determined at the 97% sequence similarity level.
The average palm surface harbors >150 distinct species-level bacterial phylotypes [a species is defined here as organisms sharing ≥97% identity in their 16S rRNA gene sequences (13)] (Table 1). Not surprisingly, this number of unique phylotypes exceeds the number of bacterial types typically cultivated from the skin surface by at least an order of magnitude (8), confirming that culture-based surveys of the skin surface, like surveys conducted in many other microbial habitats (14), dramatically underestimate the full extent of bacterial diversity. The average phylotype richness observed on a single palm surface was also >3 times higher than the richness observed in a molecular survey of forearm skin (6) and elbow skin (7). Although we would expect the hand surface to have higher levels of diversity than other skin surfaces because of the more frequent contact with potential inocula from the environment, this discrepancy in observed bacterial diversity is more likely a result of the depth of our sampling, which allowed us to survey even those rare bacterial taxa present on the skin surface. However, despite the depth of our surveys, our diversity estimates still represent only the lower bounds of phylotype richness on individual hands; the rarefaction curves for individual palm surfaces do not asymptote [supporting information (SI) Fig. S1], indicating that the true diversity is likely even higher. The total diversity of bacteria on the hand surface appears to match or exceed the levels of bacterial diversity found in other human-associated microbial habitats, including the esophagus, the mouth, and at specific sites within the lower intestine (15–17), but this may be a function of the depth of our sequencing. If we compare our results with those obtained by Andersson et al. (18) where a similar pyrosequencing-based approach was used to survey human-associated bacterial communities, we find that skin bacterial communities appear to be more diverse on average than those communities found in throat, stomach, and fecal environments.
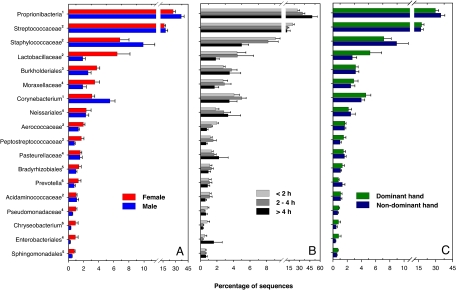
Although diversity on palm surfaces is high at both the phylotype and phylum levels (sequences from >25 phyla were detected), 3 phyla (Actinobacteria, Firmicutes, and Proteobacteria) accounted for 94% of the sequences (Fig. 1 and Table S1). The most abundant genera (Proprionibacterium, 31.6% of all sequences; Streptococcus, 17.2%; Staphylococcus, 8.3%; Corynebacterium, 4.3%; and Lactobacillus, 3.1%) were found on nearly all palm surfaces sampled. These genera have previously been found to be abundant in other molecular surveys of skin bacteria (6, 19) and are considered to be common skin residents (5), yet they still represented <65% of all of the identified sequences (Fig. 1 and Table S1). The average palm surface has a large number of rare taxa that may be either transient, short-term colonizers of skin or more persistent, longer-term residents of the skin surface that are simply present at relatively low abundances or whose abundance is determined by specific characteristics of individual hand surfaces.
Fig. 1.
Relative abundances of the most abundant bacterial groups on the hand surfaces, with the hand samples divided into categories of sex (A), time since last hand washing (B), and the dominant versus the nondominant hand (C). Error bars are 1 standard error of the mean. For the number of sequences and number of samples included in each category and the full taxonomic description of the hand surface bacterial communities see Table S1. Superscripts on the taxon name indicate the phylum or subphylum: 1, Actinobacteria; 2, Firmicutes; 3, Betaproteobacteria; 4, Gammaproteobacteria; 5, Alphaproteobacteria; 6, Bacteroidetes.
Qualitatively, the bacterial communities found on the hand surfaces (Fig. 1 and Table S1) appear to be more similar to the communities found on forearm skin (6) than to the communities found on the forehead (19) or inner elbow (7), suggesting that skin bacterial communities are not uniform across the body and that skin surfaces closer in proximity may harbor more similar bacterial communities. Additional research mapping the distribution of bacterial taxa across a wide range of skin surfaces would allow us to specifically test this hypothesis.
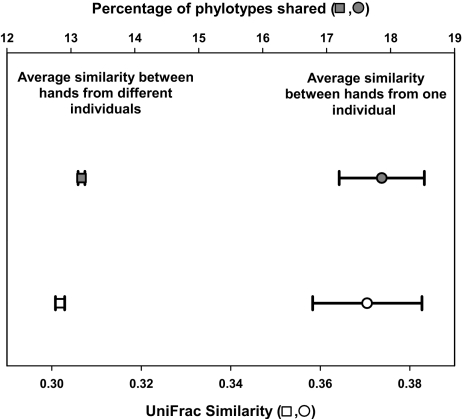
Although some bacterial taxa were cosmopolitan and were found on essentially all of the hand surfaces sampled, bacterial communities on individual hand surfaces were strikingly different. We observed a total of 4,742 distinct bacterial phylotypes across the 102 palm surfaces sampled (Table 1), and only 5 phylotypes were shared across all of the hands sampled. On average, the communities found on any pair of palm surfaces shared only 13% of their phylotypes (Fig. 2). The bacterial communities found on the skin surface, like those communities found in other human-associated microbial habitats (2, 15, 20, 21), exhibit an enormous amount of interindividual variability.
Fig. 2.
Average pairwise bacterial community similarity between left and right hands from the same individual (circles) and between hands from different individuals (squares) as measured by using the unweighted UniFrac similarity index (bottom axis, open symbols) or the percentage of phylotypes that are shared between pairs (top axis, filled symbols). Average pairwise values and 95% confidence intervals are shown. For these analyses, 2,500 sequences were randomly selected per sample, and only those samples represented by >2,500 sequences were included (n = 51 and 5,100 pairwise comparisons for intraindividual comparison and interindividual comparisons, respectively).
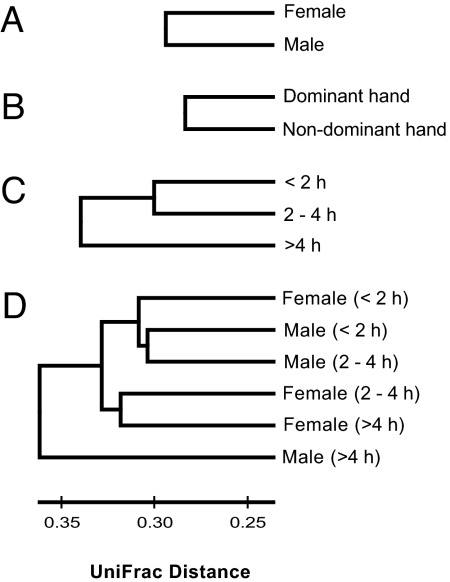
The observed differentiation in bacterial communities between hand surfaces is not determined solely by stochastic factors. For example, handedness has a significant influence on bacterial communities (P < 0.001). Dominant hands (i.e., the right hand on right-handed individuals) have similar overall levels of diversity as nondominant hands (Fig. S1), but the composition of the bacterial communities on the dominant and nondominant hands from the same individual was significantly different (Fig. 3). Taxa with relative abundances >50% greater on the dominant hand than the nondominant hand included members of the Enterobacteriales, Lactobacillaceae, Peptostreptococcaceae, and Xanthomonadales groups (Fig. 1 and Table S1). The influence of handedness on palm bacterial communities is likely due either to differences in skin environmental conditions (e.g., sebum production, salinity, hydration) or to the dominant hand coming into contact with different types of environmental surfaces than the nondominant hand. Although dominant and nondominant hands harbor distinct bacterial communities, the communities on left and right hands from the same individual were more similar than we would expect by chance (Fig. 2). However, these communities still shared only 17% of their phylotypes on average, indicating that there is an enormous amount of heterogeneity in skin bacterial communities within an individual. This intraindividual differentiation between the bacterial communities on left and right hands was not significantly affected by handedness, sex, or hand hygiene (P > 0.05 in all cases).
Fig. 3.
Differentiation in hand-surface communities between sexes (A), dominant versus the nondominant hands (B), time since last hand washing (C), and time since last hand washing for each sex (D) determined by using the unweighted UniFrac algorithm. The length of the branches corresponds to the degree of differentiation between bacterial communities in each category. All of the branch nodes shown here were found to be significant (P < 0.001), indicating that each of these categories harbored distinct bacterial communities.
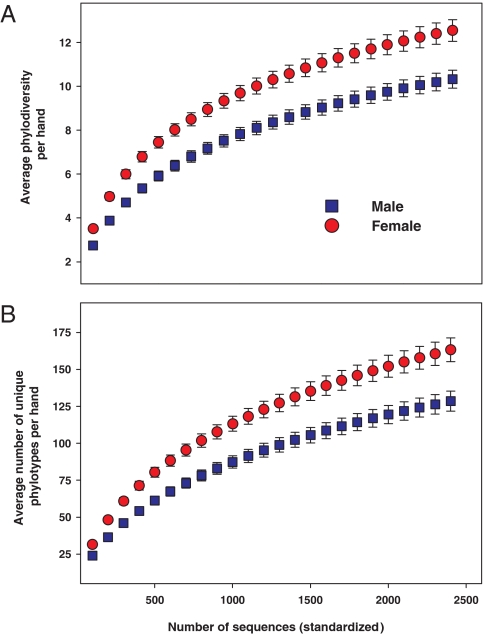
Men and women harbor significantly different bacterial communities on their hand surfaces (P < 0.001; Fig. 3). Taxa that were shared by both men and women but were more abundant on the skin of 1 sex included members of the following groups: Proprionibacterium (37% more abundant on men), Corynebacterium (80% more abundant on men), Enterobacteriales (400% more abundant on women), Moraxellaceae (180% more abundant on women), Lactobacillaceae (340% more abundant on women), and the Pseudomonadaceae (180% more abundant on women) (Fig. 1 and Table S1). Interestingly, the palms of women were also found to harbor significantly greater bacterial diversity than those of men, whether diversity was assessed by examining the overall phylogenetic structure on each hand (Fig. 4A) or the average number of phylotypes per hand (Fig. 4B). We do not know what drives these differences in overall diversity, but differences in skin pH may be influential. Men generally have more acidic skin than women (22, 23), and work from other microbial habitats has shown that microbial diversity is often lower in more acidic environments (24–26). Other explanations for why men and women appear to harbor distinct hand bacterial communities may include differences in sweat or sebum production, frequency of moisturizer or cosmetics application, skin thickness, or hormone production (4, 23). Without detailed information on the skin characteristics of the individuals sampled for this study, we can only speculate on the causes of the apparent sex differences in hand bacterial communities. Additional studies are required to determine whether differences in skin pH, or other factors, are directly related to the apparent sex differences in skin bacterial communities.
Fig. 4.
Rarefaction curves showing differences in bacterial diversity on palm surfaces from men and women. (A) Phylogenetic diversity estimated by measuring the average total branch length per sample after a specified number of individual sequences have been observed (36). (B) Diversity estimated by determining the average number of unique phylotypes per hand. For these analyses, we randomly selected 2,400 sequences per hand sample, and thus the average number of phylotypes per hand is lower than for the full dataset (Table 1). Bars indicate 95% confidence intervals.
Time since last hand washing also had a significant effect on skin community composition (P < 0.001), and this effect was slightly more pronounced than the sex differences (Fig. 3). Most notably, bacteria belonging to the Proprionibacteria, Neisseriales, Burkholderiales, and Pasteurellaceae taxa were relatively more abundant with time since last hand washing, whereas other bacteria in the Staphylococcaceae, Streptococcaceae, and Lactobacillaceae groups showed the opposite pattern and were relatively more abundant on hands that had been recently washed (Fig. 1 and Table S1). Although hand washing altered community composition, overall levels of bacterial diversity were unrelated to time since last hand washing (Fig. S1). Either the bacterial communities rapidly reestablish after hand washing, or washing (as practiced by the students included in this study) does not remove the majority of the bacterial taxa found on the skin surface.
The observed differences in skin bacterial communities between men and women may be partly related to sex differences in washing frequency because women reported having washed their hands more recently than men (Table S1). However, disparities in hygiene cannot explain all of the sex differences between men and women, as some of the taxa that were more abundant (or did not change appreciably in abundance) with time since last hand washing were less abundant on men than on women (Fig. 1). Likewise, even if we separate out the samples by both sex and by hand-washing categories, we still observe significant differences between the sexes (Fig. 3D).
To further resolve the effects of sex and hand washing on the palm bacterial communities, we conducted a smaller study of 4 men and 4 women to explicitly examine the temporal dynamics of skin bacterial communities after hand washing. This study was more controlled because we did not rely on a self-reported estimate of the time since last hand washing, but swabbed the palms of each of these 8 individuals every 2 h for a 6-h period after hand washing. Also, unlike the larger study, we used only 1 swab to sample the communities on both the left and right hands of each individual, and the volunteers were sampled during a normal work day, not immediately after taking an examination where student anxiety may have influenced their bacterial communities. However, we found very similar patterns in the 2 different studies. Specifically, we found that changes in bacterial community composition with time since last hand washing were significant and nearly identical to those described above for the larger study (Table S2 and Fig. S2). We also confirmed that men and women harbor distinct bacterial communities, even when controlling for hand hygiene, and that these differences between the sexes become more apparent with time since hand washing (Fig. S3). Likewise, we confirmed that women do harbor higher levels of bacterial diversity on their hands than men (Fig. S4).
Together these results demonstrate the utility of using new sequencing technologies to survey microbial communities at an unprecedented level of detail. There appears to be a core set of phylotypes present on the skin of the adult human palm, and the genomes of representatives of these organisms should be prioritized for sequencing to make sense of deeper metagenomic studies. However, the noncore phylotypes appear to exhibit a “long tail” effect—most phylotypes are rare—suggesting that exhaustive sampling is not a reasonable goal. Furthermore, the significant heterogeneity in community composition between left and right hands from the same individual suggests that careful sampling strategies will be required to obtain usable data for the International Human Microbiome Project. Determining the relative numbers of core and noncore lineages in different skin habitats, their variability, and the relationships between intrinsic physiological or consistent physical states (e.g., sex, handedness) and external environmental characteristics or behaviors (e.g., hand washing) is critical for establishing a healthy baseline from which to detect and understand microbial community differences associated with a wide variety of human diseases.
Methods
Sample Collection.
Approximately 85 undergraduate students were asked to participate in this study over a 1-h period in November 2007 after the students exited a room where they had all spent the previous hour taking an examination. Of the 85 students approached 51 volunteered, and samples were collected from the palm surfaces of these students. Each subject provided information on their handedness and the time since last hand washing. All individuals were made aware of the nature of the experiment and gave verbal informed consent to participate in accordance with the sampling protocol approved by the University of Colorado Human Research Committee (protocol 1007.39). The palm surfaces of both hands were swabbed separately (102 samples total) with cotton tipped swabs moistened with solution of 0.15 M NaCl and 0.1% Tween 20 (27). Swabbing has previously been shown to be as effective as other skin sampling methods for surveying bacterial diversity (7). The entire palm surface was swabbed in 2 perpendicular directions to ensure that the maximum surface area of each palm was represented in the sample. A fresh pair of sterile gloves was worn by the person sampling each individual palm surface to minimize sample cross-contamination. Sample blanks consisted of swabs that had been moistened and placed directly in 15-mL polypropylene tubes. The tubes were stored at −20 °C for <72 h before DNA extraction.
A smaller-scale study focusing on the effects of hand washing was conducted in April 2008 by sampling the palm surfaces from 8 individuals (4 men and 4 women). Each individual washed his/her hands for 30 s with a standard bar of antibacterial-free soap (Ivory; Procter & Gamble) followed by rinsing with tap water and drying with paper towels. Immediately after the hand washing and every 2 h over a 6-h period, palm surfaces were swabbed in the exact same manner as described above, except that both left and right hands from each individual were swabbed with the same cotton swab. DNA extraction, amplification, and pyrosequencing were conducted in the same manner for all of the swabs collected from this study and the larger-scale study.
DNA Extraction.
DNA was extracted from the swabs by using the Mobio UltraClean Plant DNA Isolation Kit (Mobio Laboratories) with modifications. The cotton tip of each swab was broken off directly into a bead tube to which 60 μL of Solution P1 had been added. Care was taken not to touch the tip of the swab to any surface except the inside of the 15-mL storage tube or the bead tube. The bead tubes were capped and heated to 65 °C for 10 min and then shaken horizontally for 2 min at maximum speed with the Mobio vortex adapter. The remaining steps were performed as directed by the manufacturer. DNA samples were stored at −20 °C until needed.
PCR Amplification and Sample Pooling.
For each sample, we amplified the 16S rRNA gene using a primer set similar to that described in Hamady et al. (12) that was found to be well-suited for the phylogenetic analysis of pyrosequencing reads (28). The forward primer (5′-GCCTTGCCAGCCCGCTCAGTCAGAGTTTGATCCTGGCTCAG-3′) contained the 454 Life Sciences primer B, the broadly conserved bacterial primer 27F, and a 2-base linker sequence (“TC”). The reverse primer (5′-GCCTCCCTCGCGCCATCAGNNNNNNNNNNNNCATGCTGCCTCCCGTAGGAGT-3′) contained the 454 Life Sciences primer A, the bacterial primer 338R, a “CA” inserted as a linker between the barcode and the rRNA primer, and a unique 12-bp error-correcting Golay barcode used to tag each PCR product (designated by NNNNNNNNNNNN; see Table S3). PCRs consisted of 0.25 μL (30 μM) of each forward and reverse primer, 3 μL of template DNA, and 22.5 μL of Platinum PCR SuperMix (Invitrogen). Samples were initially denatured at 94 °C for 3 min, then amplified by using 35 cycles of 94 °C for 45 s, 50 °C for 30 s, and 72 °C for 90 s. A final extension of 10 min at 72 °C was added at the end of the program to ensure complete amplification of the target region. All samples were amplified in triplicate. Negative controls (both no-template and template from unused swabs) were included in all steps of the process to check for primer or sample DNA contamination. All aliquoting and diluting of primers, as well as assembly of PCRs, were done in a PCR hood in which all surfaces and pipettors had been decontaminated with DNA Away (Molecular BioProducts) and exposed to UV light for 30 min.
A composite sample for pyrosequencing was prepared by pooling approximately equal amounts of PCR amplicons from each sample. The replicate PCRs for each sample were combined and cleaned with the Mobio UltraClean-htp PCR Clean-up kit (Mobio Laboratories) as directed by the manufacturer. Each sample (3 μL) was then quantified by using PicoGreen dsDNA reagent (Invitrogen) in 1× Tris-EDTA (pH 8.2) in a total volume of 200 μL on black, 96-well microtiter plates on a BioTek Synergy HTP microplate reader (BioTek Instruments) using the 480/520-nm excitation and emission filter pair. Once quantified, the appropriate volume of the cleaned PCR amplicons was combined in a sterile, 50-mL polypropylene tube and precipitated on ice with sterile 5 M NaCl (0.2 M final concentration) and 2 volumes of ice-cold 100% ethanol for 45 min. The precipitated DNA was centrifuged at 7,800 × g for 40 min at 4 °C, and the resulting pellet was washed with an equal volume of 70% ethanol and centrifuged again at 7,800 × g for 20 min at 4 °C. The supernatant was removed, and the pellet was air-dried for 7 min at room temperature, then resuspended in 100 μL of DNA-nuclease free water. The sample was sent to the Environmental Genomics Core Facility at the University of South Carolina (Columbia) for pyrosequencing on a 454 Life Sciences Genome Sequencer FLX (Roche) machine.
Phylogenetic Analyses.
Sequences were processed and analyzed following the procedure described in Hamady et al. (12). Only those sequences >200 bp in length with an average quality score >25 and no ambiguous characters were included in the analyses (29). Sequences were assigned to samples by examining the 12-bp barcode. Phylotypes were identified by using megablast to identify connected components (nearest neighbor) sets of similar sequences (parameters: E value, 1e-8; minimum coverage, 99%; minimum pairwise identity, 97%). A representative sequence was chosen from each phylotype by selecting the most highly connected sequence, i.e., the sequence that had the most hits more significant than the BLAST threshold to other sequences in the dataset (12). The set of all representative sequences was aligned by using NAST (30) (parameters: minimum alignment length, 190; sequence identity, 70%) with a PH lanemask (http://greengenes.lbl.gov/) to screen out hypervariable regions of the sequence. A relaxed neighbor-joining tree was built by using Clearcut (31), employing the Kimura correction. Unweighted UniFrac (32, 33) was run by using the resulting tree and the sequences annotated by environment type. Taxonomic identity of the phylotypes was assigned with BLAST against the Greengenes (34) database by using an E value cutoff of 1e-10 and the Hugenholtz taxonomy. The statistical significance of differences in microbial community composition between sample categories was determined by using the G test on relative phylotype abundances (35).
Supplementary Material
Acknowledgments.
We thank the undergraduate students for allowing us to sample their hands; J. Gordon, N. Pace, D. Nemergut, and E. Costello for helpful comments on the manuscript; J. Jones at the University of South Carolina Environmental Genomics Core Facility; and J. Zaneveld, M. Robeson, H. Hamilton, A. Vu, V. McKenzie, K. Ramirez, E. Costello, J. Widmann, R. Bowers, K. Morliengo-Bredlau, and A. Redford for their assistance with the sample collection. Analyses were run by using the Keck RNA Bioinformatics Facility at the University of Colorado. The work was supported by National Institutes of Health Molecular Biophysics Training Program Grant T32GM065103 (to M.H.), National Science Foundation East Asia and Pacific Summer Institutes Fellowship OISE0812861 (to M.H.), National Institutes of Health Grant P01DK078669 (to R.K.), and National Science Foundation Grant MCB0610970 (to N.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The bacterial 16S rDNA sequences reported in this paper have been deposited in the GenBank Short Read Archive (accession no. SRR006061.1).
This article contains supporting information online at www.pnas.org/cgi/content/full/0807920105/DCSupplemental.
References
- 1.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, et al. The Human Microbiome Project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fredricks DN. Microbial ecology of human skin in health and disease. J Invest Dermatol Symp Proc. 2001;6:167–169. doi: 10.1046/j.0022-202x.2001.00039.x. [DOI] [PubMed] [Google Scholar]
- 4.Roth RR, James WD. Microbial ecology of the skin. Annu Rev Microbiol. 1988;42:441–464. doi: 10.1146/annurev.mi.42.100188.002301. [DOI] [PubMed] [Google Scholar]
- 5.Cogen AL, Nizet V, Gallo RL. Skin microbiota: A source of disease or defence? Br J Dermatol. 2008;158:442–455. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Z, Tseng CH, Pei ZH, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grice E, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiller K, Selkin BA, Murakawa GJ. Skin microflora and bacterial infections of the skin. J Invest Dermatol Symp Proc. 2001;6:170–174. doi: 10.1046/j.0022-202x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 9.Pittet D, et al. Evidence-based model for hand transmission during patient care and the role of improved practices. Lancet Infect Dis. 2006;6:641–652. doi: 10.1016/S1473-3099(06)70600-4. [DOI] [PubMed] [Google Scholar]
- 10.Boyce JM, et al. Guideline for hand hygiene in health-care settings. Recommendations of the Healthcare Infection Control Practices Advisory Committee and the HICPAC/SHEA/APIC/IDSA Hand Hygiene Task Force. Am J Infect Control. 2002;30:S1–S46. doi: 10.1067/mic.2002.130391. [DOI] [PubMed] [Google Scholar]
- 11.Larson E. Hygiene of the skin: When is clean too clean? Emerging Infect Dis. 2001;7:225–229. doi: 10.3201/eid0702.010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamady M, Walker J, Harris J, Gold N, Knight R. Error-correcting barcoded primers allow hundreds of samples to be pyrosequenced in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stackebrandt E, Goebel BM. A place for DNA-DNA reassociation and 16S ribosomal-RNA sequence-analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 14.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–739. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 15.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei ZH, et al. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci USA. 2004;101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson AF, et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dekio I, et al. Detection of potentially novel bacterial components of the human skin microbiota using culture-independent molecular profiling. J Med Microbiol. 2005;54:1231–1238. doi: 10.1099/jmm.0.46075-0. [DOI] [PubMed] [Google Scholar]
- 20.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology—Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 21.Zhou X, et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 2007;1:121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 22.Kim MK, et al. Evaluation of gender difference in skin type and pH. J Dermatol Sci. 2006;41:153–156. doi: 10.1016/j.jdermsci.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Dao H, Kazin RA. Gender differences in skin: A review of the literature. Gender Med. 2007;4:308–328. doi: 10.1016/s1550-8579(07)80061-1. [DOI] [PubMed] [Google Scholar]
- 24.Fierer N, Jackson R. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fierer N, Morse J, Berthrong S, Bernhardt ES, Jackson RB. Environmental controls on the landscape-scale biogeography of stream bacterial communities. Ecology. 2007;88:2162–2173. doi: 10.1890/06-1746.1. [DOI] [PubMed] [Google Scholar]
- 26.Lindstrom ES, Kamst-Van Agterveld MP, Zwart G. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl Environ Microbiol. 2005;71:8201–8206. doi: 10.1128/AEM.71.12.8201-8206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulino L, Tseng C, Strober B, Blaser M. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006;44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, Lozupone C, Hamady M, Bushman F, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huse SM, Huber JA, Morrison HG, Sogin ML, Mark Welch D. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:r143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeSantis T, et al. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheneman L, Evans J, Foster JA. Clearcut: A fast implementation of relaxed neighbor joining. Bioinformatics. 2006;22:2823–2824. doi: 10.1093/bioinformatics/btl478. [DOI] [PubMed] [Google Scholar]
- 32.Lozupone C, Hamady M, Knight R. UniFrac—An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sokal R, Rohlf F. Biometry: The Principles and Practice of Statistics in Biological Research. New York: Freeman; 1994. p. 880. [Google Scholar]
- 36.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.