Abstract
Ribonucleotide reductase provides deoxynucleotides for nuclear and mitochondrial (mt) DNA replication and repair. The mammalian enzyme consists of a catalytic (R1) and a radical-generating (R2 or p53R2) subunit. During S-phase, a R1/R2 complex is the major provider of deoxynucleotides. p53R2 is induced by p53 after DNA damage and was proposed to supply deoxynucleotides for DNA repair after translocating from the cytosol to the cell nucleus. Similarly R1 and R2 were claimed to move to the nucleus during S-phase to provide deoxynucleotides for DNA replication. These models suggest translocation of ribonucleotide reductase subunits as a regulatory mechanism. In quiescent cells that are devoid of R2, R1/p53R2 synthesizes deoxynucleotides also in the absence of DNA damage. Mutations in human p53R2 cause severe mitochondrial DNA depletion demonstrating a vital function for p53R2 different from DNA repair and cast doubt on a nuclear localization of the protein. Here we use three independent methods to localize R1, R2, and p53R2 in fibroblasts during cell proliferation and after DNA damage: Western blotting after separation of cytosol and nuclei; immunofluorescence in intact cells; and transfection with proteins carrying fluorescent tags. We thoroughly validate each method, especially the specificity of antibodies. We find in all cases that ribonucleotide reductase resides in the cytosol suggesting that the deoxynucleotides produced by the enzyme diffuse into the nucleus or are transported into mitochondria and supporting a primary function of p53R2 for mitochondrial DNA replication.
Keywords: DNA precursors, immunofluorescence, mitochondrial DNA, p53R2, subcellular localization
DNA replication and repair require a balanced supply of the four common deoxynucleoside triphosphates (dNTPs). In mammalian cells DNA synthesis occurs in two separate compartments: nucleus and mitochondria. The complete nuclear DNA is replicated only in cycling cells during S-phase, whereas cycling and quiescent cells replicate mitochondrial DNA and repair damaged DNA during their whole existence. Thus cycling cells require during a limited period a large supply of dNTPs in the nucleus. Outside S-phase cells consume much smaller amounts of dNTPs, mainly in the cytosol for mitochondrial (mt) DNA replication. In all cells the major supply of dNTPs comes from the de novo reduction of ribonucleoside diphosphates to deoxyribonucleoside diphosphates by the enzyme ribonucleotide reductase (RNR) (1).
In cycling cells, the dominant form of mammalian RNR consists of two proteins called R1 and R2. The activity of the R1/R2 enzyme is exquisitely regulated by allosteric mechanisms involving nucleoside triphosphates and also by S-phase-specific transcription and proteasome-mediated degradation of R2 in late mitosis (2). Thus postmitotic cells are completely devoid of protein R2. How do these cells synthesize dNTPs for mitochondrial DNA replication and DNA repair? Until recently the answer to this question was by salvage of deoxynucleosides but the picture changed suddendly with the discovery of a p53 inducible small RNR subunit, called p53R2 (3, 4). Mouse p53R2 displays 81% identity to mouse R2 at the amino acid level. It forms an active R1/p53R2 complex (5) but lacks the KEN box required for R2 degradation in late mitosis. On account of its p53-regulated expression, p53R2 was originally attributed the function of supplying dNTPs for DNA repair during the p53-orchestrated recovery of cells after DNA damage. The first publications on p53R2 reported a translocation from the cytosol to the nucleus in response to DNA damage (3, 6) supporting the idea that p53R2 provides cells with the precursors for DNA repair at the actual repair site. No corresponding nuclear translocation of the R1 subunit was reported (3) even though p53R2 in the absence of R1 is inactive. Furthermore, the amino acid sequence of p53R2 was proposed to contain putative nuclear localization signals (3). However, these sequences do not fulfill the requirements for a classical nuclear signal (7) and a similar sequence is present in the R2 protein.
The idea of a movement of RNR from the cytosol to the nucleus during DNA replication is not new. Also the canonical R1/R2 complex some time ago was suggested to undergo this transfer during S-phase (8). According to the “replitase” model RNR together with other enzymes of dNTP synthesis and DNA polymerase forms a large protein complex that at the site of DNA replication provides and directly uses dNTPs. Recent work introduced a more complicated version of the “replitase” model involving p53 (9). However, early immunochemical studies with highly specific monoclonal antibodies did not support this view (10, 11).
A common theme in the above models is that RNR is regulated by an additional mechanism besides allosteric control of activity and substrate specificity, cell-cycle related expression and protein R2 stability, i.e., translocation of subunits from the cytosol to the nucleus to deliver deoxynucleotides at the site of their use for DNA synthesis. Also in budding and fission yeast regulation by translocation was proposed, but by a mechanism that almost reverses the “replitase” model. During S phase and after DNA damage RNR activity would depend on the export of the small subunit from the nucleus to the cytosol where the large subunit is localized (12, 13). In fission yeast the low molecular weight inhibitor Spd1p would anchor the small subunit R2 in the nucleus. However, Spd1 has no affinity to R2 (Suc22p) but instead specifically binds and inhibits R1 (Cdc22p) (14). In budding yeast, the Wtm1 protein instead was reported to act as a nuclear anchor for the small subunit (15, 16).
Although originally considered an element of the DNA damage response, more recently p53R2 was found expressed in quiescent cells in the absence of DNA damage, at a level 30-fold lower than R2 in S phase (17). Outside S-phase the only active form of RNR is R1/p53R2 that, similar to the R1/R2 complex in cycling cells, supplies dNTPs for DNA synthesis in the nucleus and mitochondria. In quiescent human fibroblasts it catalyzes ribonucleotide reduction at 2 to 3% of the rate of R1/R2 in cycling cells (18) and together with deoxynucleoside kinases of the salvage pathway (19) provides dNTPs for DNA repair and mitochondrial DNA replication.
Whereas an involvement of p53R2 in DNA repair has been difficult to demonstrate unequivocally, the discovery that children with severe mitochondrial DNA depletions carry functionally important mutations in p53R2 (20) clearly demonstrates the importance of p53R2 for mitochondrial DNA replication. Therefore it is now established that in vivo p53R2, in conjunction with R1, is required for mitochondrial DNA synthesis in differentiated tissues.
Where in the cell is p53R2 itself located during catalysis? The purported nuclear translocation of p53R2 would speak in favor of a principal function during DNA repair. It is difficult to reconcile with a primary function for mitochondrial DNA replication. To find a solution to an overt contradiction we decided to reopen the question of the subcellular localization of all three subunits of RNR in control and DNA-damaged human and mouse cells. We used three independent methods. Firstly, cell fractionation and quantitation of R1, R2 and p53R2 in nuclei and cytosol by Western blotting with specific antibodies. Secondly we followed by immunofluorescence the localization of the three proteins during the cell cycle and after DNA damage. Thirdly, we transiently transfected COS-7 and NIH 3T3 cells with expression vectors encoding green fluorescent protein-tagged mouse R1 and DsRed-Monomer-tagged mouse p53R2 and then studied the subcellular localization of the fusion proteins by fluorescence microscopy before and after DNA damage. Irrespective of the used methodology, we could only observe a cytosolic localization of all three RNR subunits. Induction of DNA damage resulted in a large increase in cytosolic p53R2 but not in its translocation to the nucleus. The amount and localization of R1 was not affected. We therefore propose that p53R2 and R2 both form active RNR complexes with R1 in the cytosol and that the produced dNTPs are imported into the nucleus or the mitochondria for DNA synthesis. In S-phase cells, the RNR complex contains R1/R2 while non-proliferating cells use R1/p53R2.
Results
We used three different approaches to determine the distribution of RNR proteins between nuclei and cytosol: (i) quantification by Western blotting of the proteins in separated nuclei and cytosol; (ii) direct visualization in fixed cells by confocal fluorescence microscopy using specific antibodies; and (iii) visualization of the proteins carrying a fluorescent tag in transfected cells.
Quantification by Western Blotting.
R1, R2, and p53R2 are cytosolic proteins.
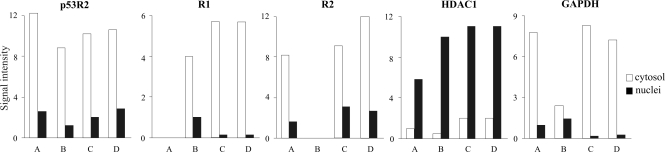
To minimize the risk of artifacts we used four different procedures to separate nuclei and cytosol as described in Materials and Methods. Our Methods A and B involved homogenization of the cells before differential centrifugation of the homogenate, whereas Methods C and D involved permeabilization of the cell membrane and removal of the cytosol from the nuclei by aspiration. After separation by denaturing gel electrophoresis of nuclear and cytosolic proteins we determined by Western blotting with specific antibodies R1, R2, p53R2, HDAC1 (histone deacetylase 1, a nuclear marker) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase, a cytosolic marker). Quantifications of the immunoblots showed that each RNR protein was almost exclusively located in the cytosol, irrespective of the method used for the separation of cytosol and nuclei and that the two marker proteins were recovered in the correct compartments (Fig. 1). Also Sp1, an additional nuclear marker, was largely found in the nuclear fraction (data not shown). These results strongly suggest that the three RNR subunits are localized in the cytosol. We obtained similar results with nuclei and cytosol from mouse 3T3 fibroblasts separated by method A (data not shown).
Fig. 1.
Localization of R1, R2, and p53R2 after cell fractionation. We separated cell nuclei and cytosol from human lung fibroblasts by four separate established methods: cell homogenization in isotonic buffer (method A) or in hypotonic buffer (method B), both followed by differential centrifugation; cell membrane permeabilization by digitonin (method C) or saponin (method D) followed by removal of the cytosolic fraction. Equal amounts of proteins from cytosol and nuclei were pairwise separated by gel electrophoresis and analyzed by Western blotting. The intensity of the bands corresponding to the three RNR proteins, the nuclear marker HDAC1 and the cytosolic marker GAPDH were quantified with Kodak one-dimensional image analysis software.
DNA damage does not translocate R1, R2, or p53R2 from cytosol to nucleus.
Treatment with adriamycin (0.1–0.5 μg/ml) during 24 h increased both p53 and p53R2 signals on Western blots more than 10-fold at the highest dose, whereas R1 remained essentially constant and R2 decreased in parallel with the exit of the cells from S-phase (data not shown). UV treatment (5–20 Joules/m2) was less effective, increasing p53R2 at most 3 times. To avoid extensive cell damage we decided to use 0.2 μg/ml of adriamycin or 5 Joules/m2 of UV for further experiments.
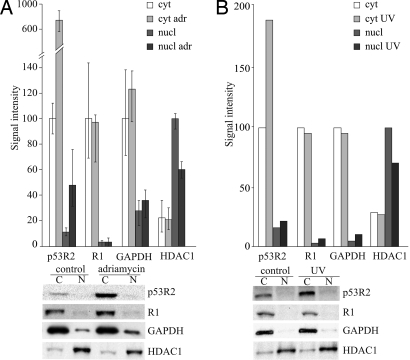
In the adriamycin experiments we used method A to separate cytosol and nuclei. The method involves the homogenization of cells by five expulsions through a fine needle. To investigate whether this procedure causes a major leakage of nuclear proteins we tested the effect of more extensive homogenization on the distribution of enzymes. Increasing the number of passages through the needle from 2 to 5 and 15 increased the percentage of the nuclear marker HDAC1 in the cytosol from 10 to 17 to 39 indicating progressive leakage from the nucleus. However, the values for the other enzymes did not change systematically suggesting that in their case no leakage had occurred. We show in Fig. 2A the average values from the three experiments for each protein with the error bars indicating the two extreme values. The most striking result is the 8-fold increase of p53R2 in the cytosol caused by adriamycin. The protein also increased in the nuclear fraction but the relative amount there remained low and actually decreased from 10 to 6% of the total. Also R1 and R2 (data not shown) remained in the cytosol. Adriamycin did not induce R1 whereas R2 was decreased due to a decline of S-phase cells from 33% to 20%. These data strongly speak against a translocation of any RNR subunit from the cytosol to the nucleus.
Fig. 2.
Effect of DNA damage on p53R2 and R1 in cytosol and nuclei. (A) After DNA damage with adriamycin we separated cytosol and nuclei from human lung fibroblasts by method A and quantitized by Western blotting the distribution of p53R2, R1 and marker proteins in the two compartments. Note the break in the scale of the ordinate. (B) We irradiated cells with UV light, separated after 24 h cytosol and nuclei by method C and determined p53R2, R1 and marker proteins in the two compartments. Quantification of proteins was as described for Fig. 1.
When we analyzed the consequences of DNA damage by UV we used method C to separate cytosol and nuclei. To validate the method, in a preliminary experiment we permeabilized control cells with 50 or 200 μM digitonin for 3, 10, or 20 min, removed the cytosol from the nuclei and determined R1, p53R2, GAPDH and HDAC1. On intensifying the digitonin treatment we found again a progressive leakage of HDAC1 without clear effects on the other proteins (data not shown). In the final UV experiment we treated the cells for 10 min with 50 μM digitonin. UV irradiation increased p53R2 almost 2-fold, but only in the cytosol, and did not affect the amount and distribution of the other proteins (Fig. 2B). After irradiation, 95% of R1 and 90% of p53R2 was in the cytosol and the two marker proteins were concentrated in the correct compartments with 77% of HDAC1 in nuclei and 90% of GAPDH in the cytosol. Thus UV gave rise to a smaller increase of p53R2 than adriamycin but the increase again occurred in the cytosol without transfer of the protein to the nucleus.
Confocal fluorescence microscopy after immunostaining.
Monoclonal antibodies against mouse R1 or R2 stain the cytosol of 3T3 cells.
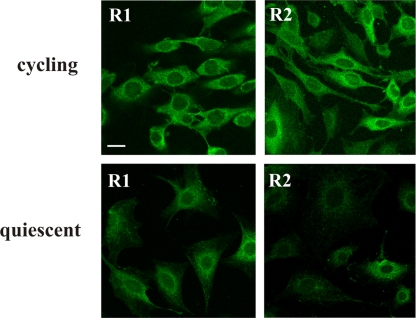
We used cultures containing either 8 or 70% S-phase cells to distinguish between different stages of the cell cycle. We used one monoclonal antibody for R1, two separate monoclonals for R2 (Fig. 3, only one R2 antibody is shown). Both proteins were visualized only in the cytosol. Cultures containing 8 or 70% S-phase cells behaved qualitatively similar. However, the fluorescence intensity of R2 was much stronger in the latter case (Fig. 3), as expected from the dependence of the cellular concentration of R2 on the cell cycle. R1 did not show this difference. We conclude that both R1 and R2 are located in the cytosol during different phases of the cell cycle.
Fig. 3.
Localization of R1 and R2 during cell growth. 3T3 cells were partially synchronized with serum and two populations with either 70% (“cycling”) or 8% (“quiescent”) cells in S-phase were immunostained with monoclonal antibodies against mouse R1 or R2. The two RNR subunits show in both instances a cytosolic localization. (Scale bar: 10 μm.)
Also p53R2 is located in the cytosol and does not move into nuclei after DNA damage.
To detect p53R2 by immunofluorescence we first used commercial polyclonal antibodies directed against peptide sequences of human p53R2 (provided by three different companies). When tested by Western blotting of an extract from human fibroblasts all antibody preparations gave a strong signal with a band located at the position of p53R2 but also reacted with other proteins revealing large variations in their specificity (data not shown). Even different lots of an antibody from the identical manufacturer varied greatly. In immunofluorescence experiments, one antibody did not stain the cells at all, the other two stained both nuclei and cytosol. The latter results were clearly at variance with our cell fractionation experiments.
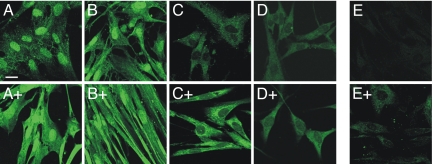
We therefore purified one of the antibodies by affinity chromatography on a column of immobilized p53R2, collected both the flow-through and eluted fractions and compared them to the original antibody preparation in immunofluorescence experiments with human lung fibroblasts. We tested cells under four different conditions (Fig. 4): (i) cycling control fibroblasts, (ii) cycling fibroblasts treated for 24 h with 0.2 μg/ml adriamycin, (iii) cycling fibroblasts irradiated with UV and (iv) fibroblasts depleted of p53R2 by siRNA transfection.
Fig. 4.
Specificity of p53R2 antibodies and effect of DNA damage on p53R2 localization. Human fibroblasts were stained with three different preparations of a commercial anti p53R2 antibody (ab8105) before or after DNA damage by adriamycin or UV (indicated by a + sign). A, antibody before affinity purification; B, not adsorbed fraction; C to E, fraction retained by the column. The antibody before purification and the not adsorbed fraction strongly stain cell nuclei, the fraction retained on the column stains only the cytosol. Adriamycin (+ in A, B, C) and UV treatment (+ in D) strongly increase the cytosolic fluorescence. E shows decreased cytosolic fluorescence in siRNA-silenced cells with (+) or without adriamycin treatment. (Scale bar: 10 μm.)
With the non-purified antibody cycling control cells showed a strong nuclear and a weaker cytosolic fluorescence (Fig. 4A). DNA damage by adriamycin increased the fluorescence, in particular in the cytosol (Fig. 4A+). The antibody that was not adsorbed by the affinity column behaved similarly with strong nuclear staining (Fig. 4B, B+). In contrast, two fractions obtained by elution from the affinity column only stained the cytosol of control cells (Fig. 4C, D). Also in this case, DNA damage by adriamycin (Fig. 4C+) or UV (Fig. 4D+) strongly increased the fluorescence, again only in the cytosol. The purified antibody showed a markedly decreased cytosolic fluorescence with cells treated with siRNAs against p53R2 indicating the specificity for p53R2 (Fig. 4E, E+). These data suggest that the nuclear staining by commercial anti-p53R2 antibodies originated from a contaminating antibody and that the cytosolic signal with the purified fraction was due to p53R2.
Transfection with R1 and p53R2 carrying a fluorescent tag.
Green fluorescent protein-tagged mouse R1 and DsRed -tagged mouse p53R2 localize to the cytosol.
Logarithmically growing COS-7 cells were transiently transfected with fluorescent protein-tagged mouse R1 or p53R2 proteins. Both the R1 and p53R2 fusion proteins showed a diffuse cytoplasmic localization and no nuclear translocation was observed after adriamycin treatment which resulted in a clear induction of the endogenous p53R2 (Fig. 5A-D). The same diffuse colocalization to the cytosol was observed independent of DNA damage after cotransfection with both plasmids in the same cells (Fig. 5E-F). The expression levels of the fusion proteins were monitored by Western blots of cell extracts (Fig. 5G). To decrease the overexpression of the fusion proteins, we transfected NIH 3T3 cells that lack the T antigen present in COS-7 cells. The NIH 3T3 cells could not be treated with adriamycin whose nuclear fluorescence interfered with the relatively weak DsRed emission. Instead we gave the cells a UV pulse which resulted in induction of the endogenous p53R2 (Fig. 6E). Again the same diffuse cytosolic localization of both the R1 and the p53R2 fusion proteins was observed with no nuclear relocalization after DNA damage (Fig. 6A-D).
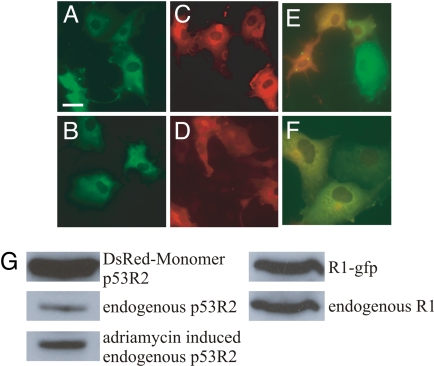
Fig. 5.
Subcellular localization of mouse R1 and p53R2 fusion proteins in transiently transfected COS-7 cells. (A and B) Localization pattern of green fluorescent protein-tagged R1 24 h after transfection. In B, adriamycin was present at 0.2 μg/ml during the last 12 h. (C and D) DsRed-Monomer-tagged p53R2 localization 24 h after transfection. In D, adriamycin was present during the last 12 h. E and F: Colocalization of green fluorescent protein-tagged R1 and DsRed-Monomer-tagged p53R2 24 h after the cotransfection. In F adriamycin was present during the last 12 h. (G) Western blots showing the endogenous and tagged protein levels in transiently transfected cells after 24 h. The level of adriamycin- induced endogenous p53R2 was measured 48 h after addition of the drug. By counting DAPI stained nuclei, the transfection efficiency for both fusion proteins was estimated to 20%. (Scale bar: 10 μm.)
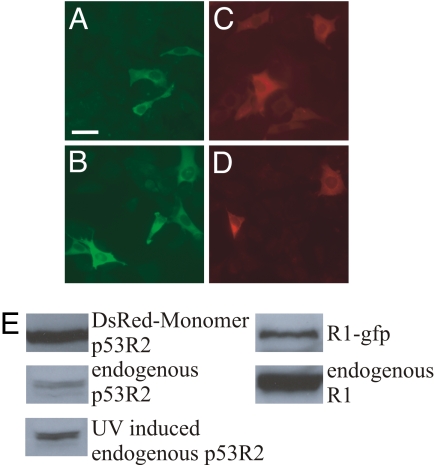
Fig. 6.
Mouse R1 and p53R2 subcellular localization in transiently transfected NIH 3T3 cells. (A and B) localization pattern of green fluorescent protein-tagged R1 24 h after transfection. In B the cells were UV irradiated 12 h posttransfection (254 nm, 10 J/m2). (C and D) DsRed-Monomer-tagged p53R2 localization 24 h after transfection. In D, cells received a UV pulse 12 h after transfection. (E) Western blots showing the endogenous and tagged protein levels in transiently transfected cells after 24 h. The levels of UV induced endogenous p53R2 was measured 48 h after irradiation. By counting DAPI stained nuclei the transfection efficiency was estimated to 9% for p53R2 and 7.5% for R1. (Scale bar: 10 μm.)
Discussion
All methods used to localize proteins to various subcellular compartments have pitfalls that need to be adressed. Fractionation of nuclei from cytosol followed by Western blotting is complicated by the possibility that the nuclear fraction includes cytosolic proteins due to incomplete cell breakage or that the cytosolic fraction contains nuclear proteins due to leakage from nuclei. The latter complication pertains especially to proteins that do not contain DNA-binding domains (21) and is particularly serious in our case when we report the absence of proteins from the nucleus. Immunofluorescence suffers from fixation artefacts and in particular from problems with antibody specificity. The latter point is amply demonstrated by our results with commercial p53R2 antibodies. All anti-p53R2 preparations contained unspecific antibodies and only after affinity purification we obtained a more specific antibody that no longer stained the nuclei. The third method, direct visualization by microscopy of proteins fused to fluorescent tags might be biased by the overexpression of the fused protein or altered intracellular trafficking due to the fusion tag.
We tried to substantiate each of the three methods in different ways. To recognize artefacts caused by nuclear leakage we applied four different methods to separate nuclei and cytosol and checked the amount of cross-contamination with marker proteins. The methods involved two separate principles which can be expected to be prone to different artefacts. With all four methods we found the three subunits of RNR in the cytosol. In two cases we intensified the treatment to increase the possibility of nuclear leakage. In both cases the nuclear marker but not the RNR proteins increased in the cytosol suggesting that the latter indeed are cytosolic proteins. For immunofluorescence we either used specific monoclonal antibodies or purified a commercial polyclonal antibody by affinity chromatography and tested separate fractions for activity. The specificity of the purified antibody was substantiated by a decrease of immunofluorescence in siRNA-treated cells and an increase after DNA damage. Transient transfection of COS-7 cells with expression vectors containing the SV40 origin resulted in high expression of fusion proteins which increased with the time of incubation. This made it easy to detect the fluorescence but especially with the DsRed-p53R2 fusion, the difference between the levels of the fusion protein and the endogenous p53R2 was quite high. However, in NIH 3T3 cells the fusion protein was expressed at levels more similar to that of the endogenous p53R2. Still, the results with both cell lines were the same.
Each method localized the three subunits of RNR to the cytosol and did not show migration of any of them to the nucleus, neither during DNA replication in S-phase, nor after DNA damage. With respect to R1 and R2, our results agree with earlier data from Engström and Rozell who reported their exclusive localization in the cytosol (11). They differ from two later reports that from experiments with polyclonal antibodies claimed that R1, R2 and p53R2 move from the cytosol to the nucleus after DNA damage (9) and during S-phase (22). We suggest that nuclear staining was caused by contaminating antibodies. A movement of p53R2 into the nucleus after DNA damage was also reported in the original publication on p53R2 (3) and further elaborated in a subsequent paper (6) from immunofluorescence and cell fractionation experiments. The specificity of the used polyclonal antibody or the quality of the cell fractionation was, however, not validated. Moreover the authors made extensive use of a purported DNA synthesis assay coupled to CDP reduction (8) to measure p53R2 activity. However, due to incomplete hydrolysis of RNA this method scores mainly incorporation of CDP into RNA (23). It is not a valid assay of ribonucleotide reduction and therefore it does not measure p53R2.
Our cells responded to DNA damage by inducing the endogenous p53R2 two to 8 fold (Fig. 2, 5G, 6E). The levels of induction were lower in mouse cells than in human cells and adriamycin was more efficient than UV irradiation. In all cases p53R2 as well as R1 and R2, when present in S and G2-phase cells, co-localized to the cytosol both before and after the induction of DNA damage. Thus our data do not support the suggestion that nuclear translocation is a new additional mechanism regulating ribonucleotide reduction in mammalian cells. The demonstration that the major function of p53R2 is to supply dNTPs for replication of mitochondrial DNA in postmitotic cells (20) has now shifted the focus from DNA repair to mitochondrial DNA synthesis. The new function of p53R2 makes its nuclear localization less likely and our present results indeed point in the same direction. Small molecules such as deoxynucleotides diffuse easily through the nuclear pores and their cytosolic synthesis should not hamper nuclear DNA polymerase activity. We conclude that ribonucleotide reduction takes place in the cytosol both by the R1/R2 and the R1/p53R2 complex. The resulting deoxyribonucleotides diffuse into the nucleus or are transported into mitochondria to support DNA replication and repair.
Materials and Methods
Cell lines, culture conditions, induction of DNA damage and siRNA treatment.
Human lung fibroblasts CCD-34Lu (CRL-1491), simian fibroblasts COS-7 (CRL-1651) and mouse fibroblasts NIH 3T3 (CRL-1658) were from the American Type Culture Collection and checked periodically for mycoplasma contamination. Cells were grown in high glucose (4.5g/l) Dulbecco's modified Eagle's medium with 10% FCS. 3T3 cells were synchronized by serum starvation followed by addition of 20% FCS (18, 24). We induced DNA damage by adding adriamycin (Sigma, Saint Louis, Mo) directly to the medium or by UV irradiation of the cell monolayer with a 254 nm UVS-11 Mineral light lamp at a fluence rate of 2 J/m2/sec after removal of the medium. To silence p53R2 in lung fibroblasts we transfected cells for 48 h with 20 nM siRNAs targeting p53R2 mRNA or with a control siRNA pool (both from Dharmacon, Chicago, IL) in fresh medium containing 0.4% DharmaFECT1. Some transfected cultures were treated with 0.2 μg/ml of adriamycin during the last 24 h. The targeting siRNAs decreased the amount of p53R2 to 27%, compared to the control. Adriamycin increased p53R2 five-fold in the control but gave no increase in the siRNA treated cells.
Antibodies.
Two rat monoclonal anti-R2 antibodies (JC4 and JB4) and one mouse monoclonal anti-R1 antibody (AD203) generated with homogeneous preparations of mouse R2 protein and calf thymus R1 protein, respectively, were available in one of our laboratories (25). Two polyclonal p53R2 rabbit antibodies (ab8105 and P49993) were from Abcam (Cambridge U.K.) and Sigma, respectively, one polyclonal p53R2 goat antibody (sc-10840) was from Santa Cruz Biotechnologies (Santa Cruz, CA). A monoclonal GAPDH antibody (MAB 372) was from Chemicon (Temecula, CA) and a polyclonal HDAC1 antibody (sc-7872) from Santa Cruz.
Affinity purification of p53R2 antibody.
After dialysis against 100 mM NaCO3/500 mM NaCl, pH 8.0, we attached 0.25 mg pure his-tagged recombinant human p53R2 (5) onto 0.1 g of CN-Br activated Sepharose resin (Sigma), washed and equilibrated the resin as recommended by the supplier and incubated it with 10 μg of p53R2 antibody (ab8105) for at least 1 h. We then transferred the slurry to a 2 ml column, collected the flow-through fraction and washed the resin with 10 ml 0.05% Tween in PBS buffer. We then eluted fractions from the resin with successive 0.3 ml portions of 100 mM glycine buffer, pH 2.5. Each emerging fraction was immediately neutralized with an equal amount of 100 mM Tris·HCl, pH 8.0, containing 1% BSA. The fractions were stored at + 4° and tested by Western blotting and immunochemistry. Material suitable for immunochemistry was usually in the second and third eluted fractions.
Separation of cytosol and nuclei and Western blotting of proteins.
For the separation of cytosol and nuclei we used altogether 4 different methods used and described in detail by various groups. In method A, developed in our laboratory (26), the cells are first scraped, then homogenized in an isotonic buffer by passing through a fine needle and finally centrifuged to separate cytosol and nuclei. Method B (6) is similar to method A but involves homogenization in a hypotonic buffer. It was used by the group that first described the movement of p53R2 into the nucleus. In methods C (27) and D (28) the cell membrane is permeabilized with digitonin or saponin, respectively, and the cytosol is removed by aspiration from the nuclei that remain attached to the dish. Thus in methods C and D the cells are not homogenized.
We separated the cytosolic and the nuclear (extracted by 1.5% SDS in PBS) proteins by gel electrophoresis on 9% denaturing gels, transferred them to Hybond-extra membranes and incubated the filters overnight at 4° with antibodies against the different proteins. After removal of the primary antibodies we incubated the filters with the appropriate horse-radish peroxidase conjugated secondary antibodies for 1 h at room temperature and quantified by Kodak one-dimensional image analysis software the bands stained with ECL-Advanced system (GE Healthcare, Milano, Italy).
Immunochemistry.
Cells were grown on coverslips for 3 days and, where indicated, treated with adriamycin or UV irradiation. After 24 h they were fixed in PBS containing 2% paraformaldehyde for 10 min and permeabilized with 0.2% Triton-X in PBS for 2 min at 4°C. The cells were blocked by gently rocking with 15% FCS in PBS at room temperature and incubated overnight with the primary antibody in humidified chambers. After washing with PBS (3 × 5 min) the cells were incubated with the appropriate secondary antibody conjugated to AlexaFluor 488 (Invitrogen, Carlsbad, CA) in 3% BSA/PBS for 1 h at room temperature. After three washings with PBS, the coverslips were mounted with UltraCruz mounting medium for fluorescence with DAPI (Santa Cruz) and analyzed with a Leica TCS SP5 confocal microscope using an immersion oil objective (63 X, N.A.1.40).
Cloning of green fluorescent protein tagged mouse R1 and DsRed-Monomer tagged mouse p53R2.
The cDNA of green fluorescent protein was excised from the pAcGFP1 vector (Clontech, Mountainview, CA) and ligated in frame to the 3′end of mouse R1 cDNA in the Okayama-Berg vector (29). To make the DsRed fusion, the cDNA of mouse p53R2 was excised from a pET3d vector (5) and ligated in frame to the 3′end of DsRed in the pDsRed-Monomer-C1 vector (Clontech).
Transient transfection of mammalian cells with fluorescent protein- tagged RNR subunits.
The DsRed-Monomer tagged mouse p53R2 and R1-green fluorescent protein fusion expression vectors were transiently transfected into COS-7 or NIH 3T3 cells. The cells were cultivated on 12 mm cover glasses (Menzel-Gläser # 0) in 10 cm cell culture plates and transfected with 8 μg DNA and 24 μl Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer. The transfected cells were fixed by 15 min incubation in 4% paraformaldehyde dissolved in PBS pH 7.0 (4°C) followed by 10 min incubation in PBS containing 0.5 μg/ml DAPI (Sigma). The cells were washed twice with PBS and finally rinsed in H2O and mounted on microscope slides in 2.5% 1,4-diazobicyclo-[2.2.2]-octane, 9% polyvinylalcohol, 23% glycerol, 0.09 M Tris·HCl pH 10 and incubated at 4°C overnight. The cells were analyzed using a Zeiss Imager Z1 microscope and AxioVision 4.6.3-SP1 software.
Acknowledgments.
This work was supported by grants from AIRC, the Italian Association for Cancer Research, Italian Telethon Grant GGP05001 and the Fondazione Cariparo (to V.B.) and from the Swedish Research Council (to L.T.) and the Kempe Foundation (to A.F.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Nordlund P, Reichard P. Ribonucleotide reduction. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 2.Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: A new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci USA. 2003;100:3925–3929. doi: 10.1073/pnas.0330774100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka H, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 4.Nakano K, Balint E, Ashcroft M, Vousden KH. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene. 2000;19:4283–4289. doi: 10.1038/sj.onc.1203774. [DOI] [PubMed] [Google Scholar]
- 5.Guittet O, et al. Mammalian p53R2 protein forms an active ribonucleotide reductase in vitro with the R1 protein, which is expressed both in resting cells in response to DNA damage and in proliferating cells. J Biol Chem. 2001;276:40647–40651. doi: 10.1074/jbc.M106088200. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi T, et al. p53R2-dependent pathway for DNA synthesis in a p53-regulated cell cycle checkpoint. Cancer Res. 2001;61:8256–8262. [PubMed] [Google Scholar]
- 7.Fontes MRM, Teh T, Jans D, Brinkworth RI, Kobe B. Structural basis for the specificity of bipartite nuclear localization sequence binding by Importin-α. J Biol Chem. 2003;278:27981–27987. doi: 10.1074/jbc.M303275200. [DOI] [PubMed] [Google Scholar]
- 8.Prem veer Reddy G, Pardee AB. Multienzyme complex for metabolic channeling in mammalian DNA replication. Proc Natl Acad Sci USA. 1980;77:3312–3316. doi: 10.1073/pnas.77.6.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue L, et al. Wild-type p53 regulates human ribonucleotide reductase by protein-protein interaction with p53R2 as well as hRRM2 subunits. Cancer Res. 2003;63:980–986. [PubMed] [Google Scholar]
- 10.Engström Y, Rozell B, Hansson HA, Stemme S, Thelander L. Localization of ribonucleotide reductase in mammalian cells. EMBO J. 1984;3:863–867. doi: 10.1002/j.1460-2075.1984.tb01897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engström Y, Rozell B. Immunocytochemical evidence for the cytoplasmic localization and differential expression during the cell cycle of the M1 and M2 subunits of mammalian ribonucleotide reductase. EMBO J. 1988;7:1615–1620. doi: 10.1002/j.1460-2075.1988.tb02987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao R, et al. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc Natl Acad Sci USA. 2003;100:6628–6633. doi: 10.1073/pnas.1131932100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C, et al. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 2003;17:1130–1140. doi: 10.1101/gad.1090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Håkansson P, Dahl L, Chilkova O, Domkin V, Thelander L. The Schizosaccharomyces pombe replication inhibitor Spd1 regulates ribonucleotide reductase activity and dNTPs by binding to the large Cdc22 subunit. J Biol Chem. 2006;281:1778–1783. doi: 10.1074/jbc.M511716200. [DOI] [PubMed] [Google Scholar]
- 15.Lee YD, Elledge SJ. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev. 2006;20:334–344. doi: 10.1101/gad.1380506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, An X, Yang K, Perlstein DL, Hicks L, et al. Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc Natl Acad Sci USA. 2006;103:1422–1427. doi: 10.1073/pnas.0510516103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Håkansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;28:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 18.Pontarin G, et al. p53R2-dependent ribonucleotide reduction provides deoxyribonucleotides in quiescent human fibroblasts in the absence of induced DNA damage. J Biol Chem. 2007;282:16820–16828. doi: 10.1074/jbc.M701310200. [DOI] [PubMed] [Google Scholar]
- 19.Eriksson S, Munch-Petersen B, Johansson K, Eklund H. Structure and function of cellular deoxyribonucleoside kinases. Cell Mol Life Sci. 2002;59:1327–1346. doi: 10.1007/s00018-002-8511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourdon A, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 21.Brini M, et al. Nuclear Ca2+ concentration measured with specifically targeted recombinant aequorin. EMBO J. 1993;12:4813–4819. doi: 10.1002/j.1460-2075.1993.tb06170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, et al. The ribonucleotide reductase subunit M2B subcellular localization and functional importance for DNA replication in physiological growth of KB cells. Biochem Pharmacol. 2005;70:1288–1297. doi: 10.1016/j.bcp.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Spyrou G, Reichard P. Ribonucleotides are not channeled into DNA in permeabilized mammalian cells. Biochem Biophys Res Commun. 1983;115:1022–1026. doi: 10.1016/s0006-291x(83)80037-0. [DOI] [PubMed] [Google Scholar]
- 24.Chabes A, Thelander L. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J Biol Chem. 2000;275:17747–17753. doi: 10.1074/jbc.M000799200. [DOI] [PubMed] [Google Scholar]
- 25.Engström Y. Monoclonal antibodies against mammalian ribonucleotide reductase. Acta Chem Scand B. 1982;36:343–345. doi: 10.3891/acta.chem.scand.36b-0343. [DOI] [PubMed] [Google Scholar]
- 26.Pontarin G, Gallinaro L, Ferraro P, Reichard P, Bianchi V. Origins of mitochondrial thymidine triphosphate: dynamic relations to cytosolic pools. Proc Natl Acad Sci USA. 2003;100:12159–12164. doi: 10.1073/pnas.1635259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Florean C, et al. High content analysis of gamma-secretase activity reveals variable dominance of presenilin mutations linked to familial Alzheimer's disease. Biochim Biophys Acta. 2008;1783:1551–1560. doi: 10.1016/j.bbamcr.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Kucera R, Paulus H. Localization of the deoxyribonucleotide biosynthetic enzymes ribonucleotide reductase and thymidylate synthase in mouse L cells. Exp Cell Res. 1986;167:417–428. doi: 10.1016/0014-4827(86)90182-5. [DOI] [PubMed] [Google Scholar]
- 29.Thelander L, Berg P. Isolation and characterization of expressible cDNA clones encoding the M1 and M2 subunits of mouse ribonucleotide reductase. Mol Cell Biol. 1986;6:3433–3442. doi: 10.1128/mcb.6.10.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]