Abstract
Transcriptional pathways controlling the development of CD44hi memory phenotype (MP) T cells with “innate-like” functions are not well understood. Here we show that the BTB (bric-a-brac, tramtrack, broad complex) domain-containing protein promyelocytic leukemia zinc finger (PLZF) is expressed in CD44hi, but not in CD44lo, CD4+ T cells. Transgenic expression of PLZF during T cell development and in CD4+ and CD8+ T cells induced a T cell intrinsic program leading to an increase in peripheral CD44hi MP CD4+ and CD8+ T cells and a corresponding decrease of naïve CD44lo T cells. The MP CD4+ and CD8+ T cells produced IFNγ upon PMA/ionomycin stimulation, thus showing innate-like function. Changes in the naïve versus memory-like subset distribution were already evident in single-positive thymocytes, indicating PLZF-induced T cell developmental alterations. In addition, CD1d-restricted natural killer T cells in PLZF transgenic mice showed impaired development and were severely reduced in the periphery. Finally, after anti-CD3/CD28 stimulation, CD4+ transgenic T cells showed reduced IL-2 and IFNγ production but increased IL-4 secretion as a result of enhanced IL-4 production of the CD44hiCD62L+ subset. Our data indicate that PLZF is a novel regulator of the development of CD44hi MP T cells with a characteristic partial innate-like phenotype.
Keywords: innate-like lymphocytes, T cell development, transgenics
Peripheral CD4+ and CD8+ T cell subsets have been traditionally divided into naïve CD44loCD62L+ and memory CD44hiCD62L− populations, while the surface expression phenotype of the latter population also resembles recently activated T cells (1, 2). In recent years it became clear that the memory T cell subset is not a population consisting only of true antigen-specific memory cells that developed in response to a foreign antigen. Rather, the memory population contains a variety of different T lymphocyte subsets: some may have acquired their memory marker phenotype through homeostatic proliferation, whereas others show immediate effector function and may play a role in the front-line defense against certain bacterial infections. These additional cells were described as memory phenotype (MP) T lymphocytes, and some subsets of the MP population are also described as innate T cells (3, 4). Certain MP or innate-like T cells are derived from double-positive (DP) thymocytes, and at least some of these cells can be selected on non-classical MHC class Ib molecules (5), achieved by interaction with hematopoietic cells rather than with thymic epithelial cells (6). Among the innate-like T lymphocyte subsets that have been characterized are natural killer T cells (NKT cells) (7–9), H2-M3 specific T cells (10), mucosal-associated invariant T cells (11), and also CD8αα expressing intraepithelial lymphocytes of the gut (12).
Important insight about signaling molecules that are involved in conventional versus innate-like T cell differentiation is provided from studies on Tec family kinase-deficient mice (4). IL2-inducible T-cell kinase (Itk) and resting lymphocyte kinase (Rlk) are essential for the development of conventional CD8+ T cells. As a consequence, Itk−/− mice and Itk−/−Rlk−/− mice are almost devoid of conventional CD8+ T cells, whereas the remaining CD8+ T cell subsets have several characteristics of innate-like T lymphocytes (13–16). Conventional CD4+ T cells are also reduced in the absence of Itk; thus, Itk−/− mice have also a relative increase in innate-like MP CD4+ T cells (17). In addition, Itk−/− mice have reduced numbers of NKT cells (18–20). Another important signaling cascade that has been shown to differentially influence conventional and innate-like MP T lymphocyte development involves the signaling lymphocyte activation molecule and signaling lymphocyte activation molecule-associated protein (21–24).
To gain more insight into the function of Itk in T cells, Affymetrix microarray experiments were performed to identify genes that are differentially expressed between WT and Itk−/− T cells (K.E.M.B., unpublished work). One of the genes identified that was up-regulated in Itk−/− CD3+ T cells was Zbtb16, which encodes for the transcriptional repressor promyelocytic leukemia zinc finger (PLZF). PLZF regulates several biological processes and has also been implicated in tumorigenesis (25). PLZF belongs to the family of BTB (bric-a-brac, tramtrack, broad complex) domain-containing zinc finger factors (BTB-ZF) (25, 26), and important functions in the T cell lineage have been described for several members of the BTB-ZF gene family (27).
In this study we investigated the role of PLZF in T cells. We show that PLZF was expressed in the CD4+CD44hi T cell lineage but not in the naïve CD4+CD44lo subset, whereas CD8+ T cells did not express detectable levels of PLZF. Enforced transgenic expression of PLZF in T lymphocytes induced a T cell intrinsic program leading to an increase in CD44hi MP T cells and a corresponding decrease of naïve CD44lo T cells. The CD44hi population produced IFNγ upon PMA/ionomycin stimulation, thus sharing characteristics with innate T lymphocytes. Transgenic CD4+ T cells showed enhanced IL-4 levels after anti-CD3/CD28 stimulation and an increase in a CD44hiCD62L+ IL-4-producing subset. Together, our data indicate that PLZF is a novel transcriptional regulator for the development of CD44hi MP T cells with innate-like features.
Results
Expression of PLZF Is Restricted to CD4+CD44hi T Cell Subsets.
We have performed Affymetrix microarray analysis to identify genes that are differentially expressed between WT and Itk−/− CD3+ T lymphocytes during T cell activation (K.E.M.B., unpublished work). One gene identified that was already expressed at higher levels in unstimulated Itk-deficient CD3+ T cells compared with WT cells was Zbtb16, which encodes for the transcriptional regulator PLZF. Immunoblot analysis confirmed the up-regulation of PLZF in Itk−/− T cells (Fig. 1A). Because T cell subsets in Itk−/− mice have a T cell memory phenotype (13–17), we investigated whether the increase in the expression of PLZF was caused by a higher percentage of MP T cells in Itk−/− mice. Therefore, WT CD4+ and CD8+ T cell subsets were sorted into CD62L+ and CD62L− fractions. Immunoblot analysis of these sorted subsets revealed that PLZF was expressed in the WT CD4+CD62L− T cell population but in neither CD4+CD62L+ nor CD8+ T cells (Fig. 1A). The majority of WT cells within the CD62L+ subset are naïve CD44lo T cells, and there is only a minor fraction of CD44hiCD62L+ cells. To test whether the CD44hiCD62L+ population expressed Zbtb16, RT-PCR analysis using RNA from sorted CD44loCD62L+, CD44hiCD62L+, and CD44hiCD62L− CD4+ T cell subsets was performed. In agreement with the protein expression data, the RT-PCR analysis revealed expression of Zbtb16 in CD44hiCD62L− cells (Fig. 1B). In addition, low-level expression of Zbtb16 was observed in CD44hiCD62L+ cells, in contrast to CD44loCD62L+ cells (Fig. 1B). Together, this set of data indicated a correlation between PLZF expression and the CD44hi MP CD4+ T cell subset. To investigate a potential role of PLZF in the generation and/or function of the CD4+CD44hi T cell subset, gain-of-function experiments were performed. Therefore, transgenic mice expressing PLZF under the control of Cd4 cis-regulatory element were generated (Fig. 1C), which direct expression from the DN3 stage on, and remain active in, CD4+ and CD8+ T cell subsets (data not shown). Further, the transgenic expression vector contained an IRES-GFP cassette to detect transgenic T cells by GFP expression. Three different founder lines (lines 1, 2, and 3) were selected for further studies. In thymocytes, PLZF was expressed at higher levels in line 2 compared with line 1, correlating with GFP levels [supporting information (SI) Fig. S1]. Furthermore, transgenic peripheral CD4+ T cells showed high levels of PLZF protein compared with WT MP CD4+CD62L− T cells, although PLZF levels were only slightly enhanced in line 2 compared with line 1 (Fig. S1).
Fig. 1.
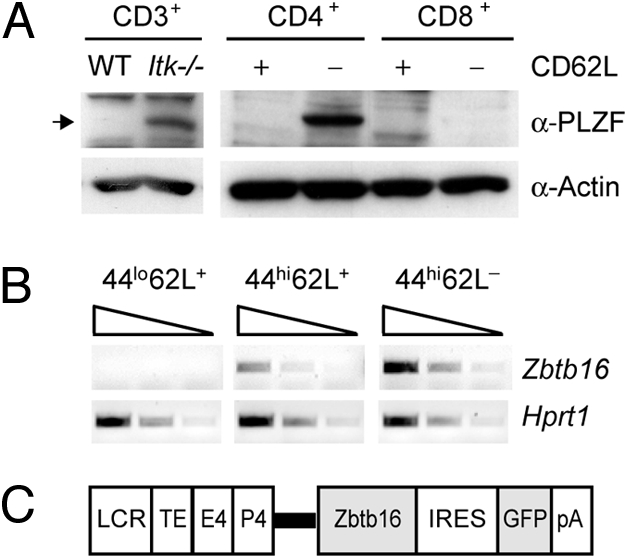
PLZF expression in peripheral T cells is restricted to CD4+CD44hi T cells (A) Protein expression levels of PLZF in WT and Itk−/− peripheral CD3+ T cells (Left), and in WT peripheral CD4 and CD8 T cell subsets that were sorted into CD62L+ and CD62L− populations (Right). The equivalent of 1.5 × 106 cells was loaded per lane. Actin was used as loading control. Data are representative of two different experiments. (B) Semiquantitative RT-PCR analysis shows Zbtb16 expression in various WT peripheral CD4+ T cell subsets. Hprt1 expression was used as loading control. The triangle indicates fivefold dilutions of input. Data are representative of two independent experiments. (C) Schematic map of the transgenic construct used for generation of Zbtb16 transgenic mice. The thick bar indicates splicing module from the Cd4 locus. (LCR/TE, locus control region and thymocyte enhancer from the Cd4 locus; E4, proximal Cd4 enhancer; P4, Cd4 promoter.)
Transgenic Expression of PLZF Induces Dose-Dependent Changes in CD44 and CD62L Expression on Peripheral T Cells.
PLZF transgenic mice displayed a moderate alteration in peripheral T cell numbers and had increased CD4/CD8 ratios (Fig. S2 A and B). Because the PLZF expression pattern suggested a specific function of PLZF in the CD44hi T cell subset, the CD44 versus CD62L expression pattern on peripheral splenic and LN T cells was determined in PLZF transgenic mice. This analysis revealed a decrease in the CD44loCD62L+ naïve CD4+ and CD8+ T cell subset and a corresponding increase of the CD44hiCD62L− population in the presence of PLZF (Fig. 2A and Fig. S3). We also noted an increase in the CD44loCD62L− population in PLZF transgenic mice compared with WT mice. Furthermore, the percentage of CD44hiCD62L+ as well as CD44hiCD62L− cells increased with increasing levels of PLZF expression, indicating a dose-dependent effect of PLZF (Fig. 2A and Fig. S2C). Transgenic CD4+ and CD8+ T cells showed no up-regulation of CD25 and CD69 activation marker expression (Fig. 2B), indicating that the presence of CD44hi T cell population is not the result of T cell activation.
Fig. 2.
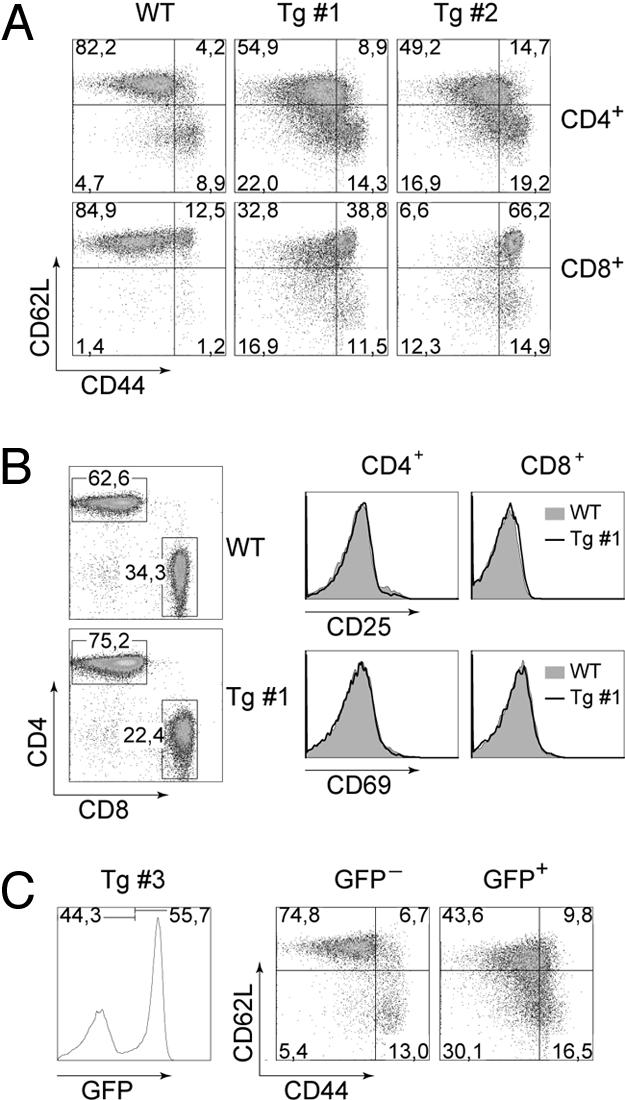
Forced expression of PLZF induces a T cell intrinsic CD44hi phenotype in CD4+ and CD8+ T cells. (A) CD62L and CD44 expression pattern on CD4+ (Upper) and CD8+ (Lower) splenic T cells from WT and PLZF transgenic mice (lines 1 and 2). Numbers in the dot plots indicate the percentage of cells in the respective quadrant. Data are representative of at least three independent experiments for each transgenic line. (B) CD69 and CD25 expression on CD4+ and CD8+ splenic T cells isolated from WT and PLZF transgenic mice. CD3-gated dot plots indicate gating regions for the histogram analysis. Data are representative of at least three independent experiments. (C) CD62L and CD44 expression pattern on GFP− (Left) or GFP+ (Right) CD4+ T cells isolated from the spleen of female PLZF mice (line 3). Numbers indicate the percentage of cells in the respective dot plot quadrants. Gating areas for GFP− and GFP+ populations are shown in the CD3+ T cells histogram on the left. Data are representative of five independent experiments.
PLZF Induces a T Cell Intrinsic Program that Leads to a Change in the CD44 and CD62L T Cell Subset Distribution.
Another transgenic founder line (line 3) had an X-chromosomal transgene insertion. Transgenic males of this line transmitted the mutation exclusively to their female offspring. Furthermore, almost all T cells of male mice of line 3 carrying the PLZF transgene were GFP+ (data not shown), whereas in hemizygous female mice of line 3, approximately only half of the T cells were GFP+ (Fig. 2C). Because there were similar numbers of GFP+ and GFP− T cells in female transgenic mice, we concluded that PLZF-expressing T cells have no major developmental advantage or disadvantage over non-transgenic T cells. Similar to transgenic lines 1 and 2, GFP+ CD4+ and CD8+ T cells in male and female mice of line 3 displayed the altered CD44/CD62L expression pattern in the spleen (Fig. 2C and data not shown), whereas the GFP− population in female transgenic mice showed a CD44/CD62L distribution similar to WT mice (Fig. 2C). Thus, PLZF induces a T cell-intrinsic genetic program in both CD4+ and CD8+ T cells that leads to a dramatic alteration in the CD44/CD62L subset distribution.
CD44hi T cells in PLZF Transgenic Mice Develop in the Thymus and Have a Memory Phenotype with Innate-Like Characteristics.
To determine whether the CD44hi T cells emerge already in the thymus, a comprehensive analysis of thymocyte development was performed. In contrast to WT cells, transgenic PLZF thymocytes had increased percentages of SP cells and reduced numbers of DP cells (Fig. 3A and Fig. S4A). However, the expression of CD3 and HSA on the various subsets was similar between WT and PLZF thymocytes (Fig. 3A). Furthermore, CD4SP and CD8SP PLZF transgenic thymocytes showed a larger fraction of cells that had down-regulated CD62L expression compared with WT SP cells, and PLZF transgenic CD8SP cells displayed an increase in CD44hi subsets (Fig. 3B). CD122 expression was similar in WT and PLZF transgenic SP cells (Fig. 3B). In agreement with the thymic emergence of PLZF MP T cells, T cells with an altered CD44/CD62L profile were already present in 14-day-old PLZF transgenic mice (Fig. S5).
Fig. 3.
MP T cells in PLZF transgenic mice develop in the thymus and have innate-like characteristics. (A) CD4 and CD8 expression on WT and PLZF transgenic (line 1) thymocytes. Histograms (Right) show HSA and CD3 expression levels on CD4SP and CD8SP cells. (B) CD44, CD62L, and CD122 expression pattern on CD4SP (Upper) and CD8SP (Lower) thymocytes. Numbers below histograms indicate the percentage of cells in the indicated region. Data in A and B are representative of at least three mice. (C and D) Intracellular IFNγ expression in ex vivo PMA/ionomycin-stimulated WT, PLZF transgenic (line 1), and Itk−/− CD4SP and CD8SP thymocytes (C) and CD4+ and CD8+ splenic T cells (D). Numbers in the dot plots indicate the percentage of cells in the respective quadrants. Data in C and D are representative of three experiments.
The CD44hi MP T cell population of WT mice is composed of several T cell subsets (3, 4). To test whether CD44hi SP cells and peripheral T cells in PLZF transgenic mice have immediate effector function, thymocytes and splenocytes were isolated and stimulated ex vivo with PMA/ionomycin. In addition, we compared the innate properties of PLZF transgenic T cells with Itk−/− SP thymocytes and splenocytes, as Itk−/− mice have a large number of innate-like T cells (13–16). Similar to the WT CD4SP and CD8SP CD44hi populations, CD4SP and CD8SP CD44hi PLZF transgenic T cells produced IFNγ, although the percentage of IFNγ-producing CD4SP cells was reduced within the CD44hi fraction (Fig. 3C and Fig. S4B). Itk−/− SP thymocytes, as reported previously (13, 14), showed increased production of IFNγ compared with WT cells. In the periphery, the percentage of IFNγ-producing CD4+ T cells within the CD44hi population was reduced in PLZF transgenic mice compared with WT controls (Fig. 3D and Fig. S4C). In contrast, there was an increase in IFNγ-producing CD44hi CD8+ T cells. The relative percentage of PLZF transgenic IFNγ-producing CD8+ T cells within the CD44hi population was moderately increased compared with WT MP T cells, but not as high as in innate-like Itk−/− CD8+ T cells (Fig. 3D and Fig. S4C). Thus, despite the dramatic increase in the numbers of MP T cells in PLZF transgenic mice, the PLZF MP population had similar innate-like functions as WT MP subsets.
Many innate T lymphocytes such as NKT cells (7) and innate-like CD8+ T cells in Itk−/− mice require IL-15 for their survival (13, 15). IL-15 signals via the IL-15 receptor complex, which is composed of several subunits including CD122 (28). PLZF transgenic and WT CD44hiCD62L+ CD8+ T cells displayed similar up-regulated CD122 expression levels, and the percentage of CD122+ cells within this transgenic subset was even slightly increased compared with the WT subset (Fig. S6). Together, these data indicate that enforced expression of PLZF leads to the development of CD44hi MP T cells with innate-like characteristics.
Impaired NKT Cell Development in PLZF Transgenic Mice.
Because NKT cells have a CD44hiCD62L− surface expression phenotype (7), we investigated whether PLZF expression led to an increase in NKT cells. To have internal staining controls, the analysis was performed in female PLZF transgenic mice of line 3, which have both GFP+ and GFP− cells within the T cell subset. CD1d tetramers (CD1d-tet) loaded with the αGalCer analogue PBS57 were used to detect invariant NKT cells. This analysis revealed that CD1d-tet+ NKT cells were severely reduced in the GFP+ T cell subset, whereas NKT cells were readily detected within the GFP− subset (Fig. 4A and Fig. S7A). Next, we investigated whether the peripheral reduction of NKT cells is caused by a (partial) block during thymic NKT cell development. Early CD1d-tet+ NKT cells are HSAhi, followed by a HSAlo stage (7, 8). Preliminary results indicated that the GFP+/GFP− ratio was approximately 50:50 in the HSAhiCD1d-tet+ fraction, whereas the percentage of GFP+ cells declined in the HSAloCD1d-tet+ fraction (data not shown). This indicated a defect in the transition to or within the HSAlo fraction. The thymic CD1d-tet+ HSAlo subset can be further divided into three stages that follow a CD44−NK1.1− (stage 1), CD44hiNK1.1− (stage 2), and CD44hiNK1.1+ (stage 3) pattern (7, 8). Our analysis revealed that the transition from stage 2 to stage 3 is impaired in PLZF transgenic mice, indicated by an increase in stage 2 and a corresponding decrease in stage 3 NKT cells (Fig. 4B and Fig. S7B). This suggests that enforced expression of PLZF impairs NKT cell development.
Fig. 4.

Impaired NKT cell development in PLZF transgenic mice. (A) NK1.1 expression and CD1d-tet binding on GFP− (Left) and GFP+ (Right) CD3+ transgenic splenocytes (line 3). Numbers in the dot plots indicate the percentage of cells in the respective quadrant. Histogram shows gating region for GFP− and GFP+ T cell populations. Data shown are representative of three mice. Similar results have also been obtained for line 1 (n = 3 mice). (B) Histogram (Upper) shows gating region for GFP− and GFP+ thymocytes (line 3). Dot plots indicate gating regions for CD1d-tet+ HSAlo NKT cells. CD44 and NK1.1 expression on CD1d-tet+ HSAlo NKT cells (Lower). Numbers in the dot plots indicate the percentage of cells in the respective quadrant. Data shown are representative of three mice. Similar results have also been obtained for line 1 (n = 3).
Transgenic Expression of PLZF Alters the Cytokine Profile of Naïve (CD44lo) and MP (CD44hi) CD4+ T Cells.
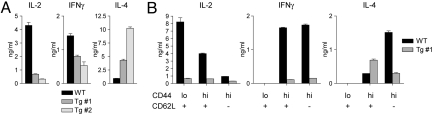
Next we analyzed whether PLZF expression changed the cytokine production of CD4+ T cells upon T cell receptor-mediated activation. Total CD4+ T cells were isolated from WT or PLZF transgenic mice and stimulated with anti-CD3/CD28. IL-2 levels were severely impaired in PLZF transgenic CD4+ T cells (Fig. 5A), and PLZF transgenic CD4+ T cells produced reduced amounts of IFNγ compared with WT CD4+ T cells. In contrast, they showed dramatically increased levels of IL-4 compared with WT controls (Fig. 5A).
Fig. 5.
PLZF alters the cytokine profile of T cell receptor-stimulated CD4+CD44lo and CD4+CD44hi T cells. (A) Diagrams show IL-2, IFNγ, and IL4 production in purified WT and PLZF transgenic (lines 1 and 2) CD4+ T cells stimulated for 48 h with plate-bound anti-CD3/CD28. For IFNγ detection, only cells with >95% purity were used to exclude “contaminating” IFNγ-producing CD8+ T cells. The cytokine levels in the supernatants were determined by ELISA in duplicates. Data shown are representative of three independent experiments. (B) IL-2, IFNγ, and IL-4 production of WT or PLZF transgenic (line 1) CD4+ T cells that were sorted into CD44loCD62L+, CD44hiCD62L+, and CD44hiCD62L− subsets. Cells were stimulated and cytokines determined as in A. Data shown are representative of four independent experiments (three with line 1 and one with line 3; male mice).
To investigate which subset is responsible for the dramatic increase in IL-4, CD44loCD62L+, CD44hiCD62L+, and CD44hiCD62L−, WT and PLZF transgenic CD4+ T cells were sorted and activated with anti-CD3/CD28. All subsets of WT and PLZF transgenic CD4+ T cells produced IL-2, whereas IFNγ was secreted by CD44hiCD62L+ and CD44hiCD62L− T cells (Fig. 5B). However, transgenic T cells showed reduced IL-2 and IFNγ production compared with WT cells. WT CD44hiCD62L− cells produced high levels of IL-4, whereas IL-4 levels in this subset of transgenic T cells were reduced (Fig. 5B). This is in contrast to the CD44hiCD62L+ subset, which showed enhanced IL-4 levels in PLZF transgenic mice. Thus, the presence of this IL-4 producing CD44hiCD62L+ subset explains why total CD4+ T cells produce elevated levels of IL-4. Although Tbx21 (encoding for T-bet) and Gata3 are known to be major transcription factors responsible for IFNγ and IL-4 expression of Th1 and Th2 cells, respectively, no major difference in the expression of Tbx21 and Gata3 in the various subsets was observed (Fig. S8).
Discussion
In this study we showed that the BTB-ZF factor PLZF is predominantly expressed in CD44hi CD4+ T cells. Enforced expression of PLZF during T cell development and in peripheral T cells led to the appearance of a large population of peripheral T cells with a CD44hi memory phenotype that were able to produce IFNγ upon ex vivo PMA/ionomycin stimulation. PLZF expressing (i.e., GFP+) T cells in female mice of line 3 showed an MP despite the presence of similar numbers of GFP− T cells with a naïve phenotype, thus indicating a T cell intrinsic defect. Together with the observations that the cells were not activated and that PLZF T cells showed no increased BrdU incorporation (data not shown), this also suggests that the CD44hi T cells are not generated via homeostatic proliferation (29). In addition, changes in the naïve versus memory-like subset distribution were already evident at the SP stage during T cell development. This indicates that the MP T cell populations in PLZF transgenic mice are at least in part derived from DP thymocytes, like other MP T cells (3, 4). We therefore propose that PLZF induces a transcriptional program that leads to the generation of CD44hi MP T cells that share certain features with innate-like T cells.
The observations that PLZF is up-regulated in the Itk−/− T cell population (which is enriched in innate-like T cells) and that PLZF expression is restricted to the CD4+CD44hi population revealed a correlation between PLZF expression and MP CD4+ T cells. A more direct link between PLZF and innate-like MP T cells is established by the CD44hi phenotype of transgenic T cells. Thus, ectopic PLZF expression leads to an increase of those T cell subsets that express PLZF in WT cells, indicating that PLZF can convert CD4+CD44lo subsets into CD4+CD44hi populations. Furthermore, endogenous PLZF was expressed in transgenic CD44hi but not CD44lo CD4+ T cells (data not shown), an expression pattern identical to the one in WT CD4+ T cell subsets. This indicates that transgenic PLZF induced the differentiation of a distinct lineage of MP T cells with innate-like characteristics. Interestingly, immunoblot analysis did not reveal expression of PLZF in innate-like CD8+ T cells. However, PLZF also induced the CD44hi phenotype in CD8+ T cells, suggesting that PLZF may induce similar transcriptional programs in CD4 and CD8 T cell lineages, and possibly indicating the existence of an endogenous factor that specifically directs the development of CD44hi MP CD8+ T cells.
We demonstrated that CD4+ and CD8+ MP T cells in PLZF mice possess innate-like characteristics. The increase in IFNγ-producing transgenic CD44hi CD8+ T cells is similar to the one observed in innate-like Itk−/− CD8+ T cells, although the percentage of IFNγ-positive cells within the CD44hi population is higher in the absence of Itk. Further, only peripheral MP CD8+ T cells and not CD8SP MP thymocytes in PLZF transgenic mice showed an increase in IFNγ-producing CD44hi cells upon PMA/ionomycin stimulation, suggesting a further maturation of innate-like functions in the periphery. In contrast, IFNγ-producing CD44hi cells were increased in the thymus and in the periphery in Itk−/− mice. These findings may indicate a different subset composition and/or differential regulation of innate-like CD8+ T cells in PLZF and Itk−/− mice. PLZF MP CD4+ T cells produced IFNγ as well; however, the percentage of IFNγ-positive CD4+ T cells within the CD44hi population was reduced. Moreover, upon anti-CD3/CD28 activation we observed reduced cytokine production in all subsets of PLZF transgenic CD4+ T cells compared with WT CD4+ T cell subsets, except for the dramatic increase of IL-4 production in the CD44hiCD62L+ subset. It is possible that PLZF may down-modulate the IL-2 and IFNγ response by binding to cytokine loci and recruiting co-repressors including nuclear receptor co-repressor and histone deacetylase (25), or by repressing the expression of an important signaling molecule or transcription factor required for cytokine expression. Alternatively, but not mutually exclusively, PLZF expression may lead to a different cellular composition of peripheral CD4+ T cells, even though we could not observe any altered expression of T-bet and Gata-3 in the various subsets of PLZF transgenic mice compared with WT mice. A “candidate” innate-like population are NKT cells that share a CD44hi expression phenotype with PLZF transgenic T cells (7–9). A recent study has shown that PLZF is primarily expressed in CD1d-restricted NKT cells (30). However, other MP T cell subsets may express PLZF as well, as PLZF expression was up-regulated in Itk−/− T cells, which have diminished numbers of NKT cell subsets compared with WT T cells (19, 20). Remarkably, NKT cell numbers in mice lacking PLZF were severely reduced, and they also displayed impaired cytokine expression patterns (30). Unexpected given this essential role of PLZF for NKT cell development, we observed a severe reduction of NKT cells in PLZF expressing mice compared with WT mice. One explanation, that both PLZF-deficient and PLZF transgenic mice have diminished numbers of NKT cells, may come from the observation that stage 1 and stage 2 NKT cells express the highest level of PLZF and down-regulate PLZF expression at stage 3 (30). Thus, enforced transgenic expression of PLZF may partially block NKT cells at this stage.
Taken together, our study gives insight into transcriptional control mechanisms that regulate conventional versus MP T cell development and suggest that PLZF is an important regulator in this process. Further studies aiming to identify and characterize the genetic program that is induced by PLZF are required to further elucidate these developmental processes.
Materials and Methods
Generation of the PLZF Transgenic Construct.
The transgene expression cassette consisted of the Cd4 Locus control region/thymocyte enhancer, the Cd4 proximal enhancer/promoter, and a splicing module from silencer-less Cd4 intron 1 sequences, followed by IRES-GFP-polyA sequences, and was provided by Ichiro Taniuchi (Riken, Yokohama, Japan). The cDNA sequence encoding for PLZF (MGI: 103222; nucleotide 249-2352) was inserted as an EcoRI fragment upstream of the IRES-GFP module.
Generation of PLZF Transgenic Mice.
The linearized construct was microinjected into a pronucleus of fertilized C57BL/6N inbred oozytes that were transferred afterward into the oviduct of pseudopregnant foster mothers according to standard protocols for generating transgenic mice (31). Transgene integrations were identified by PCR of tail DNA with GFP-specific primers. Of several identified transgenic founders, we selected lines C57BL/6N-Tg(Cd4-Zbtb16,GFP)141–143 Biat (lines 1–3 for brevity) based on the GFP expression levels.
Mice.
Animals used in this study were bred and maintained in the animal facility of the Medical University of Vienna. Analyzed mice were 6 to 8 weeks of age unless indicated otherwise. All animal experiments were approved by the Federal Ministry for Science and Research.
Flow Cytometric Analysis and Antibodies.
The following antibodies were used for the staining: PE-anti-CD62L, PE-anti-CD122, PE-Cy7-anti-CD4, PE-anti-IFNγ, and APC-anti-IL-4 from BD Pharmingen; APC-anti-CD8α (53–6.7), APC-anti-CD62L, PE-Cy7-anti-CD44, APC-anti-CD25, APC-anti-NK1.1, PE-anti-CD3e, and Pb-anti-CD3e from eBioscience; and A647-anti-CD3 from Caltag. PBS57-loaded and unloaded CD1d tetramers (conjugated with PE) were obtained from the National Institutes of Health tetramer facility and used according to the instructions. Flow cytometry was performed on LSRII (BD Biosciences) and data analyzed using FlowJo software.
Purification of T Cells and Cell Sorting.
CD3+ T cells were isolated by incubating cell suspensions of spleens (in PBS solution supplemented with 2% FCS) with biotinylated anti-CD11b, anti-CD11c, anti-CD45R, anti-Ly-6G, anti-Ter119, and anti-NK antibodies, followed by negative depletion using streptavidin beads (BD Pharmingen) according to the manufacturer's instructions. For the isolation of CD4+ T cells, biotinylated anti-CD8α antibodies were added to the depletion antibody mixture. The purity of either CD3+ or CD4+ T cells was assessed by flow cytometry and was routinely >90%–95%. For cell sorting, CD4+ T cells were incubated with anti-CD4, anti-CD44 and anti-CD62L, and CD4+ T cells subsets were sorted into CD44loCD62L+, CD44hiCD62L+, and CD44hiCD62L− populations with a FACSAria cell sorter (Becton Dickinson).
Activation of T Cells and Cytokine Measurement.
Purified or FACS-sorted CD4+ T cells (5 × 105) or T cell subsets (5 × 105) were stimulated with plate-bound anti-CD3ε (1 μg/ml) and plate-bound anti-CD28 (3 μg/ml) on 48-well plates. CD4+ T cells were grown in 1 ml RPMI1640 Glutamax-I (Invitrogen), supplemented with 10% FCS (Invitrogen), antibiotics, and 2-mercaptoethanol (Invitrogen). The supernatants for cytokine quantification were collected 48 h later. IL-2, IL-4, and IFNγ cytokine levels were determined by ELISA (BD Pharmingen).
For PMA/ionomycin stimulation, thymocytes or splenocytes were incubated for 5 h with PMA (50 ng/ml) and ionomycin (500 ng/ml) (Sigma) in the presence of GolgiSTOP (BD Biosciences). Surface staining for CD3, CD4, CD8, and CD44 was performed, and subsequently cells were fixed and permeabilized with Cytofix/Cytoperm solution (BD Pharmingen) and further stained with PE-anti-IFNγ.
Statistical Analysis.
All data are expressed as mean ± SEM. Statistical analysis was performed by using a Student t test. The P values were defined as following: *P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
Note Added in Proof.
While this article was in revision, Savage et al. (32) reported similar data as described in our study. They observed that transgenic expression of PLZF induced CD4SP thymocytes and CD4+ T cells to acquire a CD44hi phenotype. In addition, Savage et al. reported that PLZF is primarily expressed in CD1d-restricted NKT cells and that PLZF-deficient mice have impaired NKT cell differentiation and effector function (in agreement with ref. 30).
Supplementary Material
Acknowledgments.
The authors thank Ludger Klein and Ichiro Taniuchi for critical reading of the manuscript, the National Institutes of Health tetramer facility for providing the PBS57-loaded and unloaded CD1d reagents, and Ichiro Taniuchi for providing the transgenic expression cassette. This work was supported by the Special Research Area SFB-F23 Project SFB-F2305 of the Austrian Science Fund, START program Grant Y-163 of the FWF and the Austrian Ministry of Science and Research, by FWF Project P-19930, and funds from the EU MCRTN project “Chromatin plasticity” (to W.E.); and the Swedish Science Council, the Swedish Cancer Fund, and European Council FP7 Grant EURO-PADnet (to C.I.E.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805733105/DCSupplemental.
References
- 1.Swain SL, et al. CD4+ T-cell memory: generation and multi-faceted roles for CD4+ T cells in protective immunity to influenza. Immunol Rev. 2006;211:8–22. doi: 10.1111/j.0105-2896.2006.00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prlic M, Williams MA, Bevan MJ. Requirements for CD8 T-cell priming, memory generation and maintenance. Curr Opin Immunol. 2007;19:315–319. doi: 10.1016/j.coi.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Surh CD, Boyman O, Purton JF, Sprent J. Homeostasis of memory T cells. Immunol Rev. 2006;211:154–163. doi: 10.1111/j.0105-2896.2006.00401.x. [DOI] [PubMed] [Google Scholar]
- 4.Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development. Nat Rev Immunol. 2007;7:479–485. doi: 10.1038/nri2091. [DOI] [PubMed] [Google Scholar]
- 5.Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5:459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- 6.Urdahl KB, Sun JC, Bevan MJ. Positive selection of MHC class Ib-restricted CD8(+) T cells on hematopoietic cells. Nat Immunol. 2002;3:772–779. doi: 10.1038/ni814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 8.Kronenberg M, Engel I. On the road: progress in finding the unique pathway of invariant NKT cell differentiation. Curr Opin Immunol. 2007;19:186–193. doi: 10.1016/j.coi.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 9.MacDonald HR, Mycko MP. Development and selection of Valpha l4i NKT cells. Curr Top Microbiol Immunol. 2007;314:195–212. [PubMed] [Google Scholar]
- 10.Colmone A, Wang CR. H2–M3-restricted T cell response to infection. Microbes Infect. 2006;8:2277–2283. doi: 10.1016/j.micinf.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 11.Treiner E, Lantz O. CD1d- and MR1-restricted invariant T cells: of mice and men. Curr Opin Immunol. 2006;18:519–526. doi: 10.1016/j.coi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Lambolez F, Kronenberg M, Cheroutre H. Thymic differentiation of TCR alpha beta(+) CD8 alpha alpha(+) IELs. Immunol Rev. 2007;215:178–188. doi: 10.1111/j.1600-065X.2006.00488.x. [DOI] [PubMed] [Google Scholar]
- 13.Atherly LO, et al. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Broussard C, et al. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Dubois S, Waldmann TA, Müller JR. ITK and IL-15 support two distinct subsets of CD8+ T cells. Proc Natl Acad Sci USA. 2006;103:12075–12080. doi: 10.1073/pnas.0605212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J, Sahu N, Walsh E, August A. Memory phenotype CD8+ T cells with innate function selectively develop in the absence of active Itk. Eur J Immunol. 2007;37:2892–2899. doi: 10.1002/eji.200737311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, August A. Naive and Innate Memory Phenotype CD4+ T Cells Have Different Requirements for Active Itk for Their Development. J Immunol. 2008;180:6544–6552. doi: 10.4049/jimmunol.180.10.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gadue P, Stein PL. NK T cell precursors exhibit differential cytokine regulation and require Itk for efficient maturation. J Immunol. 2002;169:2397–2406. doi: 10.4049/jimmunol.169.5.2397. [DOI] [PubMed] [Google Scholar]
- 19.Au-Yeung BB, Fowell DJ. A key role for Itk in both IFN gamma and IL-4 production by NKT cells. J Immunol. 2007;179:111–119. doi: 10.4049/jimmunol.179.1.111. [DOI] [PubMed] [Google Scholar]
- 20.Felices M, Berg LJ. The tec kinases itk and rlk regulate NKT cell maturation, cytokine production, and survival. J Immunol. 2008;180:3007–3018. doi: 10.4049/jimmunol.180.5.3007. [DOI] [PubMed] [Google Scholar]
- 21.Veillette A, Dong Z, Latour S. Consequence of the SLAM-SAP signaling pathway in innate-like and conventional lymphocytes. Immunity. 2007;27:698–710. doi: 10.1016/j.immuni.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Griewank K, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W, et al. The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity. 2007;27:763–774. doi: 10.1016/j.immuni.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horai R, et al. Requirements for selection of conventional and innate T lymphocyte lineages. Immunity. 2007;27:775–785. doi: 10.1016/j.immuni.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly KF, Daniel JM. POZ for effect−POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Collins T, Stone JR, Williams AJ. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilic I, Ellmeier W. The role of BTB domain-containing zinc finger proteins in T cell development and function. Immunol Lett. 2007;108:1–9. doi: 10.1016/j.imlet.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Ma A, Koka R, Burkett P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu Rev Immunol. 2006;24:657–679. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- 29.Stockinger B, Kassiotis G, Bourgeois C. Homeostasis and T cell regulation. Curr Opin Immunol. 2004;16:775–779. doi: 10.1016/j.coi.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008 doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rulicke T. Pronuclear microinjection of mouse zygotes. Methods Mol Biol. 2004;254:165–194. doi: 10.1385/1-59259-741-6:165. [DOI] [PubMed] [Google Scholar]
- 32.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.