Abstract
Minor group human rhinoviruses (HRVs) attach to members of the low-density lipoprotein receptor family and are internalized via receptor-mediated endocytosis. The attachment of HRV2 to the cell surface, the first step in infection, was characterized at the single-molecule level by atomic force spectroscopy. Sequential binding of multiple receptors was evident from recordings of characteristic quantized force spectra, which suggests that multiple receptors bound to the virus in a timely manner. Unbinding forces required to detach the virus from the cell membrane increased within a time frame of several hundred milliseconds. The number of receptors involved in virus binding was determined, and estimates for on-rate, off-rate, and equilibrium binding constant of the interaction between HRV2 and plasma membrane-anchored receptors were obtained.
Keywords: force spectroscopy, molecular recognition, single virus binding, very low density lipoprotein receptor, picornavirus
Human rhinoviruses (HRVs), members of the Picornaviridae family, are the most frequent cause of colds. Their icosahedral capsid (30 nm in diameter) is built from 60 copies each of 4 viral proteins VP1, VP2, VP3, and VP4 that surround the RNA genome. Of the 99 so far characterized serotypes, 12 (the minor receptor group) bind low-density lipoprotein receptor (LDLR), very-LDLR (VLDLR), and LDLR-related protein (LRP) (1, 2). This receptor family functions in endocytosis and signal transduction recognizing a variety of ligands (3). LDLR and VLDLR possess 5 domains (4), including an N-terminal ligand-binding domain composed of 7 (LDLR; L1-L7) and 8 (VLDLR; V1-V8) modules, a region similar to the EGF-precursor and a β-propeller with YWTD motifs that is implicated in low pH-induced release of the ligands in endosomes (5). Adjacent to the plasma membrane is a domain carrying O-linked oligosaccharides followed by the transmembrane anchor and the carboxyl terminus carrying a NPXY clathrin localization signal. The ligand binding modules are ≈40 amino acid residues in length. They are stabilized by a Ca ion and 6 highly conserved cysteines forming disulfide bridges (6). Differences in the types and numbers of repeats allow for recognition of a large variety of structurally and functionally diverse ligands.
For infection, HRV2 attaches to LDLR and/or LRP at the cell membrane. It can be released with EDTA immediately after attachment to the cell but within some minutes becomes tightly bound and not dissociable (7). This finding was taken to indicate either recruitment of multiple receptors, thus enforcing an initial bond with a single receptor, and/or engulfment within membranes as the virus enters in clathrin coated vesicles (8). Subsequently, it presumably dissociates from its receptors upon arrival in the mildly acidic milieu (pH 6.5–6.0) of early endosomes (9); finally, the virus is delivered to endosomal carrier vesicles and late endosomes from where its RNA genome is released into the cytosol.
Performing single-molecule force spectroscopy with an atomic force microscope (AFM), we demonstrate a time-dependent transition of single to multiple virus receptor bonds. By conducting many sequential measurements, our single-molecule force trajectories provide dynamic and statistical real-time information of the distribution of molecular receptor bonds to single virus particles.
Results
Anchoring Virus Particles to the AFM Tip.
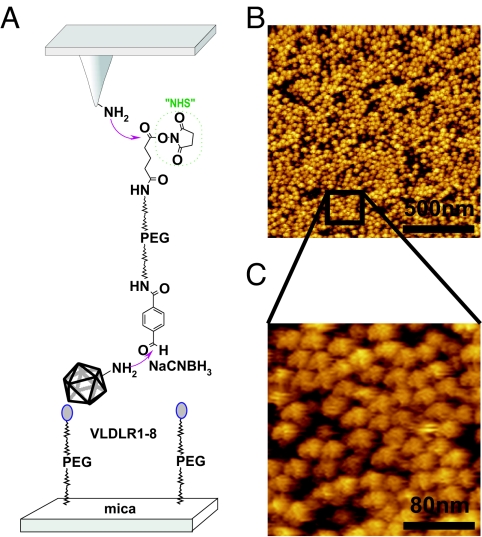
For force spectroscopy experiments, single molecules are frequently bound to the AFM tip by flexible poly(ethylene glycol) (PEG) linkers (10). Recently, a heterobifunctional cross-linker (aldehyde-PEG-NHS) that couples native proteins via their endogenous lysine residues to AFM tips was synthesizedin our laboratory at the University of Linz (11, 12). With this method, virions were flexibly tethered to the AFM tip. Fig. 1A shows a sketch of the stepwise protocol used for covalent virus immobilization on amino-functionalized tips. Topographical AFM images of the lower side of the cantilever were acquired to visualize the virus particles on the cantilever chip surface. A dense packing of virus particles with ≈570 virions·μm−2 was obtained (Fig. 1B), and single virus particles with 30-nm diameter were resolved (Fig. 1C). Assuming a tip radius of 20 nm, this density corresponds to 1 single virus particle on the tip apex, confirming that single particle measurements can be indeed performed. To show that the virus particles were covalently bound via the PEG cross-linker, the aldehyde groups of PEG were inactivated before addition of HRV2, resulting in a significantly reduced density of virus particles (≈100 virions·μm−2; data not shown).
Fig. 1.
Tethering of virions to AFM tips and receptor proteins to mica. (A) Schematic representation of the immobilization scheme used to bind HRV2 to the tip and MBP-V1-8 to the mica and control of attachment. Tip and mica were functionalized with ethanolamine hydrochloride and aldehyde-PEG-NHS before incubation with virus and receptor, respectively, and the analytes were fixed by simultaneous application of NaCNBH3. (B) Tethering of the virus particles was tested by imaging the surface of a cantilever chip modified as described above. (C) Single viruses with 30-nm diameter were resolved by the AFM.
Single Virus–Receptor Force Measurements.
First, virus–receptor interactions were studied in vitro, using surface-tethered VLDLR1-8 (a soluble native-like recombinant VLDLR fragment encompassing the entire ligand binding domain fused to maltose binding protein (MBP) at the N terminus and to His6 at the C terminus), which binds with higher affinity to HRV2 than recombinant LDLR (13, 14). Based on experiments with concatemers of V3 it is assumed that 1 virion can bind up to 12 receptor molecules (15). Following the same protocol as for virus-tip attachment, a densely packed receptor layer was prepared on amino-functionalized mica. Force distance cycles were acquired by approaching the virus-modified tip to the receptor-modified surface followed by its retraction. Interaction between virus and receptor was represented as a characteristic nonlinear force signal, arising from the elastic extension of the distensible PEG cross-linker (Fig. 2A). The characteristic shape of the cross-linker stretching allows distinguishing specific unbinding events from unspecific adhesion (10). To prove that the measured force signals are indeed a consequence of the virus–receptor interaction, the receptor was inactivated by complexation of Ca2+. Within each ligand binding module a Ca ion is tightly coordinated and is essential for virus binding (16, 17). Perfusion of the chamber with EGTA-containing buffer resulted in a drop of the binding probability from ≈10% to <1% (the binding probability is defined as the percentage of the curves displaying the characteristic nonlinear unbinding signal; see Fig. 2A Inset). After back-addition of Ca2+, the binding probability returned to its original level, indicating that the receptor inactivation was reversible (data not shown).
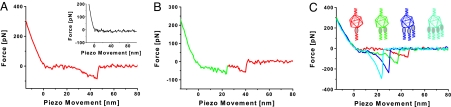
Fig. 2.
Force traces of the model system acquired with the AFM in force spectroscopy mode. (A) A typical force distance cycle showing the nonlinear stretching of the tether, allowing for the discrimination of specific and unspecific events. (Inset) A force distance cycle after specifically inactivating the receptor via removal of Ca2+ ions. This inactivation is reversible as readdition of Ca2+ restored the specific binding events. (B) An example for serial rupture of 2 receptors bound to the virus. (C) Force distance cycles showing instantaneous rupture of single (red), double (green), triple (blue), and quadruple (cyan) bonds are shown. Note the higher stiffness of the tethers with higher number of bonds (springs in parallel produce higher stiffness).
In most cases, single virus–receptor unbinding events were observed with unitary unbinding force value of 82 ± 14 pN at a retraction velocity of 300 nm·s−1. Occasionally, force signals with multiple unbinding force values were observed; these are attributed to the rupture of the bonds between 2 and more receptor molecules and 1 virus particle. Two force distance profiles were observed: (i) a sequential rupture of the receptor-virus bonds (Fig. 2B) and (ii) a simultaneous rupture of all receptor–virus bonds (Fig. 2C). The mode of rupture might depend on the binding geometry. The stepwise rupture profile was found only with a frequency of <10% (Fig. 2B). In this mode, the AFM tip initially stretches both receptor tethers until one of the bonds breaks. The remaining slack in the second tether allows the applied force to drop, followed by a short region corresponding to the loading of the remaining tether, which finally results in the rupture of the second bond. However, in the majority of cases a single rupture event with a broad range of observed forces was found (Fig. 2C), which can be explained by taking into account that breaking the first bond leads to a redistribution of force among the remaining bonds, and the abrupt increase in the applied load reduces the bond lifetime to nanosecond levels (18). Therefore, the remaining bonds rupture very shortly after the first one is broken and the finite bandwidth of the AFM (10 kHz) causes the multiple bond breakages to be registered as a single rupture event.
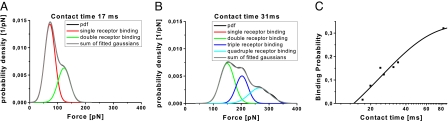
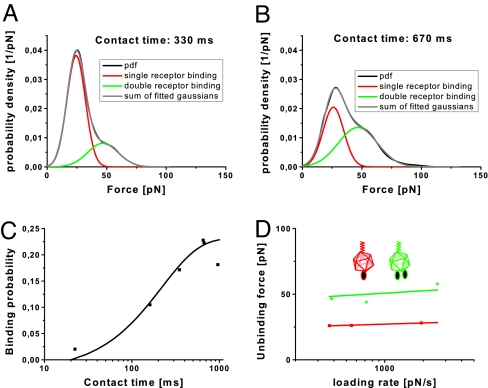
The specific force signals were used to determine rupture forces and their accuracy estimates of the HRV2/VLDLR1-8 interaction by calculating empirical probability density functions (PDFs) (19) (see Materials and Methods). PDFs are preferable to histograms, because the data are weighted by their reliability (i.e., standard deviation). Fig. 3A shows a PDF when tip and surface were in contact for 17 ms (encounter time). A characteristic force spectrum shows bimodal distribution with the main peak at 69 pN, and a second peak at 123 pN was obtained. The first and the second peak are interpreted as the simultaneous unbinding between 1 and 2 surface-immobilized receptors from a single tip-bound virus particle, respectively. Increasing the contact time, i.e., the time during which the virus resides close enough to the receptors for interaction, increases the probability of multiple receptor attachment. A multimodal force spectrum with main peaks at 149, 203, and 273 pN was observed at 31-ms encounter time (Fig. 3B), corresponding to the simultaneous unbinding of 2, 3, and 4 receptor molecules from a single virus particle, respectively. The increase of the number of bonds with contact time is a clear indication that several single receptors sequentially bind to the virus.
Fig. 3.
Distribution of rupture forces (black line) as a function of the contact time. (A and B) Extension of the contact times shifts the forces toward higher values, indicating binding of >1 receptor molecule. Sums of Gaussians were fitted to the measured curves, yielding an almost perfect reproduction of the experimental data. (C) The binding probability is plotted as a function of the contact time. The solid line is the result of a least-squares fit of a monoexponential decay (0.33 exp((t − 15)/24)).
Kinetic On-Rate Constant from Force Measurements.
Assuming that the virus–receptor interaction can be approximated with pseudo first-order kinetics, estimation of the kinetic on-rate constant kon from single molecule unbinding force measurements requires the determination of the interaction time τ and the effective concentration ceff, for kon = (τceff)−1 (20). The interaction time was calculated from the binding probability at different encounter times by using P = A (1 − exp(−(t − t0)/τ)) (21), with t0 being the lag time and A the maximum observable binding probability. A equals 1 if unbinding events are found in every force distance cycle (i.e., binding probability 1). A least-square fit resulted in τ = 24 ± 1 ms (Fig. 3C). The effective concentration describes the number of binding partners (receptor molecules) within the effective volume Veff accessible for free equilibrium interaction (20). Veff can be described as a sphere with radius reff, with the latter being the sum of the equilibrium cross-linker length (3 nm) and the diameter of HRV2 (30 nm). Assuming 4 possible (compare Fig. 3B) binding partners kon can be estimated to 1 × 106 M−1·s−1.
Force Spectroscopy of Single Virus–Receptor Bonds.
Because molecular interaction forces depend on the time scale of the measurements (22), we performed unbinding force experiments at different pulling speeds. For retraction velocities in the range between 150 and 3,000 nm·s−1, the force values for the unbinding of single virus–receptor bonds (compare to first peak of Fig. 3A) were determined, and the peak maxima were plotted against the loading rate. The loading rate was calculated from the retraction velocity v times the effective spring constant keff, where keff is the slope of the force distance curve at rupture (23). According to the theory that a single energy barrier is crossed in the thermally activated regime, a linear rise of the unbinding force with respect to a logarithmically increasing loading rate is expected (22, 24). Indeed, such a dependency was seen in the force spectroscopy for the rupture of a single virus–receptor bond. From the plot in Fig. 4A, the separation of the energy barrier from the equilibrium position xβ and the kinetic off-rate constant koff can be determined by using Eq. 2 (see Materials and Methods) (22). The values were xβ = 0.40 ± 0.16 nm and koff = 0.05 ± 0.15 s−1. With the kinetic on-rate as determined above, the dissociation constant KD = koff/kon was calculated to 24 nM.
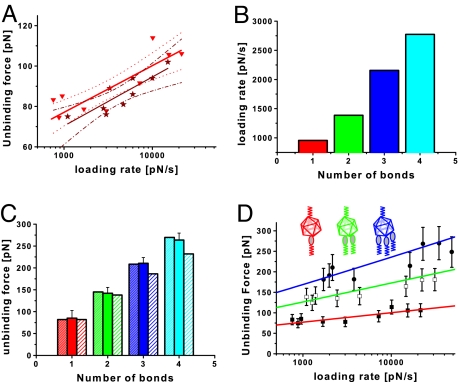
Fig. 4.
Force spectroscopy of the interaction between HRV2 and MBP-V1-8. (A) Dynamic force spectra of HRV2 interacting with MBP-V1-8 using the immobilization scheme shown in Fig. 1 (red triangles), and the reversed scheme in which the receptor protein (MBP-V1-8) was bound to the tip and HRV2 was immobilized on the surface (dark red stars). The resulting fit using Eq. 2 is shown by the solid triangles (red line) with error boundaries (red dotted line) and by the stars (dark red line) with error boundaries (dark red dash-dotted line). (B) Average of instantaneous loading rate acquired for the different numbers of receptor bound to a virus particle. (C) The mean unbinding force from the fitted Gaussian versus the number of receptors bound. The experimental data (solid fill) can be reproduced with Eq 1 (densely striped bars), using the average loading rate determined for the respective number of bonds. The slight striped bars show the results of Eq. 1 using an uncorrected loading rate. (D) Dynamic force spectra of 1 (solid squares), 2 (empty squares), and 3 (solid circles) MBP-V1-8 molecules bound to HRV2. The results are very well represented by a Markov binding model (Eq. 1) for the double bond (green) and triple bond (blue) interaction using parameters derived from the single-barrier model fit (red). The error bars account for the uncertainty in the determination of the spring constant and the uncertainty in finding the most probable rupture force.
Inverting the immobilization procedure of virus and receptor, i.e., binding a single receptor molecule to the tip and the virus to the surface, resulted in a configuration where only 1 receptor molecule can bind to the virus. Indeed, the resulting PDFs displayed only a single force peak, and no multiple unbinding force values were obtained. Force spectroscopy data acquired with this configuration were compared with the data above (Fig. 4A). According to a t test, the 2 force spectroscopy curves were not significantly different within a 95% confidence level, indicating that the virus–receptor unbinding force is independent of the arrangement of the binding partners. The values for the separation of the energy barrier from the equilibrium position and the kinetic-off rate constant were determined to be 0.41 ± 0.2 nm and 0.09 ± 0.07 s−1, respectively.
Rupture of Multiple Bonds.
Analyzing the behavior of multiple bonds requires a careful consideration. Our experimental system represents a configuration for multiple-bond attachments, where load is shared between all of the bonds (“parallel attachment”). In addition to this connection architecture, rupture dynamics of multiple bonds depends on the failure mode. In the correlated mode all bonds are closely coupled, and failure of 1 bond implies failure of the rest of the bonds. In the uncorrelated system all attachments can fail independently, and the load force is redistributed among the surviving bonds. Because the long tethers used in this study do not cause close coupling, the analysis for uncorrelated failure was used (25).
The uncorrelated failure mode implies no particular mechanical coupling between individual bonds. Williams (26) showed that the force-induced rupture of multiple uncorrelated bonds can be described as a Markovian sequence. The measured unbinding force, i.e., most probable rupture force, F* scales with the number of bonds, NB, and the measured loading rate, rf, as
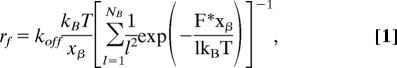 |
where xβ and koff are the corresponding parameters derived from a single bond analysis using Eq. 2 (see Materials and Methods). This equation was recently verified by using single-molecule force spectroscopy experiments of multiple antibody–antigen interactions (18, 25). Critical to the analysis of multiple bond rupture is the proper treatment of the loading rate rf = keffv, determined by the pulling velocity v and the effective spring constant keff. For springs in parallel the effective spring constant keff,NB is the sum of the individual spring constants keff,1 with forces equally distributed to the bonds keff,NB = NBkeff,1(F*/NB).
A direct consequence of this relationship is that rf is a function of the number of bonds NB. Accordingly, proper loading rates were calculated for each number of bonds involved, resulting in higher loading rates for unbinding events with more receptors involved (Fig. 4B).
The unbinding force as a function of the number of bonds is shown in Fig. 4C. At this particular retraction velocity of 300 nm·s−1, up to 4 bonds were observed. Using Eq. 1 and the loading rates shown in Fig. 4B the observed unbinding forces could be well reproduced. The dependence of the loading rate on the number of bonds is important, otherwise the unbinding force would saturate with increasing number of attached receptor molecules (Fig. 4C). Fig. 4D shows this analysis applied to a full force spectroscopy experiment for double and triple bonds. The single-bond interaction was fitted by using the single barrier model (Eq. 2). Using the acquired xβ and koff from this fit together with Eq. 1, dynamic force spectra for multiple bond interactions were calculated. A good agreement with the measured dynamic force spectra for double and triple bond interaction was found (Fig. 4C).
Virus Binding to Living Cells.
Finally, in vivo experiments with HRV2 immobilized on an AFM cantilever and cells grown in a tissue culture dish were performed (Fig. 5A). In these experiments M4-LDLR cells were used. This line is an SV40 large-T antigen-immortalized mouse fibroblast line overexpressing human LDLR; the endogenous LDLR and LRP genes were disrupted (27, 28).
Fig. 5.
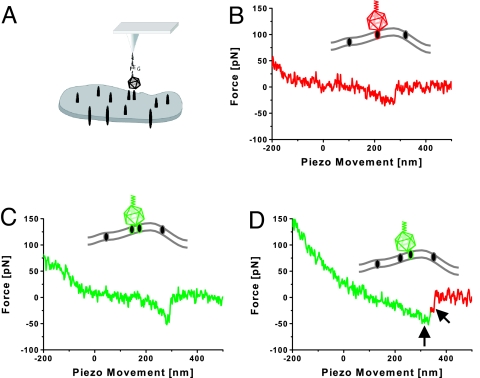
Force traces of virus–cell interaction acquired by the AFM in force spectroscopy. (A) Schematic representation of the experimental procedure. HRV2 was attached to the tip as above. Human LDLR-expressing cells were grown on glass substrates. (B) Virus interacting with a single receptor. (C) Virus interacting with 2 receptor molecules. (D) Serial rupture of 2 receptors from the virus. The arrows indicate the individual rupture events.
In most cases, rupture forces of ≈22 pN at a retraction velocity of 1,500 nm·s−1 were found (Fig. 5B). Occasionally events with higher force values were recorded (Fig. 5C). Rare events with a stepwise rupture (Fig. 5D) strongly indicate that these force values correspond to the unbinding of individual receptor molecules. Indeed, the spectrum of rupture forces showed a significant component of higher forces, which can be attributed to the virus being bound to multiple receptors (Fig. 6A). Increasing the contact time leads to a redistribution of rupture forces to higher values (Fig. 6B). This finding is taken to indicate that HRV2 was more often bound to multiple receptors. Perfusion of the chamber with an EGTA-containing medium resulted in a drop of the binding probability from ≈22% to ≈4%. The same binding probability was found in experiments with M4 cells, which were not transfected to overexpress human LDL receptor.
Fig. 6.
Force spectroscopy of HRV2 binding to tissue culture cells overexpressing human LDLRs. (A and B) Distribution of rupture forces (black line) as a function of contact time. As in Fig. 3, extension of the contact time increased the number of rupture events with higher forces, indicating that >1 receptor is bound to the virus. The sums of Gaussians were fitted to the experimental distribution of rupture forces, yielding an almost perfect reproduction of the experimental data. (C) The binding probability is plotted as a function of the contact time. The solid line is the result of a least-squares fit of a monoexponential decay (0.23 exp((t − 20)/220)). (D) Dynamic force spectra of 1 (squares) and 2 (stars) cell receptors bound to HRV2. The results are very well represented by a Markov binding model (Eq. 1) for the double bond (green line) interaction by using parameters derived from the single-barrier model fit (red line).
As above, the kinetic on rate constant kon can be estimated as (τceff)−1. The characteristic interaction time τ was found from fitting the binding probability as a function of the contact time to P = A (1 − exp(−(t − t0)/τ)), leading to a characteristic interaction time of 220 ms. To estimate the effective concentration, the number of binding partners must be divided through the effective volume. Because the configuration of the tip did not change, the effective volume also is the same as above. In this experiment only force values corresponding to 2 bound receptors were found, therefore the number of binding partners was set to 2, yielding kon = 2 × 105 M−1s−1.
Varying the pulling velocity v leads to a full force spectroscopy plot (Fig. 6D) for 1 and 2 bound receptors. As above, the loading rate rn for n bound receptors was keff,nv and keff,n was the average slope at rupture for n receptors interacting with HRV2. Fitting the case of 1 receptor–HRV2 interaction to Eq. 2 (see Materials and Methods) allowed the determination of the kinetic off-rate constant koff to be 5 × 10−7 s−1. The dissociation constant was determined to be KD = koff/kon = 2 pM. Using the parameters obtained by fitting the force spectroscopy plot of 1 bound receptor, together with Eq. 1, a theoretical dynamic force spectrum for 2 bound receptors was calculated. This theoretical expected spectrum agreed very well with the measured data (Fig. 6D).
Discussion
We investigated the first step of viral infection, the attachment of a virus to a cell, with single-molecule force spectroscopy. Such single-molecule experiments offer several advantages. First, by conducting many sequential measurements they allow the distribution of molecular properties of inhomogeneous systems to be determined. Second, being direct records of the system's fluctuations, single-molecule trajectories provide dynamic and statistical information, which is often hidden in ensemble-averaged results. Finally, they permit real-time observation of rarely populated transients, which are difficult or impossible to capture with conventional methods (29). In this particular study a HRV was immobilized to an AFM tip via a flexible polyethylene glycol linker, and the unbinding forces dominating virus–receptor interaction were measured, allowing counting for the number of receptor molecules interacting with the virus and estimation of kinetic rates.
As proof of principle we used a model system with VLDL receptors bound to a mica surface with the same chemistry as used to immobilize the virus to an AFM cantilever. This process resulted in a dense layer of receptors, able to freely rotate and diffuse within the volume defined by the length of the tether, allowing for multiple virus–receptor interactions. The distribution of rupture forces showed multimodal behavior corresponding to multiple virus–receptor interactions. Perfusion of the liquid cell with Ca2+-free buffer containing EGTA resulted in a drop of the binding probability. The same low binding probability was found in experiments with M4 cells, which were not transfected to overexpress human LDL receptor, verifying that the forces indeed arise from virus–receptor interactions.
With increasing contact time, the force spectrum changed. Higher rupture forces were measured, indicating binding of more receptors to the virus. Moreover, the binding probability increased with longer contact time. This behavior was modeled with a pseudo first-order kinetics, yielding the kinetic on-rate constant kon. In this particular case we found kon = 1 × 106 M−1s−1, which is very close to the 1.5 × 106 M−1s−1 as measured by surface plasmon resonance (SPR) (30). Nevertheless, it should be kept in mind that knowledge of the effective volume, i.e., the effective radius, is critical for a correct determination of this value from the AFM data.
Varying the contact time of virus and receptor allowed calculating kon, and varying the retraction speed led to the value of the kinetic off-rate constant koff as determined from fitting the most probable rupture force for a single virus–receptor interaction as a function of the loading rate according to the single barrier model (Eq. 2), yielding koff ≈0.05 s−1. In addition, the kinetic off-rate constant of the same virus–receptor interaction, this time in inverted configuration, i.e., receptor immobilized on the tip and virus on the surface, was determined. The 2 values were not significantly different. Knowing the kinetic rate constants, we found that the dissociation constant KD = koff/kon was 24 nM.
Whereas the kinetic on-rate compared very favorably with the data obtained from SPR measurements, the koff derived from the AFM measurements were by ≈200 times higher as compared with those from SPR (30). Consequently, the equilibrium binding constant was also 200 times higher. In AFM experiments the measured off-rate is governed by the prominent barriers traversed in the energy landscape along the force-driven bond-dissociation pathway. In this force-induced dissociation outer energy barriers can be suppressed, and for this case the off-rate is higher than in the thermal case (22). In addition, it should be noted that SPR measurements have a well-known problematic of rebinding and equilibrium binding constants determined by SPR are at least 1 order of magnitude higher than equilibrium binding constant measured with fluorescence correlation spectroscopy (31, 32).
In contrast to the binding parameters measured via methods such as SPR (30), capillary electrophoresis (15), or fluorescence correlation spectroscopy (32), the single-molecule experiments reported here are suited to investigate processes occurring at the cell surface during virus attachment.
Multiple virus–receptor interactions cannot be described with the single-barrier model. Instead, it can be considered as an uncorrelated Markov chain, but with the parameters determined from the fit of a single virus–receptor interaction to the single-barrier model.
To address our main aim of characterizing the virus–receptor interaction during viral entry, forces governing virus attachment to the cell were investigated with force spectroscopy. HRV2 was immobilized to AFM tips and the native LDL receptors were anchored in the cell membrane. To increase the probability of virus–receptor contact we used a mouse fibroblast overexpressing human LDLR (27, 28, 33). Like in our model system we found a bimodal behavior in the force spectrum of rupture forces, strongly suggesting that the virus binds to >1 receptor upon attaching to the plasma membrane. The multimodal behavior can be explained with an uncorrelated Markov chain, indicating lack of cooperativity between the individual receptors, which suggests that the process is only diffusion controlled with no additional driving force. Multiple-receptor binding characteristically occurred within 200 ms for this particular system, and it is possible that this time is much longer at lower receptor density. In a previous study, transition from weakly bound to tightly bound virus was found to happen after minutes (7), whereas our data suggest that after seconds several receptors are bound to the virus. This discrepancy might come from the different setups: in the first case the measurement of an ensemble compared with single-molecule measurements done here. Nevertheless, we cannot exclude that the transition represents an early stage of internalization rather than multiple binding of the receptors. However, it is unlikely that a virus particle becomes internalized without receptor clustering and when bound to only a single receptor molecule.
In addition, we estimated kon = 2 × 105 M−1s−1, koff = 5 × 10−7 s−1, and KD = 2 pM. The value for kon is ≈1 order of magnitude lower than the on-rate constant obtained for VLDLR1-8, whereas the value for koff is many orders of magnitudes lower than the value obtained for VLDLR1-8. Consequently, the dissociation constant KD is also lower than that of VLDLR1-8. This finding contradicts data on in vitro interactions between LDLR and VLDLR fragments with HRV2, indicating much stronger binding of VLDLR. It is thus possible that the particular environment of the LDLR within the plasma membrane strongly increases its affinity for the virus. It is possible that the glycocalyx decreases the probability of encounter with LDLR but that other components of the plasma membrane help to keep the virus in place once it has bound to its receptor.
Materials and Methods
Tip Chemistry.
Commercially available AFM cantilevers (Veeco Instruments) were amine-functionalized as described, by using the room temperature method for reaction with ethanolamine hydrochloride (34). A heterobifunctional PEG linker (aldehyde-PEG-NHS) (12) was attached by incubating the tip for 2 h in 0.5 mL of chloroform containing 6.6 mg·mL−1 aldehyde-PEG-NHS and 0.5% triethylamine, resulting in acylation of surface-linked ethanolamine by the N-hydroxysuccinimide (NHS) group (Fig. 1). After rinsing with chloroform and drying, the tips were incubated in a mixture of 50 μL of ≈0.05 mg·mL−1 HRV2 in HBS (10 mM Hepes, 150 mM NaCl, pH 7.4) and 2 μL of 1 M NaCNBH3 (freshly prepared by dissolving 32 mg of solid NaCNBH3 in 500 μL of 10 mM NaOH) for 50 min. Then, 5 μL of 1 M ethanolamine hydrochloride (adjusted to pH 9.6 with 20% NaOH) was added and incubation was continued for 10 min to block unreacted aldehyde groups. The specificity of the coupling was checked by blocking the cantilever-bound PEG-aldehyde for 30 min with 50 μL of 1 M ethanolamine (pH 9.6).
Surface Preparation.
Soluble VLDLR fragments encompassing the entire ligand binding domain i.e., all 8 binding repeats fused to MBP at the N terminus and to His6 at the C terminus (VLDLR1-8), were prepared as described (35). They were attached to mica by using a protocol analogous to that described above. Freshly cleaved sheets of mica were amino-functionalized, and aldehyde-PEG-NHS was subsequently attached. The receptor was bound by incubating the derivatized mica sheet with 200 μL of ≈0.2 mg·mL−1 VLDLR1–8 and 8 μL of freshly prepared 1 M NaCNBH3. Unreacted aldehyde groups were blocked with ethanolamine.
Force Measurements.
All measurements were carried out in HBS with 2 mM CaCl2 by using a PicoPlus (Molecular Imaging) AFM. Force distance cycles were performed at room temperature by using HRV2-coated tips with 0.01–0.03 Nm−1 nominal spring constants and mica-immobilized VLDLR1-8. By varying the maximum contact force, different contact times were achieved. The contact time was defined as the time the virus particle was close enough to the surface to allow for contacting a receptor. To exclude applied force/contact area effects the contact time was also varied by leaving the contact force constant and changing the retraction speed. Both methods showed similar results. The spring constants of the cantilevers were determined by using the thermal noise method (36, 37). Empirical force distributions of the rupture forces of the last unbinding event (PDF) were calculated as described (19). The PDFs were fitted with the equation
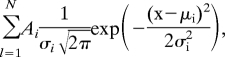 |
including the boundary condition
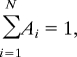 |
taking the probability density property of the PDFs into account, where Ai is a prefactor, μi is the position of the peak, and σi is the width of the peak. These measurements were performed at 300-nm sweep range and 2-Hz sweep rate.
For force spectroscopy a sweep range of 150 nm and sweep rate of 0.5–10 Hz was used, resulting in loading rates from 1,000–24,000 pNs−1. The loading rates were determined by multiplying the pulling velocity with the effective spring constant, i.e., the mean slope at rupture. In the single-barrier model (22), the most probable rupture force F* is given as function of the loading rate
where the force fβ is the ratio of the thermal energy kBT and xβ with the latter marking the thermally averaged projection of the transition state along the direction of the force. The parameters xβ and koff were determined by fitting F* against ln r. The accuracy of the parameters was calculated by using propagation of errors (38) assuming that the standard error of F* is ≈15% (10% for the spring constant determination and 5% to account for the uncertainty in determining the most probable rupture force).
Cell Preparation.
M4-LDLR cells (an SV40 large-T antigen-immortalized mouse fibroblast line overexpressing human LDLR) (25, 26) were grown in DMEM containing 1% glutamine, 1% penicillin-streptomycin, and 10% FCS in Petri dishes. For AFM measurements the growth medium was exchanged for HBS with 2 mM CaCl2. All measurements were done at room temperature.
Acknowledgments.
This work was supported by Austrian Science Fund Projects P-14549, P-18693-B09, and N-00104 and the European Union Project Tips4Cells (Grant LSHG-CT-2005-512101).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Hofer F, et al. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc Natl Acad Sci USA. 1994;91:1839–1842. doi: 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vlasak M, et al. The minor receptor group of human rhinovirus (HRV) includes HRV23 and HRV25, but the presence of a lysine in the VP1 HI loop is not sufficient for receptor binding. J Virol. 2005;79:7389–7395. doi: 10.1128/JVI.79.12.7389-7395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider WJ, Nimpf J. LDL receptor relatives at the cross-road of endocytosis and signaling. Cell Mol Life Sci. 2003;60:892–903. doi: 10.1007/s00018-003-2183-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nykjaer A, Willnow TE. The low-density lipoprotein receptor gene family: A cellular Swiss army knife? Trends Cell Biol. 2002;12:273–280. doi: 10.1016/s0962-8924(02)02282-1. [DOI] [PubMed] [Google Scholar]
- 5.Rudenko G, et al. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 6.Daly NL, Scanlon MJ, Djordjevic JT, Kroon PA, Smith R. Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. Proc Natl Acad Sci USA. 1995;92:6334–6338. doi: 10.1073/pnas.92.14.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noble Harvey J, Lonberg Holm K. Sequential steps in attachment of human rhinovirus type 2 to HeLa cells. J Gen Virol. 1974;25:83–91. doi: 10.1099/0022-1317-25-1-83. [DOI] [PubMed] [Google Scholar]
- 8.Snyers L, Zwickl H, Blaas D. Human rhinovirus type 2 is internalized by clathrin-mediated endocytosis. J Virol. 2003;77:5360–5369. doi: 10.1128/JVI.77.9.5360-5369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prchla E, Kuechler E, Blaas D, Fuchs R. Uncoating of human rhinovirus serotype 2 from late endosomes. J Virol. 1994;68:3713–3723. doi: 10.1128/jvi.68.6.3713-3723.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc Natl Acad Sci USA. 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonanni B, et al. Single molecule recognition between cytochrome C 551 and gold-immobilized azurin by force spectroscopy. Biophys J. 2005;89:2783–2791. doi: 10.1529/biophysj.105.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebner A, et al. A new, simple method for linking of antibodies to atomic force microscopy tips. Bioconjugate Chem. 2007;18:1176–1184. doi: 10.1021/bc070030s. [DOI] [PubMed] [Google Scholar]
- 13.Gruenberger M, et al. Avian homologs of the mammalian low-density lipoprotein receptor family bind minor receptor group human rhinovirus. J Virol. 1995;69:7244–7247. doi: 10.1128/jvi.69.11.7244-7247.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marlovits TC, Abrahamsberg C, Blaas D. Very-low-density lipoprotein receptor fragment shed from HeLa cells inhibits human rhinovirus infection. J Virol. 1998;72:10246–10250. doi: 10.1128/jvi.72.12.10246-10250.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konecsni T, et al. Twelve receptor molecules attach per viral particle of human rhinovirus serotype 2 via multiple modules. FEBS Lett. 2004;568:99–104. doi: 10.1016/j.febslet.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 16.Kienberger F, et al. Visualization of single receptor molecules bound to human rhinovirus under physiological conditions. Structure {London) 2005;13:1247–1253. doi: 10.1016/j.str.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Lonberg-Holm K, Whiteley NM. Physical and metabolic requirements for early interaction of poliovirus and human rhinovirus with HeLa cells. J Virol. 1976;19:857–870. doi: 10.1128/jvi.19.3.857-870.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sulchek T, Friddle RW, Noy A. Strength of multiple parallel biological bonds. Biophys J. 2006;90:4686–4691. doi: 10.1529/biophysj.105.080291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baumgartner W, Hinterdorfer P, Schindler H. Data analysis of interaction forces measured with the atomic force microscope. Ultramicroscopy. 2000;82:85–95. doi: 10.1016/s0304-3991(99)00154-0. [DOI] [PubMed] [Google Scholar]
- 20.Baumgartner W, Gruber HJ, Hinterdorfer P, Drenckhahn D. Affinity of trans-interacting VE-cadherin determined by atomic force microscopy. Single Mol. 2000;1:119–122. [Google Scholar]
- 21.Atkins PW. Physical Chemistry. Oxford: Oxford Univ Press; 1998. [Google Scholar]
- 22.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans E, Ritchie K. Strength of a weak bond connecting flexible polymer chains. Biophys J. 1999;76:2439–2447. doi: 10.1016/S0006-3495(99)77399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 25.Sulchek TA, et al. Dynamic force spectroscopy of parallel individual Mucin1-antibody bonds. Proc Natl Acad Sci USA. 2005;102:16638–16643. doi: 10.1073/pnas.0505208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams PM. Analytical descriptions of dynamic force spectroscopy: Behavior of multiple connections. Anal Chim Acta. 2003;479:107–115. [Google Scholar]
- 27.Willnow TE, Herz J. Genetic deficiency in low density lipoprotein receptor-related protein confers cellular resistance to Pseudomonas exotoxin A. Evidence that this protein is required for uptake and degradation of multiple ligands. J Cell Sci. 1994;107:719–726. [PubMed] [Google Scholar]
- 28.Ishibashi S, et al. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J Clin Invest. 1993;92:883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nevo R, et al. A molecular switch between alternative conformational states in the complex of Ran and importin β1. Nat Struct Biol. 2003;10:553–557. doi: 10.1038/nsb940. [DOI] [PubMed] [Google Scholar]
- 30.Moser R, et al. Neutralization of a common cold virus by concatemers of the third ligand binding module of the VLDL-receptor strongly depends on the number of modules. Virology. 2005;338:259–269. doi: 10.1016/j.virol.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 31.Schubert F, Zettl H, Hafner W, Krauss G, Krausch G. Comparative thermodynamic analysis of DNA–protein interactions using surface plasmon resonance and fluorescence correlation spectroscopy. Biochemistry. 2003;42:10288–10294. doi: 10.1021/bi034033d. [DOI] [PubMed] [Google Scholar]
- 32.Wruss J, et al. Attachment of VLDL receptors to an icosahedral virus along the 5-fold symmetry axis: Multiple binding modes evidenced by fluorescence correlation spectroscopy. Biochemistry. 2007;46:6331–6339. doi: 10.1021/bi700262w. [DOI] [PubMed] [Google Scholar]
- 33.Herz J, Clouthier DE, Hammer RE. LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell. 1992;71:411–421. doi: 10.1016/0092-8674(92)90511-a. [DOI] [PubMed] [Google Scholar]
- 34.Riener CK, et al. Simple test system for single molecule recognition force microscopy. Anal Chim Acta. 2003;479:59–75. [Google Scholar]
- 35.Ronacher B, Marlovits TC, Moser R, Blaas D. Expression and folding of human very-low-density lipoprotein receptor fragments: Neutralization capacity toward human rhinovirus HRV2. Virology. 2000;278:541–550. doi: 10.1006/viro.2000.0636. [DOI] [PubMed] [Google Scholar]
- 36.Butt HJ, Jaschke M. Calculation of thermal noise in atomic force microscopy. Nanotechnology. 1995;6:1–7. [Google Scholar]
- 37.Hutter JL, Bechhofer J. Calibration of atomic force microscopy tips. Rev Instr Sci. 1993;64:1868–1873. [Google Scholar]
- 38.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in C: The Art of Scientific Computing. 2nd ed. Cambridge, UK: Cambridge Univ Press; 1992. [Google Scholar]