Abstract
We report a simple and effective method for the high-throughput purification of a variety of nucleic acids (NAs) from whole cell lysates or whole blood using a photactivated polycarbonate solid-phase reversible immobilization (PPC-SPRI) microfluidic chip. High-throughput operation was achieved by placing 96 purification beds, each containing an array of 3800 20 µm diameter posts, on a single 3″ × 5″ polycarbonate (PC) wafer fabricated by hot embossing. All beds were interconnected through a common microfluidic network that permitted parallel access through the use of a vacuum and syringe pump for delivery of immobilization buffer (IB) and effluent. The PPC-SPRI purification was accomplished by condensation of NAs onto a UV-modified PC surface in the presence of the IB comprised of polyethylene glycol, NaCl, and ethanol with a composition dependent on the length of the NAs to be isolated and the identity of the sample matrix. The performance of the device was validated by quantification of the recovered material following PCR (for DNA) or RT-PCR (for RNA). The extraction bed load capacity of NAs was 206 ± 93 ng for gDNA and 165 ± 81 ng for TRNA from Escherichia coli. Plate-to-plate variability was found to be 35 ± 10%. The purification process was fast (>30 min) and easy to automate, and the low cost of wafer fabrication makes it appropriate for single-use applications.
The output of most clinical or forensic nucleic acid-based assays is highly dependent on the quality of the input sample as are other techniques typically used in biological discovery such as cloning, gene expression analysis and DNA sequencing. It is not surprising that the development of new purification protocols for a variety of nucleic acid (NA) samples continues to be a subject of intense interest. Factors typically considered when evaluating novel NA purification procedures include DNA/RNA yield and purity, the labor-intensiveness of the protocol, carryover of contaminants that may interfere with downstream processing (e.g., PCR inhibitors), potential handling and disposal of toxic materials, and the overall cost of the procedure.
Many NA purification methods involve selective precipitation using salts or organic solvents. However, surface binding techniques have recently found great favor due to their ease of use and high recoveries of the target NAs. Surface binding techniques (e.g., solid-phase extraction, SPE) use selective adsorption of the NAs onto a solid surface followed by a solvent or surface-property change for elution of the purified target. Often, a wash step is used to remove coadsorbing impurities and further improve sample purity. Various solid-phase capture media are commercially available (i.e., silica, polymers, pH sensitive polymer coatings) in different formats, such as flow through filter membranes, spin columns, magnetic microparticles, or surface-coated microtubes.
SPE methods utilize different NA-selective adsorption mechanisms. For example, chemical agents can cause selective NA condensation via modification of the electrostatic interactions between NA segments. Condensation can also occur through localized distortion of the helical structure. Condensing agents typically decrease electrostatic repulsion between NA segments through the use of mono/multivalent cations that can neutralize the negative charge on the phosphate backbone or cause reorientation of the water dipoles near the NA surface.1,2 Neutral polymers, such as polyethylene glycol (PEG), when used at proper concentrations and in the presence of adequate amounts of salt, provoke NA condensation through an excluded volume mechanism.3 Accordingly, biomolecules are excluded from the regions of the solvent occupied by the inert synthetic polymer and thus, are concentrated until their solubility is exceeded and precipitation occurs. Due to the nonspecific repulsive interactions between the chains of two different polymers (i.e., PEG and DNA) present in the same solvent, they both compete for the solvent space, which results in their separation into two phases.1,4
The incorporation of SPE protocols into microfluidic formats is a relatively new research approach and is appealing because microfluidics offer the ability to process samples in a high-throughput and automated format. In addition, because the capture occurs in a closed architecture, the potential risk of cross-contamination is significantly reduced. By combining NA purification with other on-chip processing steps, fully integrated lab-on-a-chip systems can be realized with a concomitant reduction in the time, complexity, and cost of carrying out the analysis.5,6
Various approaches for microfluidic-based purification of NAs have been reported. For example, DNA capture, purification, and preconcentration on microchips have been achieved using SPE with silica and chaotropic salts.7 The elution of DNA from the silica support is accomplished with water or low ionic strength buffer with extraction efficiencies of ~50%.7 Other studies have employed an amino-silane modified open channel microchip to extract DNA from blood with adsorption/desorption of DNA controlled through pH changes in the solvent.8 Cao et al.9 demonstrated a DNA SPE method based on changes in the charge of chitosan as the purification medium. Tian et al.10 evaluated the use of silica resins for the direct extraction of DNA from biological samples in a miniaturized format and reported a recovery of ~70%, while ~80% of cellular proteins were removed. Recently, Wen et al.11 addressed protein co-adsorption during DNA purification by designing a dual-phase microchip for retention of protein followed by a second phase containing monoliths for the selective extraction of DNA. The authors observed a DNA extraction efficiency of 69% from human blood.11
We have previously demonstrated the use of a photactivated polycarbonate solid-phase reversible immobilization (PPC-SPRI) microchip for the purification of genomic DNA (gDNA) from whole cell lysates.12 gDNA was selectively captured on this photoactivated surface in an immobilization buffer (IB), which consisted of 3% PEG, 0.4 M NaCl, and 70% ethanol. The recovery of DNA following the purification was estimated to be 85 ± 5%. The extraction bed consisted of a series of micro-posts that were hot-embossed into fluidic channels in the polymer substrate to increase the available load of DNA and thus, did not require subsequent processing steps to create a high-surface area extraction bed. Using a post-to-post distance of 10 µm, as opposed to 1–3 µm pore size in polymer monoliths,11 extraction of DNA could be achieved without the risk of clogging the device from cellular debris or other sample matrix components. This PPC-SPRI method has also been reported to possess the ability to purify Sanger sequencing products by selecting the appropriate IB conditions.13
While the aforementioned reports were directed toward DNA purifications, widespread work on RNA requires microfluidic methods for RNA purification as well. RNA constitutes the bulk of the NA content in a cell; it is ~10 times more abundant than DNA with messenger RNA (mRNA) making up ~5% of total cellular RNA (TRNA).14 The 3′-ends of some prokaryotic and all eukaryotic mRNAs are polyadenylated, which makes this structural characteristic the most utilized feature for mRNA isolation using an oligo-dT coupled to a support medium (e.g., cellulose, polystyrene, polyacrylamide, or magnetic beads). Oligo-dT based capture of mRNA has been applied also to a microfluidic format.15 mRNA preconcentration onto magnetic beads incorporated in a chip was shown by Jiang et al.16 with a load of ~60 ng of mRNA. Recently, Satterfield et al. published mRNA isolation on high capacity porous polymer monoliths.17
The removal of ribosomal and transfer RNA (rRNA and tRNA, respectively) from TRNA samples is very beneficial, especially in the construction of random-primed cDNA libraries, where the presence of rRNA can dilute out the cDNAs of interest. However, rRNA is becoming a standard of choice in many RT-PCR and qRT-PCR applications. Therefore, it becomes necessary to consider techniques that can isolate and purify TRNA, which oligo-dT extraction phases cannot. Also for bacterial mRNA isolation, oligo-dT methods may not be appropriate because it has been shown that while 100% of eukaryotic mRNA possess poly(A) tails, it is only present in 2–60% of mRNA in bacteria.18
Another attractive aspect of microfluidics is the potential to produce wafers for high-throughput processing by densely packing analytical functions onto a single chip. Examples of such devices include 96- or 384-channel microchip capillary electrophoresis wafers, protein crystallization, or microarrays.19–23 Unfortunately, no report has appeared that describes the ability to use SPE to purify different types of NAs (DNA or RNA) using a single chip format in a high throughput format. Recently, we have reported on the fabrication of a 96-well, disposable microfluidic titer plate for the high-throughput purification of NAs.24 The UV-LiGA process was used for the fabrication of a nickel large area mold insert (LAMI) for hot embossing microfluidic structures into PC that could be used for the purification of genomic DNAs. While specifics on the fabrication of the device were reported, no information was provided as to the analytical performance of the device for purifying a variety of NAs, especially TRNA.
Herein, we report on the application of a high-throughput PPC-SPRI microchip for the purification of gDNA and TRNA of different origins (i.e., bacterial, mammalian) when found in complex matrices, including blood and blood containing high concentrations of sodium polyanetholesulfonate (SPS), used as an anticoagulant. The purification of gDNA and TRNA was accomplished using the same microchip with NA extraction specificity provided by selecting different IB cocktails tailored for the target to be analyzed. The extraction efficiency was evaluated by performing PCR (DNA) or RT-PCR (RNA) on the purified NAs with sorting via gel electrophoresis and fluorescence spectroscopy for securing quantitative information.
EXPERIMENTAL SECTION
Fabrication of PPC-SPRI 96-Well Microfluidic Plate
The microfluidic plate fabrication procedure involved three major steps: (1) Mold insert fabrication using a UV-LiGA process; (2) hot embossing of the microfluidic structures in a PC wafer; and (3) postprocessing of the microfluidic plate including drilling of sample reservoirs and interconnecting holes, UV-photoactivation of the immobilization beds and cover plate assembly. A detailed protocol for the entire process has been reported24 and a brief description can be found in the Supporting Information.
Design and Operation of PPC-SPRI 96-Well Microfluidic Plate
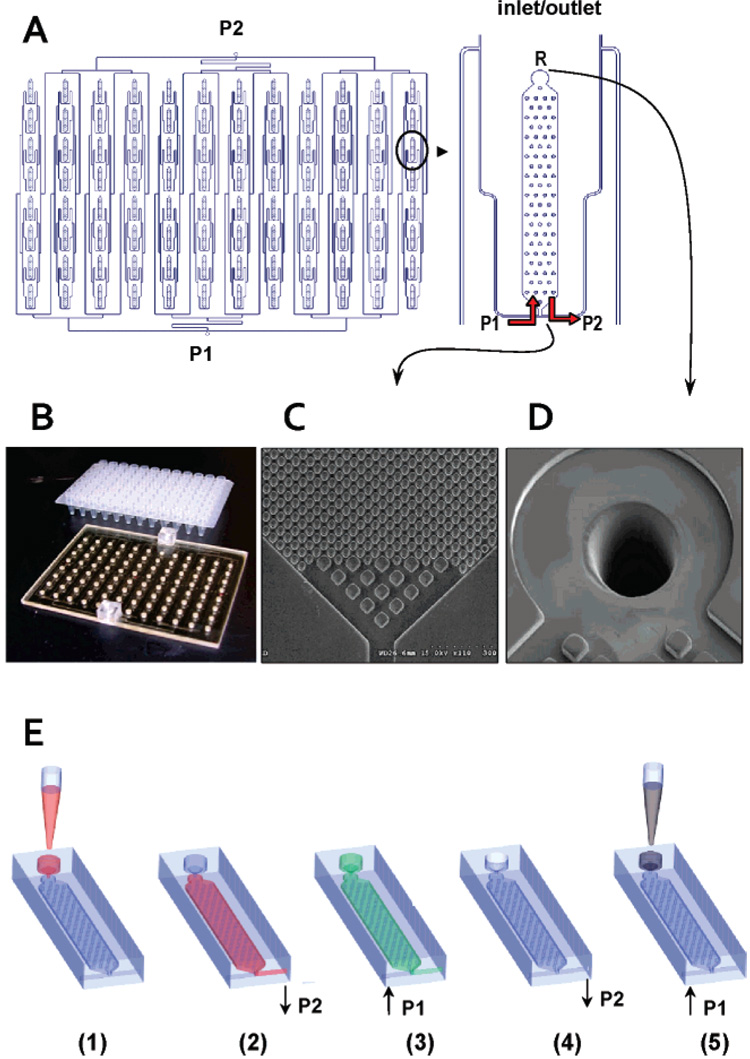
A schematic of the fluidic network of the 96-well plate is shown in Figure 1A. Also shown are pictures of the assembled device (B), SEM images of the posts in the purification bed (C), and laser-drilled 150 µm diameter reservoir-to-chip interconnecting holes (D). Operation of the device is schematically presented in Figure 1 E and consists of 5 steps: (1) Samples are dispensed into all 96 reservoirs by either manual or robotic loading, which is preceded by mixing the sample with the appropriate IB. (2) A vacuum pump (port P2) is activated and all samples are simultaneously drawn from the inlet/outlet reservoir (R), through the extraction bed and out to waste. (3) The vacuum pump is turned off and ethanol from a syringe connected to port P1 is dispensed through the microfluidic network to wash each purification bed. Both the vacuum and syringe pumps act as end-of-the-line stop valves when not activated. (4) After a preset amount of ethanol is dispensed, it is drawn out of the microchip through port P2 to waste by activating the vacuum pump. The vacuum pump also draws air through the entire microfluidic network to remove ethanol and dry the extraction beds. Ethanol removal is critical, because residual amounts of ethanol may serve as a PCR inhibitor.25 (5) A syringe with water or elution buffer is connected to port P1 and effluent pushed through the channels, which is collected from the extraction beds into the appropriate reservoir from where the purified sample could be collected.
Figure 1. Layout of the 96-well PPC-SPRI titer plate-based microfluidic platform.
(A) Schematics of the individual immobilization beds and microfluidic control ports, P1 and P2. R is the sample inlet/outlet reservoirs. (B) Brightfield image of the 96-well PPC-SPRI microfluidic plate and a commercial 96-well titer plate. (C–D) SEMs of the entrance/exit section of the plate showing 20 µm posts and 50 µm entrance posts along with a ~150 µm bed entrance hole drilled using a KrF excimer laser. (E) Schematic showing the operation of the 96-well plate: (1) sample is introduced into the reservoir (R); (2) sample is pulled toward port P2 using a vacuum pump, and DNA/RNA is immobilized onto the extraction bed surface; (3) ethanol is pushed from port P1 to R using a syringe pump to wash the extraction bed; (4) ethanol in the extraction beds and in R is evacuated by using a vacuum pump through P2, and then the beds are dried by pulling air through P2; (5) effluent (water) is pushed from port P1 to R and purified NAs are collected.
Chemicals and Reagents
All bacterial cell lines used herein (see Table S1 in Supporting Information) were purchased from Sigma-Aldrich (Milwaukee, WI). Rat liver and Escherichia coli TRNAs were purchased from Ambion (Austin, TX). λ-DNA and M13mp18 was purchased from New England BioLabs, Ipswich, MA. Whole blood was purchased from Colorado Serum Company (Denver, CO). Polyethylene glycol (PEG, Mw = 8000), NaCl, sodium polyanetholesulfonate (SPS), ethanol, and 2-propanol were all purchased from Sigma and used as received.
Preparation of Inoculated Blood and Cell Lysis
Lyophilized bacteria were added to blood containing SPS (1 mg/mL) to achieve a final cells concentration of ~5 µg/µL. Thermal lysis was performed at 94 °C for 15 min, while mechanical lysing was performed at 14 000 rpm (3500 × g) for 20 min. Lysed cells in blood were suspended in the appropriate immobilization buffer (1/1, vol/vol).
Preparation of Various Immobilization Buffers
Stock solutions of different immobilization buffers were prepared with nuclease-free water. Final immobilization buffer compositions were as follows; Buffer A: 3% PEG/0.5 M NaCl; Buffer B: 3% PEG/ 0.5 M NaCl/63% EtOH; Buffer C: 6% PEG and 0.5 M NaCl; Buffer D: 5% PEG/0.5M NaCl/63% EtOH. DNA- or RNA-containing samples were suspended in the immobilization buffer of choice and processed using the PPC-SPRI microchip as described above.
Polymerase Chain Reaction (PCR) and Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
All PCRs and RT-PCRs were performed under a positive flow hood (AirClean Systems 600 Workstation, Raleigh, NC). PCR was carried out on the purified gDNA samples using 1–2 µL of the PPC-SPRI purified material taken from the wells of the microfluidic plate. The purified TRNA was transcribed to cDNA using RT. PCR was performed using a GeneAmp PCR reagent kit with AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA). RT of RNA was accomplished using a SuperScript First-Strand Synthesis System (Invitrogen). Details on the PCR, RT-PCR and primer sequences used for PCR can be found in the Supporting Information.
Slab Gel Electrophoresis
RT-PCR and PCR products were electrophoresed using a 3% agarose gel (Bio-Rad Laboratories, Hercules, CA) prestained with ethidium bromide. Amplicons were indexed against a DNA sizing ladder (50–1000 bp, Molecular Probes, Eugene, OR). The quality of the purified TRNA was evaluated using a 1% agarose gel and indexed against a sizing ladder (Ambion). Separation was performed at 4.8 V/cm in 1 × TBE (Tris/Boric Acid/EDTA, Bio-Rad Laboratories). After separation, the gels were imaged using a Logic Gel imaging system (Eastman Kodak).
UV-Vis Spectroscopy
SPS concentrations in all samples were determined using an Ultraspec 4000 Spectrometer (Amersham Pharmacia Biotech, Piscataway, NJ) at the absorption maximum of 284 nm for SPS. Calibration curves were constructed using a concentration range of SPS from 1 to 500 µg/mL. All measurements were performed in quartz cuvettes.
Fluorescence Spectroscopy
Native and purified DNAs were stained with PicoGreen (excitation 480 nm/ emission ~520 nm, Molecular Probes) with the resulting emission spectra used for determining the concentration of DNA in the samples (see Supporting Information for details). RNA concentration was determined by staining with RiboGreen (excitation 500 nm/ emission ~525 nm, Molecular Probes). Calibration curves were constructed using concentrations ranging from 5 pg/ µL to 1000 pg/µL for DNA/RNA standards. Correlation coefficients for standard curves of both materials were ≤0.998, however, the sensitivity of the DNA-PicoGreen curve was 3 × higher than that for RNA-RiboGreen.
RESULTS AND DISCUSSION
PPC-SPRI 96-Well Titer Plate Performance
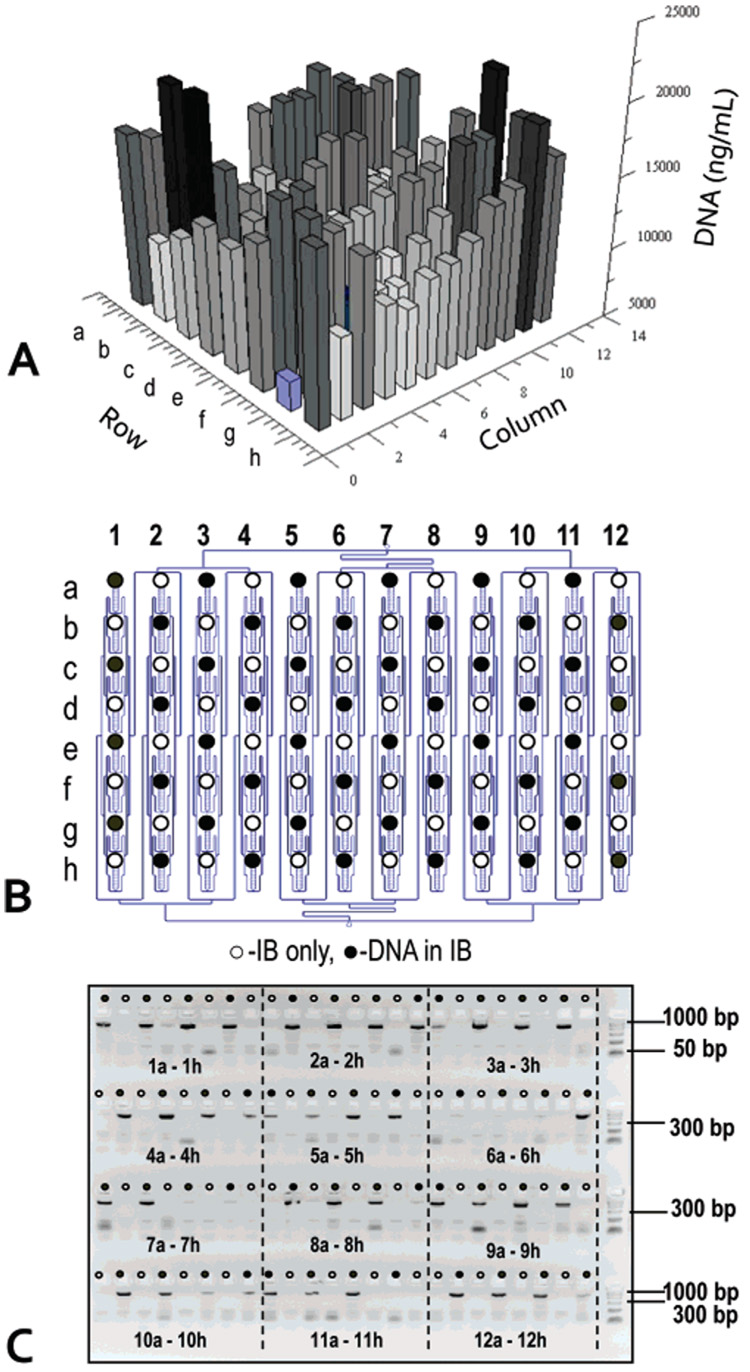
We characterized the performance of the 96-well PPC-SPRI plate by monitoring the extraction of gDNA from E. coli whole cell lysates. The lysates were mixed with an immobilization buffer (Buffer B) and 20 µL was dispensed into each of the 96 sample reservoirs of the plate. The samples were drawn through the purification beds at an average volumetric flow rate of 1.8 ± 0.7 µL/min, which was found to produce the optimal extraction efficiency. Ethanol (85%) was used to wash the beds and finally, the isolated DNA was released from the PPC-SPRI surface using deionized water. Figure 2A presents the well-to-well reproducibility of extracted gDNA. At surface saturation, the concentration of extracted gDNA was determined to be 13.7 ± 6.2 µg/mL, yielding the mass of isolated gDNA to be 206 ± 93 ng with a surface density of 724 ± 327 ng/cm2. From plate-to-plate (n = 4), the variability for gDNA recovery was determined to be 35 ± 10%. The average surface loading density of gDNA corresponded to the previously reported value of 790 ng/cm2 for E. coli gDNA captured in a single bed format.12 The RSD value for the intrachip recovery was, however, higher than that reported for the single bed format.12 We attributed this observation to the variability of the flow rate across the chip during operation since the extraction efficiency of targets to solid phases is dependent on the contact time, which is inversely related to the sample flow rate through the extraction bed.17 Although the chip was designed to possess a uniform pressure drop along the entire channel network (connecting channel branches were designed to be of the same length and have the same cross sectional dimensions, see Figure 1B), the manufacturing tolerances of the replication process will change the hydraulic resistance for fluid delivery to each of the extraction beds. For example, a measured manufacturing variability for both the height and width of the channels was of the order of 5%. Based on these measurements and the chip architecture, we estimated a flow rate variation through each extraction bed to be 37%.24 For a given sample and immobilization buffer, the NA recovery efficiency would also depend on the extraction bed characteristics, including the active surface area and the inter-post distance and post layout. Using the architecture associated with this 96 well chip design, we calculated a recovery efficiency of gDNA (intrachip) to be 63 ± 24% when averaged over all 96 extraction beds.
Figure 2. Performance evaluation of the recovery efficiency for the 96-well PPC-SPRI plate using genomic DNA from E. coli.
(A) Bar graph showing the recovered DNA (ng/mL) from each of the 96 extraction beds poised on the plate. The DNA concentration was evaluated using the PicoGreen assay. (B) Schematic of the 96-well plate showing the position of positive DNA samples (●) and blanks (○) containing only the immobilization buffer. (C) Fluorescence image of the agarose electrophoresis gel showing the presence of 48 amplicons at positions corresponding to (●) in (B). The separation was performed on a 3% agarose gel (4 × 24 wells) using a field strength of 4 V/cm in 1 × TBE. The sample volume loaded onto the gel was 5 µL along with 1 µL of loading dye. The DNA was immobilized using an immobilization buffer (Buffer B) at a flow rate of 1.8 ± 0.7 µL/min and eluted from the bed at a flow rate of 6.3 ± 0.6 µL/min using water.
We investigated potential cross-contamination between extraction beds by loading every other sample reservoir with gDNA. Cross-contamination could result from the fact that the extraction beds poised on the plate share a common fluidic network, which uses common ports (P1 and P2, see Figure 1) to pump wash and elution buffers through the extraction beds. Wells labeled with black dots in panels B and C of Figure 2 contained E. coli gDNA with the immobilization buffer (Buffer B), while the remaining wells contained only the immobilization buffer. After processing the plate, samples and blanks were interrogated by PCR by probing, using the appropriate sequence-specific primers, the 16s-RNA gene. The results of this experiment are presented in Figure 2C. After 30 PCR cycles, no amplicon was evident in the gel for beds that were loaded with immobilization buffer only, providing direct evidence for the lack of cross-contamination in this fluidic chip.
Parallel Purification of gDNA Samples from Whole Blood
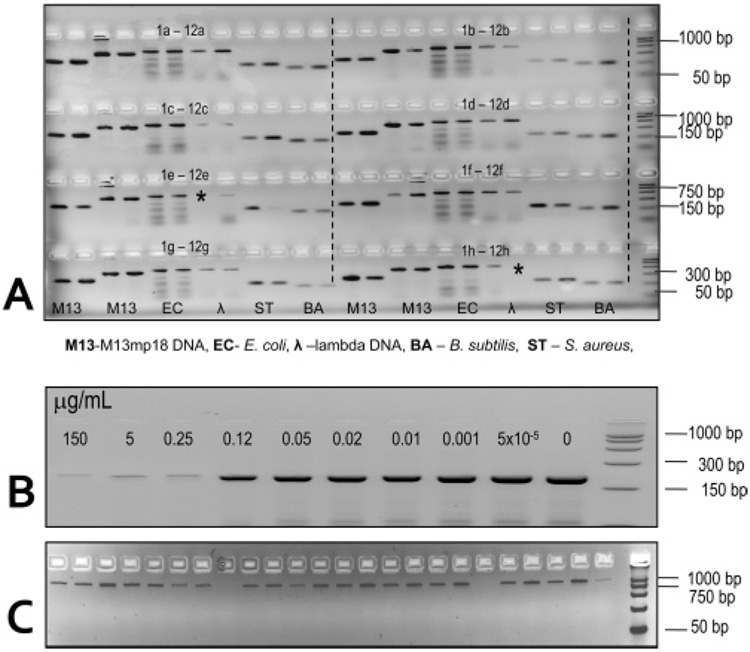
We and others have shown that the ability to generate successful amplicons via PCR from whole blood depends intimately on the ability to remove potential PCR inhibitors, such as hemoglobins.12 To test the ability of the 96-well PPC-SPRI plate to purify DNA of a variety of sizes from blood samples, whole blood was spiked with 3 different bacterial species that possess a diverse range of molecular sizes. These included Bacillus subtilis (4.2 Mbp genome) and Staphylococcus aureus (2.8 Mbp genome), which are gram-positive bacteria, and a gram-negative bacteria, E. coli with a genome size of 4.8 Mbp. In the same experiment, we also tested two other DNA lengths, λ-DNA with a genome size of 48.5 kbp and the single-stranded (ss) M13mp18 composed of 7.2 knt.
Blood was seeded with bacterial cells and mixed with Buffer A followed by thermal lysing of the sample. λ-DNA template and ssM13mp18 used Buffer C. The higher content of PEG used for λ-DNA and M13mp18 was required to promote condensation of these shorter DNAs onto the solid support.26 Figure 3A shows a fluorescence image of the PCR products generated following amplification with a DNA sizing ladder contained in lane m. Amplicons with sizes of 159, 204, 600, 500 bp for B. subtilis, S. aureus, E. coli, and λ-DNA were clearly seen indicating the successful recovery of the DNA from these targets using the PPC-SPRI chip. In addition, no evidence of failed PCRs was observed due to the inability to remove potential PCR inhibitors by the PPC-SPRI purification process or the inability to recover the DNA from whole blood. In addition, two products of 381 and 272 bps for the ssM13mp18 sample were also observed in the gel indicating that the PPC-SPRI device could also recover and purify ssDNAs. Careful inspection of Figure 3A indicated that successful PCRs were obtained for 94 of the 96 capture beds (~98% efficiency). All 96 PCRs were performed at the same thermal conditions and as such, were not necessarily the optimal temperature profiles for the various targets and their primers used herein. For example, the E. coli PCR amplifications produced nonspecific product bands (see lane c of Figure 3) due to the rather high Tm’s of these primers (see Table S1 in the Supporting Information) and the rather low temperatures used in the annealing step. By increasing the annealing temperature to 68 °C, we were able to observe only a single 600 bp product for E. coli (data not shown).
Figure 3.
(A) Fluorescence image of an ethidium-stained 3% agarose gel showing the presence of amplicons with sizes of 159, 204, 600, 500 bp for the B. subtilis, S. aureus, E. coli, λ-DNA, respectively, and 381 and 272 bp for M13mp18. The PCR products were generated from bacterial cells or gDNA seeded into whole blood and purified using the PPC-SPRI microchip (2 µL of purified sample was taken for each PCR). Prior to purification, the samples were thermally lysed and mixed with the appropriate immobilization buffer. In all cases, uniplex PCRs were performed on each sample purified using the PPC-SPRI microchip. (B) Fluorescence images of a 3% agarose gel with amplicons generated via PCR from 24 ng of S. aureus template containing the SPS anticoagulant at different concentrations (0–150 µg/mL, concentrations of SPS are indicated in the image). (C) Twenty-two purified S. aureus samples from blood containing SPS using the PPC-SPRI plate. Separation conditions and sample volumes loaded into the gel are the same as those reported in Figure 2. Lane m designates the DNA length markers used for PCR fragment sizing.
We next evaluated the elution volume required to remove the surface-immobilized gDNA from the extraction bed and the recovery efficiency of the gDNA. In these studies, we collected 30 1-µL increments of eluted material and estimated DNA concentration in that 1 µL effluent. In the first microliter of effluent, no DNA was detected. Approximately 56% of the input DNA was recovered in the next 3 µL and 83% was recovered in a total of 8 µL of elution medium when measured from a single extraction bed. No detectable amount of eluted DNA was found after introducing 8 µL of the elution medium. The elution profile indicated the process of reversible immobilization was fairly fast. With flow velocities of ~6 µL/min, optimized to reduce processing time without sacrificing DNA recovery, it took approximately 1.3 min to release the DNA from the bed surface (total volume = 8 µL). We speculate that the limiting factor for desorption of DNA from the PPC surface is hydration of the DNA and DNA segments regaining negative charge on the phosphate groups along with possible reorientation of water dipoles allowing for “detachment” from the PPC surface.1
Purification of DNA from Blood Containing SPS
Blood cultures contain anticoagulants, such as SPS. Typical concentrations of SPS in blood cultures are ~0.5 mg/mL. Unfortunately, SPS is a PCR inhibitor and tends to co-purify with DNA using for example, silica or ethanol-based extraction techniques.27 SPS is a high MW polyanion that is soluble in water but insoluble in alcohol. Fredricks et al. showed that phenol-chloroform fails to remove SPS.27 Alcohol precipitates SPS along with DNA. In addition, SPS binds to silica in the presence of chaotropes and elutes with water, similar to DNA. Washing cell pellets by centrifugation and their resuspension was shown to be unsuccessful in removing SPS, mainly because SPS binds to hemoglobin and erythrocyte membranes.28 Successful DNA purification from SPS was achieved by extraction with benzyl alcohol in an organic phase.27 A solid-phase extraction method demonstrating the ability to remove SPS from blood culture would be very beneficial and provide microfluidic-based protocols for processing cultured blood.
Figure 3B demonstrates the PCR-inhibitory effect of SPS at various concentrations when the anticoagulant was added to 24 ng of a solution containing an S. aureus template. Results indicated that an SPS concentration of 0.25 µg/mL (5 ng for a 20 µL PCR) was inhibitory under the PCR conditions used herein, whereas the presence of SPS at 0.12 µg/mL showed only slightly lower numbers of amplicons as compared to a control containing no SPS.27 Requiring an SPS concentration of <0.12 µg/mL for successful PCR indicated that unprocessed blood cultures would require dilution by more than 4000-fold in order to successfully amplify microbial DNA.
We next introduced S. aureus into inoculated blood samples containing 0.5 mg/mL SPS, typical clinical amounts used, and purified the sample using the PPC-SPRI chip. The final procedure for purification of DNA from blood containing SPS required Buffer A. Additional wash steps with 5–10 µL of immobilization buffer was also used to remove residual SPS. In the final step, the extraction bed was rinsed with 85% ethanol, dried, and the SPE-purified DNA was eluted with nuclease free DI water. Figure 3C presents the fluorescence image of PCR products generated from 22 separate purified samples subjected to PCR using S. aureus sequence-specific primers targeting the 16s-RNA gene of S. aureus. The gel electrophoresis confirmed the presence of the PCR products in all cases, suggesting the concentration of SPS in the PCR had been significantly reduced to less than 0.25 µg/mL following PPC-SPRI purification.
Purification of Prokaryotic and Eukaryotic TRNA
We evaluated the immobilization buffer composition and its effectiveness for the SPE of TRNA using the PPC-SPRI microchip. We evaluated 9 different IBs containing either TEG, PEG, and NaCl or MgCl2. The amount of recovered TRNA was estimated using the RiboGreen assay. The highest recovery of TRNA was achieved using IB Buffer D. The amount of RNA captured using the 96-well PPC-SPRI chip was determined to be, at surface saturation, 165 ± 81 ng (surface concentration = 582 ± 286 ng/cm2) when operated at a flow rate of 1.8 ± 0.7 µL/min. The estimated efficiency for TRNA recovery was 73 ± 15% (n = 4).
Preliminary RT-PCR results with TRNA purified from the PPC-SPRI chip for bacterial TRNA suggested the presence of trace amounts of gDNA. The PPC-SPRI was unable to completely remove gDNA from the sample. However, in the case of mammalian samples, the amount of gDNA interferences found in the PPC-SPRI purified samples was negligible (data not shown). Large bp differences between the mammalian RNA and its gDNA as compared to bacterial RNA and its gDNA allowed for better solid-phase selectivity between these species.29 Therefore, it was necessary to treat bacterial samples with DNase prior to the PPC-SPRI purification to improve the purity of the TRNA in terms of gDNA contamination, while the mammalian TRNA samples did not require DNase treatment.
The quality of TRNA following PPC-SPRI microchip purification was assessed by monitoring the electrophoretic band pattern for rRNAs. This method relies on the assumption that rRNA quality and quantity reflect that of the underlying mRNA population.31 The rRNA species used for this assessment included the 18S and 28S rRNAs for mammalian samples and the 16S and 23S rRNAs for bacteria. Ratios of these band intensities of ~2 are indicative of intact TRNA, while for degraded RNA samples, these bands appear less pronounced with increasing numbers of lower molecular weight components producing smearing in the electrophoretic bands.30
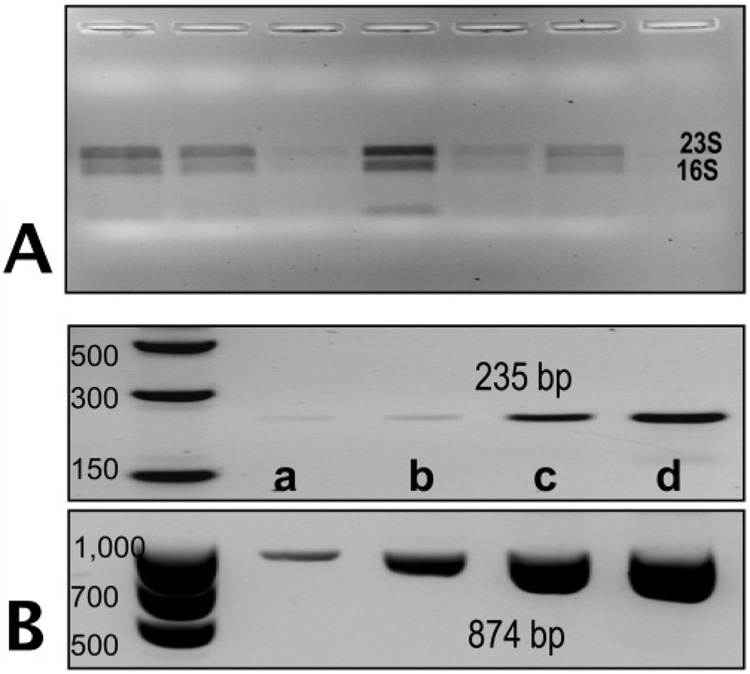
Control samples of rRNA not subjected to PPC-SPRI purification produced electrophoretic bands before PPC-SPRI purification that were sharp and intense with the 28S rRNA band (4.7 kb) for rat and the 23S rRNA band for E. coli (2.9 kb) being more intense than the 18S rRNA band (1.9 kb) for rat and the 16S rRNA (1.5 kb) for E. coli. The average ratio for the 28S/18S pair was 1.93 ± 0.23 (n = 6) and for the 23S/16S ratio it was 1.84 ± 0.16 (n = 5). Results of RNA integrity following PPC-SPRI purification are shown in Figure 4A for E. coli RNA. The entire volume collected from the PPC-SPRI chip (minus 4 µL for RT-PCR) was loaded into the gel. The ratio of the 23S/16S was found to be 1.79 ± 0.12 (n = 6) for the E. coli TRNA indicating that the TRNA extracted was predominately intact and not degraded due to shear following hydrodynamic transport through the microfluidic or RNase activity. For the mammalian TRNA, this ratio was found to be 1.87 ± 0.25 (n = 6), indicating high quality of the isolated RNA (data not shown).
Figure 4.
(A) Agarose gel electrophoresis fluorescence image (1%) of E. coli rRNA purified using the PPC-SPRI plate for 6 replicate measurements. The bands represent the 16s and 23s rRNA of E. coli. (B) Fluorescence images of RT-PCR products for mRNA generated from the 16s-RNA (874 bp) gene and the gyrB (235 bp) gene separated using a 4% agarose gel. The concentration of TRNA recovered from the PPC-SPRI microchip was quantified using the RiboGreen assay and various amounts of the purified TRNA was introduced for the RT-PCR, which were: (a) 0.1 ng, (b) 0.5 ng, (c) 2 ng, and (d) 4 ng. Samples were immobilized to the PPC-SPRI bed using immobilization buffer Buffer D. Separation conditions for the agarose gel electrophoresis and sample load volumes are given in Figure 2.
RT-PCR was performed on the PPC-SPRI purified TRNA isolated from E. coli to confirm the presence of mRNA, which is typically used for gene expression analysis. A two-step RT-PCR with a pdT23 RT-primer was used for these reactions. Two genes were probed by PCR following RT; the highly abundant mRNA transcribed from the small-subunit 16s_RNA gene and the low abundant mRNA transcribed from the gyrB gene (~120 copies per cell) that encodes the subunit B protein of DNA gyrase, which regulates the supercoiling of dsDNA.6,31 Negative controls performed (-RT) suggested the samples were free of DNA contamination after DNase treatment. Figure 4C shows fluorescence images of the gels for the RT-PCR products of 16s RNA (874 bp) and gyrB (254 bp), confirming the presence of the mRNA in the purified sample. The PCR product concentration varied with the amount of isolated TRNA loaded into the RT-PCR assays (see Figure 4B).
Houskeeping Gene Abundance in Rat Liver TRNA
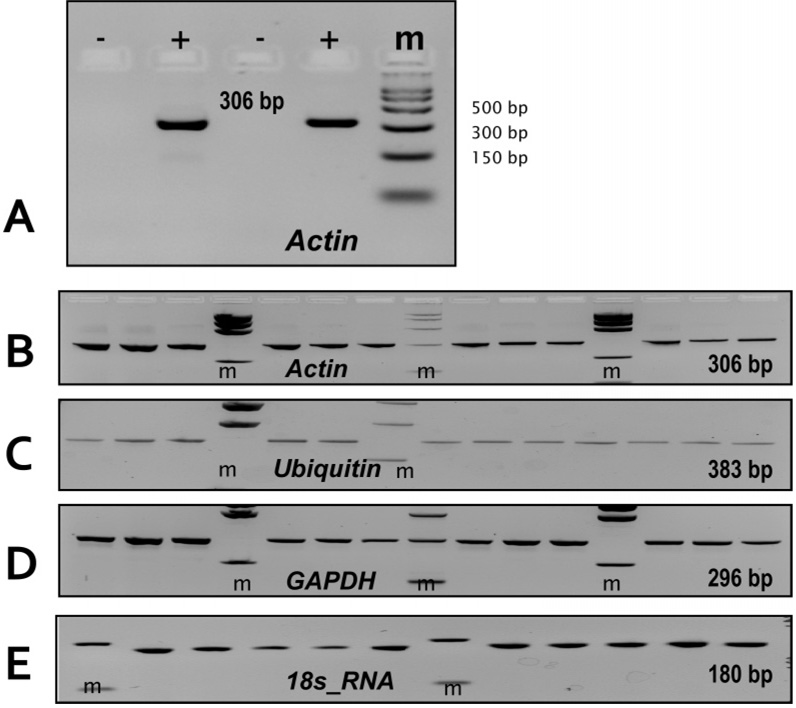
We next evaluated the ability to perform expression analysis of mRNAs using as a model system several housekeeping genes in rat liver including β-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ubiquitin and 18s_RNA genes following TRNA isolation via the PPC-SPRI microchip. As can be seen from Figure 5A, for PCRs not preceded by a reverse transcription step (−), no 306 bp products for actin mRNA was observed in the gel indicating that the purification protocol adopted removed most of the gDNA probably due to better selectivity for eukaryotic mRNAs due to their smaller size compared to the gDNA. In the cases where the PCRs were preceded by reverse transcription, product bands were clearly visible in the gel indicative of the presence of actin mRNA in the purified sample. Figures 5B–E shows fluorescence images of the RT-PCR products for actin, ubiquitin, GAPDH, and the 18s_RNA genes following PPC-SPRI microchip purification. Based on the size of these products, they could be definitively ascribed to the 18s_RNA, β-Actin, ubiquitin and GAPDH gene mRNAs suggesting the presence of intact mRNA following purification using the PPC-SPRI microchip. Based on these measurements, it was found that the highest abundant mRNA was from the 18s_RNA gene, followed by actin, GAPDH, and ubiquitin, consistent with the expression levels reported for these genes in the literature.32
Figure 5.
Fluorescence images of ethidium-stained 3% agarose gels showing the presence of RT-PCR products generated from PPC-SPRI purified rat liver RNA. The RNA was immobilized to the SPE bed using Buffer C. The probed genes were (RT-PCR product sizes are given in parentheses): 18s_RNA (180 bp), Ubiquitin (383 bp), Actin (306 bp), GAPDH (296 bp). (A) PPC-SPRI purified rat liver RNA subjected to PCR with (+RT) and without (−RT) a reverse transcription step. (B–E): RT-PCR of 12 PPC-SPRI purified samples evaluating the expression of; (B) Actin, (C) Ubiquitin, (D) GAPDH, and (E) 18s_RNA genes. Separation conditions and the sample volumes loaded into the gel were the same as those described in Figure 2.
CONCLUSIONS
A 96-well titer plate-based polymer microfluidic chip was designed, micro-replicated from a UV-LiGA generated master, and evaluated for the SPE purification of a variety of NAs, including DNA of various sizes and TRNA. The receiving/output wells were arranged with a 9 mm spacing similar to conventional titer plates, which permitted loading/unloading of the plate with standard laboratory fluid handling equipment. The type of NAs purified using the chip was controlled by simply changing the composition of the immobilization buffer used for the SPE. The chip provided a load of 206 ± 93 ng of gDNA from E. coli per bed and 165 ± 81 ng of TRNA. The purification chip was demonstrated to provide high quality samples that could be used for a variety of molecular assays, such as the identification of different pathogens isolated from whole blood containing SPS serving as an anticoagulant or expression profiling of mRNAs.
Using the 96-well plate, gDNA could be purified and recovered with an efficiency of approximately 63%. TRNA could also be purified using the same PPC-SPRI chip with a recovery of 73%. RT-PCRs of the purified mRNAs were successful with the amounts of mRNA isolated shown to agree with the relative activity of the genes from which they were generated allowing the PPC-SPRI wafer to be appropriate for expression analysis. Unlike other methods that utilize polydT-modified supports for mRNA capture through interactions with the polyA tail, the PPC-SPRI extracts TRNA, making it attractive for analyzing a variety of different forms of RNA. Additionally, the rRNA isolated using the PPC-SPRI chip can serve as an internal standard for the normalization of gene expression levels.
The use of the PPC-SPRI device is particularly advantageous for NA purification requiring disposable or one-time use formats for minimizing potential sample carryover contamination due to chip fabrication method, which used micro-replication from a master (hot embossing or injection molding), which can provide high-volume and low-cost production of devices.33,34 Also, by utilizing injection molding as the replication protocol, better control of chip manufacturing tolerances can be realized, which could improve the uniformity of fluid distribution through the immobilization beds, producing less variable NA recovery. In addition, the surface activation protocol via UV-exposure is extremely simple in its execution and does not require extensive post-fabrication processing to create the NA selective isolation surface, such as the formation of chemically modified polymer or monolithic supports or the addition of functionalized beads to the extraction bed. The use of a photomask can provide regio-selective modification of the PC surface to ensure that the NA isolation occurs only within the lithographically defined extraction bed.
ACKNOWLEDGMENT
This work was generously supported by the National Institutes of Health (R01-HG001499), the National Science Foundation (EPS-0346411) and the State of Louisiana Board of Regents Support Fund. The authors would also like to thank Dr. Proyag Datta and the Center for Advanced Microstructures and Devices (CAMD) at Louisiana State University for sharing expertise in microfabrication and embossing of the plates. The first two authors contributed equally to this work.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bloomfield VA. Curr. Opin. Struct. Biol. 1996;6:334–341. doi: 10.1016/s0959-440x(96)80052-2. [DOI] [PubMed] [Google Scholar]
- 2.Budker VG, Slattum PM, Monahan SD, Wolff JA. Biophys. J. 2002;82:1570–1579. doi: 10.1016/S0006-3495(02)75508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lerman LS. Proc. Natl. Acad. Sci. U.S.A. 1971;68:1886–1890. doi: 10.1073/pnas.68.8.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kombrabail M, Krishnamoorthy G. J. Fluoresc. 2005;15:741–747. doi: 10.1007/s10895-005-2982-8. [DOI] [PubMed] [Google Scholar]
- 5.Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP, Landers JP. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toriello NM, Liu CN, Mathies RA. Anal. Chem. 2006;78:7997–8003. doi: 10.1021/ac061058k. [DOI] [PubMed] [Google Scholar]
- 7.Christel LA, Petersen K, McMillan W, Northrup MA. J. Biomech. Eng. 1999;121:22–27. doi: 10.1115/1.2798037. [DOI] [PubMed] [Google Scholar]
- 8.Nakagawa T, Hashimoto R, Maruyama K, Tanaka T, Takeyama H, Matsunaga T. Biotechnol. Bioeng. 2006;94:862–868. doi: 10.1002/bit.20904. [DOI] [PubMed] [Google Scholar]
- 9.Cao W, Easley CJ, Ferrance JP, Landers JP. Anal. Chem. 2006;78:7222–7228. doi: 10.1021/ac060391l. [DOI] [PubMed] [Google Scholar]
- 10.Tian H, Huhmer AFR, Landers JP. Anal. Biochem. 2000;283:175–191. doi: 10.1006/abio.2000.4577. [DOI] [PubMed] [Google Scholar]
- 11.Wen J, Guillo C, Ferrance JP, Landers JP. Anal. Chem. 2007;79:6135–6142. doi: 10.1021/ac0703698. [DOI] [PubMed] [Google Scholar]
- 12.Witek MA, Llopis SD, Wheatley A, McCarley RL, Soper SA. Nucleic Acids Res. 2006;34:e74/71–e74/79. doi: 10.1093/nar/gkl146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Vaidya B, Patel AB, Ford SM, McCarley RL, Soper SA. Anal. Chem. 2003;75:2975–2984. doi: 10.1021/ac030031n. [DOI] [PubMed] [Google Scholar]
- 14.Alberts B, Alexander J, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4th ed. New York: Garland Science; 2004. [Google Scholar]
- 15.Lien K-Y, Lee W-C, Lei H-Y, Lee G-B. Biosen. Bioelectron. 2007;22:1739–1748. doi: 10.1016/j.bios.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Jiang G, Harrison DJ. Analyst. 2000;125:2176–2178. doi: 10.1039/b005999o. [DOI] [PubMed] [Google Scholar]
- 17.Satterfield BC, Stern S, Caplan MR, Hukari KW, West JAA. Anal. Chem. 2007;79:6230–6235. doi: 10.1021/ac0709201. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar N. Microbiol. 1996;142:3125–3133. doi: 10.1099/13500872-142-11-3125. [DOI] [PubMed] [Google Scholar]
- 19.Anderson MJ, Hansen CL, Quake SR. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16746–16751. doi: 10.1073/pnas.0605293103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang-Yen DA, Myszka DG, Gale BK. JMEMS. 2006;15:1145–1151. [Google Scholar]
- 21.Paegel BM, Emrich CA, Wedemayer GJ, Scherer JR, Mathies RA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:574–579. doi: 10.1073/pnas.012608699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada M, Hirano T, Yasuda M, Seki M. Lab. Chip. 2006;6:179–184. doi: 10.1039/b514054d. [DOI] [PubMed] [Google Scholar]
- 23.Yeung SHI, Greenspoon SA, McGuckian A, Crouse CA, Emrich CA, Ban J, Mathies RA. J. Forens. Sci. 2006;51:740–747. doi: 10.1111/j.1556-4029.2006.00153.x. [DOI] [PubMed] [Google Scholar]
- 24.Park DS-W, Hupert ML, Witek MA, You BH, Datta P, Guy J, Lee JB, Soper SA, Nikitopoulos DE, Murphy MC. Biomed. Microdevices. 2008;10(1):21–33. doi: 10.1007/s10544-007-9106-y. [DOI] [PubMed] [Google Scholar]
- 25.Wilson IG. Appl. Environ. Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lis JT. Methods Enzymol. 1980;65:347–353. doi: 10.1016/s0076-6879(80)65044-7. [DOI] [PubMed] [Google Scholar]
- 27.Fredricks DN, Relman DA. J. Clin. Microbiol. 1998;36:2810–2816. doi: 10.1128/jcm.36.10.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edberg SC, Edberg MK. J. Clin. Microbiol. 1983;18:1047–1050. doi: 10.1128/jcm.18.5.1047-1050.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lis JT, Schleif R. Nucleic Acids Res. 1975;2:383–389. doi: 10.1093/nar/2.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voet D, Voet JG. Biochemistry. 2nd ed. New York: John Wiley & Sons; 1995. [Google Scholar]
- 31.Yamamoto S, Harayama S. Appl. Environ. Microbiol. 1995;61:3768. doi: 10.1128/aem.61.10.3768-3768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu L-j, Altmann SW. Anal. Biochem. 2005;345:102–109. doi: 10.1016/j.ab.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 33.Becker H, Gartner C. Electrophoresis. 2000;21:12–26. doi: 10.1002/(SICI)1522-2683(20000101)21:1<12::AID-ELPS12>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 34.Becker H, Locascio LE. Talanta. 2002;56:267–287. doi: 10.1016/s0039-9140(01)00594-x. [DOI] [PubMed] [Google Scholar]