Summary
BLISS 2.0 is a web-based application for identifying conserved regulatory modules in distantly related orthologous sequences. Unlike existing approaches, it performs the cross-genome comparison at the binding site level. Experimental results on simulated and real world data indicate that BLISS 2.0 can identify conserved regulatory modules from sequences with little overall similarity at the DNA sequence level.
1 Introduction
Identifying functional Transcription Factor Binding Sites (TFBSs) in the regulatory region of a DNA sequence is essential for understanding gene regulation at the transcription level. In eukaryotes, distinct TFBSs are often grouped together into regulatory modules to control a specific aspect of gene expression. The composition of a particular regulatory module can be identified by experimental approaches; for example, through enhancer region dissection, DNase hypersensitivity assay, DNA foot printing, etc. However, most of these approaches are laborious and costly. More importantly, many of these approaches, such as the DNase hypersensitivity assay, can only be applied to short DNA sequences (a few hundred base pairs long). They are intractable if the relative location of the module cannot be narrowed to an experimentally testable region.
With the emergence of bioinformatics, computational approaches are being developed to predict regulatory modules that are in turn helping the design and verification of laboratory experiments. A number of such computational approaches (Aerts et al., 2005; Sandelin et al., 2004; Loots and Ovcharenko, 2004; Sharan et al., 2003; Sinha et al., 2004) employ cross-genome comparison. The assumption underlying these approaches is that DNA sequences encoding functional TFBSs are conserved during evolution due to selective pressure, whereas non-functional DNA sequences evolve (mutate) much faster. The search for conserved regulatory modules could therefore be narrowed to regions with high DNA similarity between (among) orthologs. While these approaches have proven very helpful in some cases, they have certain limitations—their success depends on identifying significant sequence level similarity between (among) the DNA regions that harbor the regulatory modules. However, TFBS sequences are degenerate in nature and they are interspersed by non-functional sequences, which are not under conservation pressure. It is therefore possible that although the regulatory modules are conserved, i.e., all of the functional TFBSs are conserved, the overall similarity of the sequences harboring the regulatory modules is only marginal and cannot be distinguished from the background. Applications based on cross-genome comparison at the DNA sequence level will fail when the orthologous DNA sequences are highly diverged. Since the conservation pressure is at the binding site level, a novel methodology named BLISS has been developed to perform the cross-genome comparison on binding site profiles (Meng et al. 2006). As a complementary tool, BLISS demonstrates the ability to identify conserved regulatory modules from diverged orthologous sequences whose overall DNA similarity is undetectable.
2 Bliss 2.0 vs. Bliss
In the first release of BLISS, we demonstrated the feasibility of identifying conserved regulatory modules through binding site level comparisons (Meng et al. 2006). However, the original BLISS algorithm was developed to identify conserved modules in a short sequence and a long sequence, with the assumption that the short sequence harbored exactly one regulatory module (Meng et al. 2006). This is applicable when a biologist has already narrowed the regulatory function to a small interval in a model organism and would like to identify the corresponding module in another organism where information regarding its location is absent. In many cases, however, biologists have no prior knowledge about the locations of the conserved regulatory modules in either of the sequences to be analyzed. To address this limitation of BLISS, we developed the BLISS 2.0 algorithm to identify conserved regulatory modules between two long sequences without pre-knowledge about the existence, location, or number of conserved module(s).
It has been observed that the distances among the cluster of TFBSs in a regulatory module are highly conserved during evolution, probably reflecting the space constraint essential for the protein-protein interactions among TFs required for combinatorial transcriptional regulation (Ludwig et al., 2005). Based on this observation, BLISS 2.0 introduces a new concept “constrained cluster”. A constrained cluster is a cluster of TFBSs, where the type (identity), number, and order of TFBSs are highly conserved during evolution, and the variation of inter-TFBS distances is within a certain range (i.e. less than 10 bps). A regulatory module may have one or multiple constrained TFBS clusters. In the later case, the distance between the clusters can vary greatly. Unlike BLISS where the TFBS profile of the shorter sequence was scanned across that of the longer sequence, BLISS 2.0 detects all conserved constrained clusters between two long sequences in two-dimensional space. The regulatory modules could then be predicted based on the identification of the constrained clusters.
3 Algorithm
Detailed explanation of the BLISS 2.0 algorithm is available at the BLISS 2.0 web site (Meng et al. 2007). In short, BLISS 2.0 takes four steps to predict constrained clusters. First, binding site profiles, which indicate potential occurrences of binding sites at each location of both sequences, are calculated based on frequency matrices collected by TRANSFAC 9.1 (Matys et al., 2003). Second, p values of binding site scores are integrated to differentiate binding sites with various occurrence frequencies. In the third step, a Gaussian smoothing is applied to each profile to account for the variation in the distances between binding sites. Finally, BLISS2_scores, which indicate the degree of conservation at the binding site level, are calculated for all pairs of positions on the two sequences. Statistically significant BLISS2_scores averaged over an optimized window size indicate conserved constrained clusters with similar binding site profiles.
4 Web Interface
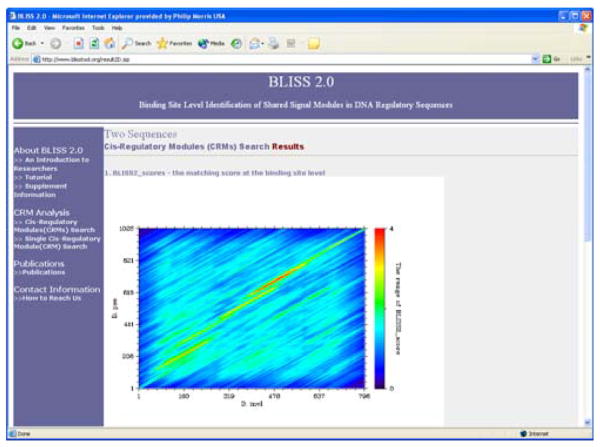
Given two orthologous DNA sequences, BLISS 2.0 is able to output all constrained clusters shared by those two sequences. The analysis in BLISS 2.0 proceeds in three major steps. To begin, users are required to input two DNA sequences. BLISS 2.0 then generates binding site profiles for each sequence based on matrices collected in the TRANSFAC database (version 9.1) (Matys et al., 2003). To strike a balance between sensitivity and selectivity at this stage, the user is prompted to choose a binding site score cutoff of either 0.75 or 0.8. Second, BLISS 2.0 measures the degree of conservation at the binding site level between the two sequences as BLISS2_scores. They are displayed and visualized as a two-dimensional color plot (Figure 1). High and sustained BLISS2_scores along diagonal segments indicate potential matches of conserved TFBS clusters between the two sequences. In order to be able to evaluate a particular BLISS2_score, we have also analyzed the distribution of BLISS2_scores using simulated sequence pairs. Based on this statistical analysis, users can choose a BLISS2_score cutoff to output all shared TFBS clusters. Finally, all shared TFBS clusters with BLISS2_score greater than the cutoff are displayed. Matched TFBSs are listed in a separate table for each shared TFBS cluster pair. Users also have the option to determine how to rank the shared TFBSs in the outputted regulatory modules. They can be ranked based on locations on the input sequences, by numeric contributions to the BLISS2_score, or by product of p-values of the matching TFBSs on both sequences. To facilitate the inspection of the results, BLISS 2.0 provides users the option to highlight in green TFBSs that they are interested in.
Fig. 1.
Web interface of BLISS 2.0. Output of BLISS2_scores, which indicates the degree of conservation at the binding site level between the two sequences. The two axes correspond to the two inputted sequences. The color reflects the BLISS2_score value at corresponding positions.
We have successfully applied BLISS 2.0 to identify the Even-skipped (eve) stripe 2 enhancer (S2E) (Ludwig et al. 2005) in D. mojavenis and D. virilis (which cannot be detected by existing tools like BLAST, rVista 2.0 and ConSite.).
Acknowledgments
This work was supported in part by a UF Genetic Institute Seed Grant (to AB and LZ) and by NIH CA095542 and AI067555 (to LZ).
Footnotes
Publisher's Disclaimer: This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/2.0/uk/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- Aerts S, Van Loo P, Thijs G, Mayer H, de Martin R, Moreau Y, De Moor B. TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. Nucleic Acids Res. 2005;33:W393–396. doi: 10.1093/nar/gki354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Banerjee A, Zhou L. BLISS: biding site level identification of shared signal-modules in DNA regulatory sequences. BMC Bioinformatics. 2006;7:287. doi: 10.1186/1471-2105-7-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Banerjee A, Zhou L. 2007 http://www.blisstool.org/doc/supplement.html.
- Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–52. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R, Ovcharenko I, Ben-Hur A, Karp RM. CREME: a framework for identifying cis-regulatory modules in human-mouse conserved segments. Bioinformatics. 2003;19 1:i283–291. doi: 10.1093/bioinformatics/btg1039. [DOI] [PubMed] [Google Scholar]
- Sinha S, Blanchette M, Tompa M. PhyME: a probabilistic algorithm for finding motifs in sets of orthologous sequences. BMC Bioinformatics. 2004;5:170. doi: 10.1186/1471-2105-5-170. [DOI] [PMC free article] [PubMed] [Google Scholar]