Dear Editor,
Reaper, Hid, Grim, and Sickle are a group of fruit fly proteins that play pivotal roles in regulating apoptosis during development and in response to cytotoxic stimuli. A rich array of structure/function analysis indicated that two short motifs shared among these proteins contribute to their proapoptotic activity. So far, only in Drosophila melanogaster have all four proteins been identified. Taking advantage of genome sequence projects, we have identified orthologs of these proteins in several divergent Drosophila species. Comparison of the two motifs within and among the ortholog groups revealed a striking distinctiveness of the Iap-binding motif, which is characteristic of each group and is strictly conserved. This finding suggests that these proteins function as ‘unique’ rather than ‘general’ keys to unlock the inhibition of Iap on caspase activation. The GH3 motif, on the other hand, shows differential conservation. Together, these findings support the hypothesis that these proteins are not simply redundant, but each has a distinct role in regulating cell death.
In the D. melanogaster genome, a region of about 300 kb on the third chromosome is essential for the proper execution of cell death. Mutant animals lacking this region are deficient for most developmental cell death and have dramatically impaired cell death response to cytotoxic stimuli.1 Four of the genes harbored in this region, reaper (rpr), head involution defective (hid), grim, and sickle (skl), have proapoptotic function. Proteins encoded by these four genes, often referred to as the RHG proteins, have varying length and share little overall sequence similarity, except for two loosely linked short motifs. The lack of significant similarity also makes it difficult to judge whether they are paralogs resulting from duplication events, or alternatively, evolved from different protein lineages.
The first motif is at the extreme N-terminal of the RHG proteins. It was termed RHG motif2 or IBM (Iap-binding motif). Structural and functional analysis indicated that IBM binds to Iap and thus releases its inhibition on the caspases.3 The activation of caspases, a family of proteases, leads to cellular destruction through a morphological and biochemical ritual termed apoptosis. It has been demonstrated that short peptides harboring IBM are capable to induce cell death.
The second motif shared by Reaper, Grim, and Sickle is termed trp-block,2 or GH3 (Grim Helix 3).4 It can induce cell death by itself in the absence of the IBM motif, with comparatively less efficiency. However, when both motifs are present, a cooperative effect in promoting cell death is observed.4 Unlike the IBM motif, the functional mechanism of GH3 is less clear with some seemingly conflicting observations.
To understand the evolution of these cell death genes, we sought to compare distant orthologs of these four proteins with a focus on the two functional motifs. Four Drosophila species were chosen for this comparison. D. yakuba (D. yaku) and D. pseudoobscura (D. pseu) are from the same subgenus (Sophophora) as D. melanogaster, and separated from D. melanogaster about 10 and 40 million years ago, respectively.5 D. mojavensis (D. moja) and D. virilis (D. viri) belong to a different subgenus, Drosophila, which separated from Sophophora about 60–65 million years ago.
Corresponding orthologs of the four Iap-antagonists were identified in all of the four selected genomes (Figure 1, and Supplementary material). Complete ORFs for these orthologs were identified with the only exception of hid in D. mojavensis. The genomic sequence of D. mojavensis has yet to be completed. However, strong homology between Hid and some segments of D. mojavensis genomic sequence left little doubt that hid also exists in this genome. In both D. yakuba and D. pseudoobscura, the four genes clustered nearby on the left arm of the third chromosome, which highly resembles their arrangement in D. mela. Chromosomal assignment of genomic contigs in D. mojavensis and D. virilis has not been finished. However, it appeared that the four genes were also clustered in these two genomes since their ORFs can be identified in the same contig or contigs overlapping with each other.
Figure 1.
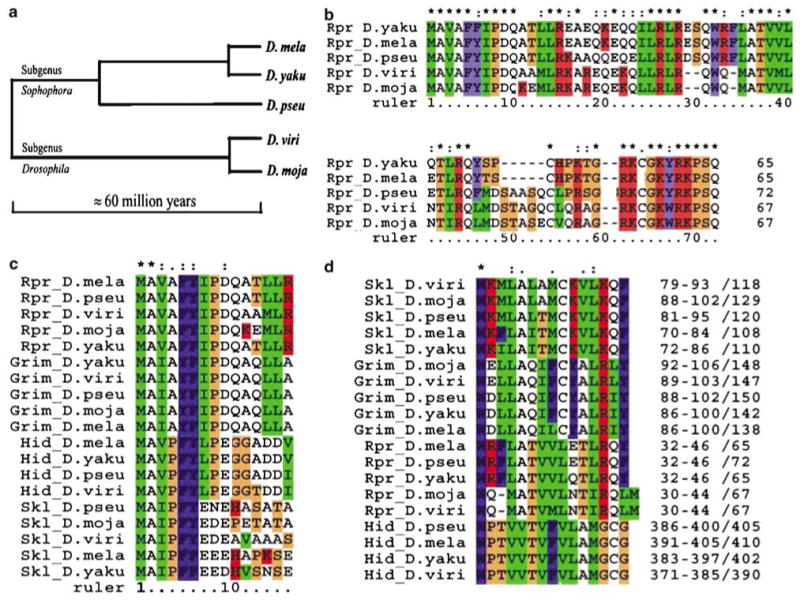
RHG proteins in distantly related species. (a) The evolutionary distance of the selected species. (b) Alignment of Rpr sequences from the five species. (*) and (:) denotes identical and conserved positions, respectively. Color scheme reflects the chemical property of amino acids. Alignments for Grim, Skl, and Hid can be found in Supplementary materials or the project website. (c) Alignment of the IBM motifs. Note that the Iap-binding key of aa 2–8 is almost identical for each ortholog group. (d) Alignment of the GH3 motif. Statistically significant motifs were identified by MEME, realigned using ClustalX. The numbers at the end indicate the relative position of the GH3 motif in each protein. Format: beginning position–end positions/length of the protein
The fact that orthologs of rpr, grim, skl, and hid exist in the distantly related D. virilis and D. mojavensis indicated that the four genes existed before the separation of the subgenera Drosophila and Sophophora about 60–65 million years ago. This finding ruled out the possibility that the four Iap-antagonists identified in D. mela arose from a recent duplication event. Rather, it indicated that the four genes are conserved through a long evolutionary period in divergent species.
Rpr, Grim, Skl, and Hid form a subgroup of Iap-antagonists that have the Iap-binding motif at the very N-terminal end. They function as active Iap-antagonists right after protein synthesis. In another group of Iap-antagonists, the Iap-interacting motif is embedded inside the protein. These proteins cannot function as Iap-antagonists until the motif is exposed through removal of preceding signal peptide or cleavage. While proteins belonging to the later group have been identified in insects as well as mammals, the first group of Iap-antagonists has so far only been identified in the Drosophilae genus. No orthologs of the RHG proteins were identified by the mosquito (Anopheles gambiae) genome project due to ‘the rapid sequence diversification’ of these proteins.6 Our finding here, however, suggested that the function of the RHG proteins is evolutionarily conserved and there is a strong likelihood of these proteins present in mosquito or even mammalian genomes. The question of whether RHG proteins are paralogs also awaits the identification of orthologs in more distantly related species.
One striking discovery of our comparison is the distinctiveness of the N-terminal Iap-binding motif and its absolute conservation within each ortholog group (Figure 1c). In eukaryotic cells, the leading Met is removed by methionine aminopeptidase if the second position is a small amino acid. In the case of the RHG proteins, the Ala2 is exposed at the extreme N-terminus shortly after protein synthesis. This free alanine is required for interaction between IBM and IAP.3 Biochemical evidence indicated that the first seven amino acids (aa 2–8) of RHG proteins are directly involved in interacting with the BIR (Baculaviral Iap Repeats) domain of Iap proteins.7 Structural analysis revealed that this heptapeptide acts as a ‘key’ to fit into a groove on the surface of the BIR.3
A potential implication of structural data is that the IBM motifs of RHG proteins function as a general key to match the surface groove of the BIR domain. When the structure of the binding complexes of the BIR2 domain of Diap1 with either Grim2–9 (AIAYFIPD) or Hid2–9 (AVPFYLPE) was resolved through X-ray crystallography, it appeared that the binding position, and interacting points of Grim2–9 overlaps with that of Hid2–9. Using a tetrapeptide library, it has been shown that while the first Ala is absolutely required for binding, the other positions enjoy much more leniency.8 Many different substitutions in these positions can have similar or even higher IAP binding efficiency. For example, when the second position was substituted with basic amino acid (R or H), an increase in binding affinity was observed.8 These findings suggested that amino acids in the IBM motif are substitutable without losing the functionality.
By identifying the orthologs described in this paper, we have dramatically increased the number of known sequences for Iap-antagonists as well as the evolutionary span they represent. Our finding indicates that over a long evolutionary period, the N-terminal IBM motif is highly conserved (Figure 1c). What is more interesting is that the distinctiveness of the IBMs in RHG proteins is strictly conserved within each ortholog group. In fact, in light of this finding, it seems appropriate to subdivide the IBM into at least four subtypes corresponding to the four orthologous groups. The distinctiveness of the four subtypes can be represented with the first four amino acids of IBM, which are invariable within each group, that is, Rpr subgroup (AVAF), Grim subgroup (AIAY), Hid subgroup (AVPF), and Skl subgroup (AIPF).
The strict conservation of the distinctiveness of IBM for each ortholog group over 60 million years of evolution indicates a strong functional constraint for each IBM subtype. This constraint on coexistence of multiple subtypes is unlikely due to alternative expression of these genes in different tissues. As all four genes are expressed in cells destined to die, furthermore, it has been documented that multiple RHG genes are required for the proper cell death pattern of the same cell lineage.9 On the other hand, it is not a surprising revelation that multiple subtypes of Iap-antagonists coexist. There are four Iap genes existing in Drosophila, each having one to three BIR domains. The existence, and the conservation of multiple subtypes of IBM, may well reflect the structural complexity and subtleness required for fine-tuning the multiplayer system controlling the die-or-live decision of the cell. Indeed, genetic and biochemical analysis showed that the mode of action of Rpr, Grim, Hid, and Skl is different.2,7 Their IBMs are not interchangeable, as swapping Grim IBM to Rpr does not transform the chimeric protein into Grim, and vice versa.2 RHG proteins display differential affinity to different BIR domains, and thus affect the activation of different combinations of caspases.7 Our finding indicates that the distinct modes of action possessed by individual proteins are essential for the proper regulation of cell death in the organism.
It is interesting to note that, comparing to Drosophila, the Iap protein family is expanded in the mosquito genome of A. gambiae, presumably due to the functional requirement of fine-tuning cell death regulation in response to parasites and viruses.6 In mammals, the number of BIR-domain-containing genes is far expanded. Thus it is reasonable, in light of our finding here, to expect that more Iap-antagonists exist than the few we currently know of in mammalian systems.
With the exception of Hid, N-terminal deleted Rpr, Grim, and Skl still have cell death -inducing capabilities, indicating the existence of another proapoptotic domain. Detailed structure/function analysis mapped the killing function of Grim-C to the GH3 motif, which is also shared by Rpr and Skl.4 The GH3 motif can induce cell death through general inhibition of protein synthesis,10,11 or via causing the release of cytochrome c from mitochondria.12
Unlike IBM, the conservation of GH3 is different among the ortholog groups. Sequences encompassing the GH3 motif in Skl and Grim are selectively conserved as indicated by the Reactive-site sequence conservation (RSC) ratio, which measures the selective conservation of functional motifs as compared to the general similarity of the whole protein (Table 1).13 In contrast, GH3 in Rpr does not appear to be under more selective pressure than the average of the whole protein. The RSC ratio between Rpr orthologs in D. mela and D. virilis was about one, indicating that there is a lack of specific selection (Table 1). Close examination revealed that the GH3 region in Rpr orthologs from D. virilis and D. mojavensis, the two species from the Drosophila subgenus, appear to have suffered a deletion event (Figure 1d). This finding suggests that either the selective pressure on the GH3 domain varies in individual ortholog groups, or the functionality of GH3 is not susceptible to single amino-acid deletion/substitution. The first alternative is more plausible as the GH3 domains in Grim and Skl ortholog groups are highly conserved (Table 1, Figure 1d).
Table 1.
RSC ratio
| D. mela/D. pseu | D. mela/D. viri | |||||
|---|---|---|---|---|---|---|
| RSC ratio | RSC ratio | |||||
| Avg. whole | IBM | GH3 | Avg. whole | IBM | GH3 | |
| Grim | 3.247 | 1.909 | 1.745 | 2.589 | 2.395 | 1.931 |
| Hid | 2.909 | 2.166 | 2.131 | 2.706 | 2.328 | 2.291 |
| Rpr | 3.681 | 1.684 | 1.485 | 2.681 | 2.313 | 1.044 |
| Skl | 2.165 | 2.494 | 2.494 | 1.812 | 2.539 | 2.796 |
The ratios are calculated, for each pair of orthologs, as (average similarity of the motif/average similarity of the whole protein (avg. whole)). A ratio greater than one indicates selective conservation due to functional constraints
Surprisingly, the MEME program also identified a conserved region in the Hid orthologs that can be compared to the GH3 domain identified in Rpr, Grim, and Skl (Figure 1d). The first two-thirds of the putative GH3 in Hid matches well with the counterpart of GH3 in Grim/Skl/Rpr. The last third appears to have diverged much further. The strict conservation of this region in Hid indicates functional constraints. However, since N-terminal-deleted Hid has no cell death inducing capability, this region in Hid is likely engaged in function(s) that does not directly cause apoptosis, but is nonetheless important for organism survival. Elucidation of the functional mechanism of this region in Hid may well shed light on the emergence and evolution of the cell death-inducing GH3 domains in the other ortholog groups.
In summary, comparing the sequences from distantly related species has revealed many interesting aspects of the structural conservation of the RHG proteins. The distinctiveness of their N-terminal IBM and the differential conservation of the GH3 domain all suggest that these proteins have unique and essential roles in the regulation of cell death. Identification of orthologs in different genera should be pursued to fully understand the emergence and evolution of these proteins and the machinery that controls the life and death decision of the cell.
Supplementary Material
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary Information:
I.) Distance Tree of RGH Proteins From The Five Species. Sequences were aligned using ClustalX and the Neighbor-Joining tree was bootstrapped for 2000 trials. The resulting distance tree were displayed and outputted with TreeView (Page 1996). Scale is for the rate of nucleotide substitution per site.
II.) Alignment of Orthologs of Grim.
III.) Alignment of Orthologs of Hid.
IV.) Alignment of Orthologs of Sickle (Skl).
Acknowledgments
I am very grateful to Dr. John R Nambu and Dr. Barbara A Osborne for their insightful suggestions and comments to this paper. I also thank Jim Mailliard, Gina Chan, and Yanping Zhang for proof reading the manuscript, and Sriram Parthasarathy for technical assistance in setting up the website. Additional data about the comparison of cell death machinery in different Drosophila species can be found at the project website http://159.178.64.61/ApoCom/start.html. This work is supported by NIH Grant CA95542.
References
- 1.White K, et al. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 2.Wing JP, et al. Mech Dev. 2001;102:193–203. doi: 10.1016/s0925-4773(01)00316-1. [DOI] [PubMed] [Google Scholar]
- 3.Wu JW, et al. Mol Cell. 2001;8:95–104. doi: 10.1016/s1097-2765(01)00282-9. [DOI] [PubMed] [Google Scholar]
- 4.Claveria C, et al. EMBO J. 2002;21:3327–3336. doi: 10.1093/emboj/cdf354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powell J. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford: Oxford University Press; 1997. [Google Scholar]
- 6.Christophides GK, et al. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 7.Zachariou A, et al. EMBO J. 2003;22:6642–6652. doi: 10.1093/emboj/cdg617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kipp RA, et al. Biochemistry. 2002;41:7344–7349. doi: 10.1021/bi0121454. [DOI] [PubMed] [Google Scholar]
- 9.Zhou L, et al. Proc Natl Acad Sci USA. 1997;94:5131–5136. doi: 10.1073/pnas.94.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoo SJ, et al. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- 11.Holley CL, et al. Nat Cell Biol. 2002;4:439–444. doi: 10.1038/ncb798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claveria C, et al. J Biol Chem. 2004;279:1368–1375. doi: 10.1074/jbc.M309819200. [DOI] [PubMed] [Google Scholar]
- 13.Das R, Gerstein M. Proteins. 2004;55:455–463. doi: 10.1002/prot.10639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary Information:
I.) Distance Tree of RGH Proteins From The Five Species. Sequences were aligned using ClustalX and the Neighbor-Joining tree was bootstrapped for 2000 trials. The resulting distance tree were displayed and outputted with TreeView (Page 1996). Scale is for the rate of nucleotide substitution per site.
II.) Alignment of Orthologs of Grim.
III.) Alignment of Orthologs of Hid.
IV.) Alignment of Orthologs of Sickle (Skl).


