Abstract
Genomic imprinting is a reversible condition that causes parental-specific silencing of maternally or paternally inherited genes. Analysis of DNA and RNA from 52 human hepatocarcinoma samples revealed abnormal imprinting of genes located at chromosome 11p15 in 51% of 37 informative samples. The most frequently detected abnormality was gain of imprinting, which led to loss of expression of genes present on the maternal chromosome. As compared with matched normal liver tissue, hepatocellular carcinomas showed extinction or significant reduction of expression of one of the alleles of the CDKN1C, SLC22A1L, and IGF2 genes. Loss of maternal-specific methylation at the KvDMR1 locus in hepatocarcinoma correlated with abnormal expression of CDKN1C and IGF2, suggesting a function for KvDMR1 as a long-range imprinting center active in adult tissues. These results point to the role of epigenetic mechanisms leading to loss of expression of imprinted genes at chromosome region 11p15 in human tumors.
Genomic imprinting is a reversible “mark,” established during gametogenesis, that causes differential expression of maternally and paternally inherited alleles. The correct contribution of imprinted genes is required for normal embryonic and fetal development, because neither androgenetic or parthenogenetic embryos are viable. Imprinted genes are often clustered, suggesting that imprinting may be under the control of long-range regulatory elements (1). The two larger known clusters of autosomal imprinted genes in the human genome are located at chromosomes 15q11–12 and 11p15; alterations of these clusters are associated with the Prader–Willi/Angelman syndromes and Beckwith–Wiedemann syndrome (BWS), respectively (2, 3). BWS is a neonatal overgrowth syndrome that predisposes to cancer (4), and the importance of a maternally active locus at chromosome 11p15 in tumorigenesis is supported by the finding that loss of constitutive heterozygosity (LOH) at chromosome 11p15 in sporadic Wilms' tumors specifically involves maternal alleles (5, 6). At least six maternally expressed genes, CDKN1C, SLC22A1L (also known as BWR1A, IMPT1, ORCTL2, and TSSC5), BWR1C (IPL and TSSC3), KvLQT1, H19, and ASCL2, and two paternally expressed genes, LIT1 (KvLQT1-AS) and IGF2, are clustered at chromosome 11p15 (7–10). Because of their function, some of these genes, CDKN1C, BWR1C, and IGF2 (7, 11–14), may play a role in BWS and tumorigenesis. However, detection of mutations in one or more genes at 11p15 that might identify the critical tumor suppressor gene in this region has remained elusive. In some studies, mutations in SLC22A1L were detected in only very few solid tumors and not in BWS (7, 15), whereas other studies reported mutations in CDKN1C in less than 5% of BWS and not in tumors (16, 17). These findings raise the possibility that epigenetic mechanisms affect 11p15 genes in tumorigenesis and BWS more frequently than mutations, especially because aberrant regulation of genomic imprinting may influence the expression of several of these genes. Direct evidence of a role for abnormal imprinting in human tumorigenesis is the detection of loss of imprinting (LOI) leading to biallelic expression of IGF2 in Wilms' tumor (18, 19). The same abnormality was later detected in BWS (20) and other neoplasms (21, 22). However, LOI represents a gain of expression of an otherwise imprinted IGF2 allele, a finding that appears to contrast with the notion that loss of maternal alleles at chromosome region 11p15 is a crucial event in tumorigenesis and BWS.
Here, we report that an epigenetic mechanism leading to loss of expression of maternally expressed genes is frequent in human hepatocarcinomas (HCCs). This mechanism has consequences similar to those caused by physical loss of genetic material, hence it does not contrast LOH at 11p15 detected in various human neoplasms and may explain the low frequency of somatic mutations in specific 11p15 genes. We also detected various methylation differences between normal liver and matched HCC DNAs. In particular, loss of maternal-specific methylation at an internal KvLQT1 differentially methylated CpG-rich region [designated KvDMR1 (9, 10)] in tumor DNA correlated with an abnormal somatic switch of imprinting in tumor cells. Loss of maternal methylation negatively influenced various genes expressed from the maternal chromosome. Unlike the LOI detected at the IGF2 locus, which causes gain of expression of the IGF2 silent allele, here we demonstrate loss of expression of various imprinted genes at 11p15 and we designated this epigenetic mechanism “gain of imprinting” or GOI.
Materials and Methods
Tumor Samples.
Histopathologically diagnosed hepatocarcinomas (HCCs) and matched normal livers of 52 unrelated patients were analyzed. All tissues were collected at the Department of Internal Medicine and Gastroenterology, University of Bologna.
Detection of DNA Heterozygosity and Allele-Specific Expression.
Genomic DNAs and RNAs were purified from tissues as described (7). All RNA samples were treated with DNase before cDNA synthesis. Absence of genomic DNA contamination was confirmed by PCR tests. Single-strand cDNA was synthesized from 1 μg of total RNA by using the SUPERSCRIPT Choice System for cDNA Synthesis (GIBCO/BRL). SLC22A1L polymorphisms were identified by single-strand conformation polymorphism (SSCP) analysis and characterized by direct sequencing. The Thr6 ↔ Ala6 (ACT ↔ GCT), Gln12 ↔ Arg12 (CAG ↔ CGG) polymorphisms have been described (23), whereas Val78 ↔ Val78 (GTG ↔ GTA) is novel. CDKN1C polymorphism was a variable number of proline-alanine (PAPA) three-nucleotide repeats in exon 2 (24). The IGF2 polymorphism was an ApaI restriction site polymorphism in exon 9 (25). Constitutive heterozygosity and allele-specific (imprinted) expression of SLC22A1L, CDKN1C, and IGF2 loci were analyzed by PCR amplification of genomic DNA or cDNA, gel electrophoresis, autoradiography, and densitometric analysis (quantity one Program, Bio-Rad). All PCRs, unless specified differently, were carried out as described (26) with 50 ng of genomic DNA or cDNA using 0.5 unit of Taq polymerase (Promega) in a 10-μl volume. Constitutive heterozygosity and LOH at the SLC22A1L locus were analyzed by amplification of matched normal/tumor genomic DNAs using the following primers: (i) 592-TOTF (5′-CCT GCT TGG ATC TCT CCT GG-3′) and 592-ESONE 2R (5′-GCA GGA GGA ACA GCG GTT CA-3′) for polymorphisms at amino acids 6 and 12 (PCR product size, 248 nt); (ii) 592-EX 3F (5′-CAG ATT CTA GGC CCT GCA GTC-3′) and 592-EX 3R (5′-GAC ACA GGA GTG CCG TCA TC-3′) for polymorphism at amino acid 78 (PCR product size, 288 nt). SLC22A1L allele-specific expression was detected semiquantitatively by amplification of matched normal/tumor cDNAs using the following primers: (i) 592-TOTF and 592-1R (5′-CCG AGA CAG GTA TGG CAC GA-3′) for polymorphisms at amino acids 6 and 12 (PCR product size, 200 nt); (ii) primers 592-2F (5′-GCT GGC CGC CAC AGA ACT TA-3′) and 592-2R (5′-AGG AGC AGG TAG AGC GCC AA-3′) for polymorphism at amino acid 78 (PCR product size, 225 nt). PCR was carried out in the presence of 12.5% glycerol for 23 cycles (LOH analysis) or 28 cycles (allele-specific expression) consisting of a denaturing step at 94°C for 15 s, annealing at 61°C for 30 s, and extension at 72°C for 30 s. SLC22A1L polymorphisms were detected by SSCP on 0.5× nondenaturing mutation detection enhancement gels (FMC). LOH and imprinting studies on CDKN1C were performed by using a seminested PCR. Genomic DNAs or cDNAs were first amplified by a 25-cycle PCR (98°C for 30 s, 58°C for 30 s, 72°C for 60 s) using primers KIP2-81F (5′-AAC CCG ACG CAG AAG AGT CC-3′) and KIP2-940R (5′-CCT GCT CGG CGC TCT CTT GAG G-3′) in a reaction containing the Expand Long template enzyme mixture (Roche Molecular Biochemicals) and 10% DMSO. One-fiftieth of the first reaction was reamplified using primers KIP2-3F (5′-TGG ACC GAA GTG GAC AGC GA-3′) and KIP2-940R for 25 cycles. Hot start was used in the first amplification. PCR products ranged from 444 to 456 nt in size. Products were electrophoresed on 4.5% denaturing polyacrylamide gels. For IGF2, genomic DNA or cDNA were both amplified using primers 3 and 4 described by Cui et al. (27). PCR product size was 292 nt. PCR was carried out for 25 cycles (LOH) or 30 cycles (allele-specific expression) and consisted of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s. IGF2 polymorphisms were detected by SSCP on 0.5× nondenaturing mutation detection enhancement gels (FMC). β-actin primers and PCR conditions were as described (28).
Analysis of Methylation at the KvDMR1 Locus.
Southern blot analysis of DNA from HCCs and matched normal livers was carried out following standard procedures. DNA was double-digested with BamHI and HpaII, which is sensitive to CpG methylation. Internal control DNAs were digested with BamHI or with MspI, an isoschizomer of HpaII that is methylation-insensitive.
Statistical Analysis.
Results were analyzed by the Fisher's exact test or the χ2 test with Yates correction. P < 0.05 was considered significant.
Results
Loss of Expression of Imprinted Genes.
Expression of three imprinted genes, IGF2, CDKN1C, and SLC22A1L, were studied in 52 HCC and matched normal liver samples. Of the 37 informative samples heterozygous for at least one of the genes, none displayed LOH at any of chromosome 11p15 loci, whereas, under the same conditions, LOH was readily detected in 3 of 7 Wilms' tumor DNAs analyzed (data not shown). Thus, no LOH-dependent allelic imbalance that might have confused our analysis of allele-specific expression of the three 11p15 imprinted genes was detected. Table 1 summarizes the results for each gene in the 37 informative samples. Analysis of β-actin expression served to normalize expression levels among matched samples, and all results were confirmed in at least two independent experiments.
Table 1.
Quantitative analysis of allele specific expression of 11p15 imprinted genes in HCC and matched normal liver samples
| Patient |
SLC22A1L
|
CDKN1C
|
IGF2
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Li | HCC | Ratio | Li | HCC | Ratio | Li | HCC | Ratio | |
| HCC 236 | 100/100 | 100/100 | Normal | 100/15 | 100/40 | Normal | 100/100 | 100/100 | Normal |
| HCC 240 | 100/100 | 100/100 | Normal | / | / | nd | / | / | nd |
| HCC 233 | 100/100 | 100/100 | Normal | hom | hom | nd | hom | hom | nd |
| HCC 244 | 100/20 | 20/20 | GOI | hom | hom | nd | hom | hom | nd |
| HCC 247 | 100/100 | 100/100 | Normal | / | / | nd | hom | hom | nd |
| HCC 249 | hom | hom | nd | hom | hom | nd | 100/35 | 30/100 | INV |
| HCC 248 | hom | hom | nd | hom | hom | nd | 100/100 | 100/100 | Normal |
| HCC 241 | 100/90 | 100/25 | GOI part | 100/100 | 100/0 | GOI | 100/30 | 100/0 | GOI part |
| HCC 232 | 100/35 | 100/35 | Normal | 100/30 | 100/30 | Normal | hom | hom | nd |
| HCC 1462 | hom | hom | nd | hom | hom | nd | 100/100 | 100/60 | GOI part |
| HCC 1430 | 100/80 | 100/80 | Normal | 100/7 | 100/15 | Normal | 100/100 | 100/20 | GOI |
| HCC 1431 | 100/75 | 100/75 | Normal | 100/10 | 0/0 | GOI | hom | hom | nd |
| HCC 1432 | hom | hom | nd | 100/15 | 100/10 | Normal | 100/25 | 25/100 | INV |
| HCC 1433 | 100/100 | 100/100 | Normal | hom | hom | nd | 100/100 | 100/100 | Normal |
| HCC 1434 | / | / | nd | hom | hom | nd | 100/80 | 100/100 | Normal |
| HCC 1435 | 100/100 | 100/100 | Normal | 100/10 | 100/100 | LOI | 100/45 | 100/10 | GOI part |
| HCC 1439 | 100/35 | 30/30 | GOI | / | / | nd | hom | hom | nd |
| HCC 1440 | 100/90 | 100/90 | Normal | 100/20 | 100/20 | Normal | hom | hom | nd |
| HCC 1441 | 100/80 | 100/80 | Normal | hom | hom | LOSS | 100/60 | 100/40 | Normal |
| HCC 1442 | 100/10 | 100/12 | Normal | hom | hom | nd | 100/35 | 35/35 | GOI |
| HCC 1443 | 100/100 | 100/10 | GOI | 100/0 | 0/0 | GOI | hom | hom | nd |
| HCC 1444 | 100/80 | 100/80 | Normal | 100/0 | 45/100 | INV | hom | hom | nd |
| HCC 1446 | / | / | nd | / | / | nd | 100/100 | 100/100 | Normal |
| HCC 1447 | 100/100 | 100/100 | Normal | hom | hom | nd | 100/100 | 100/100 | Normal |
| HCC 1449 | 100/100 | 100/100 | Normal | hom | hom | nd | 100/100 | 100/100 | Normal |
| HCC 1451 | 100/30 | 30/100 | INV | hom | hom | nd | hom | hom | nd |
| HCC 1452 | 100/100 | 100/100 | Normal | 100/12 | 100/15 | Normal | hom | hom | nd |
| HCC 1453 | 100/100 | 100/100 | Normal | / | / | nd | hom | hom | nd |
| HCC 1454 | 100/90 | 100/90 | Normal | hom | hom | LOSS | hom | hom | nd |
| HCC 1456 | hom | hom | nd | 100/20 | 10/10 | GOI | 100/100 | 100/0 | GOI |
| HCC 1457 | 100/80 | 100/80 | Normal | 100/25 | 100/35 | Normal | 100/100 | 100/0 | GOI |
| HCC 1458 | 100/90 | 100/90 | Normal | hom | hom | nd | hom | hom | nd |
| HCC 1459 | 100/100 | 100/100 | Normal | / | / | nd | 100/15 | 100/10 | Normal |
| HCC 1460 | 100/90 | 100/95 | Normal | 100/40 | 100/35 | Normal | hom | hom | nd |
| HCC 1463 | / | / | nd | / | / | nd | 100/80 | 100/0 | GOI |
| HCC 1464 | hom | hom | nd | hom | hom | nd | 100/100 | 100/0 | GOI |
| HCC 1466 | hom | hom | nd | 100/0 | 100/10 | Normal | 100/50 | 100/50 | Normal |
| Total | 52 | 52 | 52 | ||||||
| Analyzed | 46 | 33 | 44 | ||||||
| Heterozygous | 27 | 15 | 21 | ||||||
| Homozygous | 19 | 18 | 23 | ||||||
| Imprinted | 5/27 | 14/15 | 8/21 | ||||||
| GOI | 4/27 | 4/15 | 9/21 | ||||||
| INV | 1/27 | 1/15 | 2/21 | ||||||
| LOI | 0/27 | 1/15 | 0/21 | ||||||
GOI, gain of imprinting; GOI part, partial gain of imprinting; LOI, loss of imprinting; INV, allele inversion; LOSS, loss of expression; hom, homozygous; nd, not determined; Li, normal liver; HCC, hepatocarcinoma.
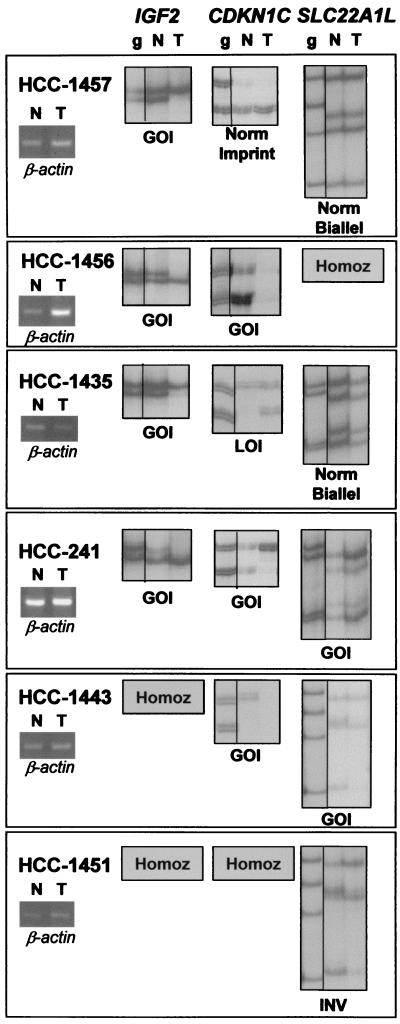
In somatic tissues, expression of an imprinted gene is detected as a difference in the expression level between the two alleles. Among the genes analyzed, SLC22A1L and CDKN1C are known to be expressed from the maternal allele and IGF2 from the paternal allele. Analysis of these genes in normal liver revealed a variable degree of imprinted expression: SLC22A1L was imprinted in 5 of 27 (18.5%) informative heterozygote samples, CDKN1C in 14 of 15 (93%), and IGF2 in 8 of 21 (38%). Densitometric analysis indicating a more than 30% difference in expression levels between alleles was considered positive for imprinted expression. Compared with their normal matched liver tissue, several HCC samples displayed abnormal allele-specific expression (Fig. 1). The most frequent abnormality was the loss of expression of one allele, GOI, observed in 15–43% of HCC samples, depending on the gene tested (Table 1). Other abnormalities, such as allelic inversion or LOI, were detected at a lower frequency in HCCs (Table 1 and Fig. 1).
Figure 1.
Abnormal patterns of allelic expression detectable in 11p15 imprinted genes in human HCCs. Gain (GOI), loss (LOI), and inversion (INV) of imprinting are shown for normal liver (N) and HCC (T). Corresponding constitutive heterozygous HCC genomic DNA (g) is shown. Analysis of β-actin served to normalize expression levels among matched normal liver and HCC samples. GOI, the most frequently detected abnormality, was considered positive when reduction of expression of one allele in HCC was greater than 70% as compared with the same allele in the matched normal liver; reduction between 25 and 70% was considered partial GOI, and a difference in allele expression less than 25% was considered negative for abnormality. When the imprinted gene was monoallelically expressed in normal liver, GOI led to complete loss of expression in matched HCC (most frequently encountered for CDKN1C). When the gene was biallelically expressed in normal liver, GOI in HCC was detected as a reduction or disappearance of one of the alleles (most frequently observed for the IGF2 gene).
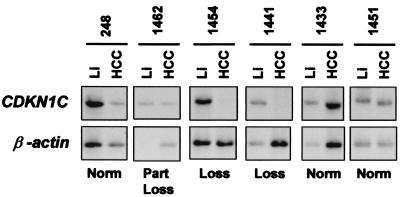
For SLC22A1L, GOI was detected in 4 of 27 informative HCCs (15%). Only two of these cases (HCC-244 and -1439) displayed an imprinted expression in the normal liver, and GOI led to reduction of expression of the most active allele, likely the maternally inherited. In the two remaining cases (HCC-241 and -1443), SLC22A1L was biallelically expressed in normal liver and therefore maintained one of the two normally active alleles in the HCC. A fifth case (HCC-1451) had an inversion in allele expression. The CDKN1C gene displayed GOI in 4 of 15 (26.7%) informative HCCs. In three of these cases (HCC-1431, -1443, and -1456), CDKN1C expression was nearly or completely extinguished. Furthermore, 3 of 10 additional cases that were not informative for allele-specific expression because of homozygosity, showed a significant reduction of CDKN1C expression in HCC as compared with the matched normal liver (Fig. 2). This result may be interpreted and explained by the same GOI demonstrated for heterozygous CDKN1C samples. Together, the analysis of CDKN1C and SLC22A1L indicates that GOI causes loss of expression of genes active on the maternally inherited chromosome. Unexpectedly, GOI was also detected for IGF2 in 9 of 21 (43%) informative HCCs. In all nine cases, however, at least one IGF2 allele remained active.
Figure 2.
Expression of CDKN1C in human HCCs. CDKN1C and β-actin expression level were determined semiquantitatively in normal liver (Li) and matched HCC (HCC) by direct cDNA amplification: 40 cycles for CDKN1C (primers KIP2-81F and KIP2-940R), and 25 cycles for β-actin. Norm, normal expression; Loss, loss of expression.
Several tumor samples (HCC-241, -1443, and -1456) revealed GOI in more than one of the 11p15-imprinted genes (Table 1 and Fig. 1), suggesting that a common mechanism, possibly alteration of one or more imprinting centers, controls all of the affected genes.
Association Between Abnormalities in HCC-Imprinted Expression and Loss of KvDMR1 DNA Methylation.
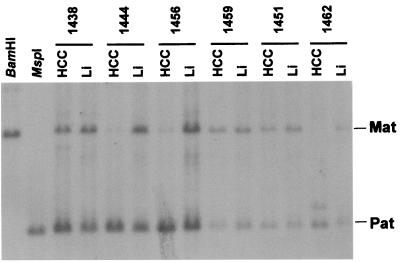
Genomic imprinting is associated with parental-specific methylation of DNA (29, 30). The KvDMR1 locus at chromosome 11p15 is differentially methylated on the paternal or maternal chromosome (9, 10). Southern blot analysis revealed abnormalities in KvDMR1 methylation patterns in HCC DNAs: loss of maternal-specific methylation was detected in tumor DNA but not in the matched normal liver (Fig. 3). Loss of maternal-specific methylation was observed in 10 of 20 randomly selected HCC tumor DNAs. This somatically acquired abnormal imprinted methylation could cause abnormal allele-specific expression. Indeed, analysis of KvDMR1 methylation status in comparison with normal or abnormal imprinted gene expression in HCC (Table 2) revealed a statistically significant association between KvDMR1 methylation status and allele-specific expression of CDKN1C and IGF2 genes, but not SLC22A1L, which is imprinted in fetal tissues and generally less in adult tissues. For CDKN1C and IGF2, loss of maternal KvDMR1 methylation was generally associated with GOI, whereas normal maternal and paternal methylation was associated with maintenance of the same pattern of expression observed in the matched normal liver. There were, however, notable exceptions: loss of KvDMR1 maternal methylation was detected in HCC-1435 in association with LOI at CDKN1C and partial GOI at IGF2, and also in HCC-1444, which displays an allelic inversion of CDKN1C. In HCC-1432, an inversion of IGF2 allelic expression was detected in the absence of KvDMR1 methylation abnormalities. Thus, whereas the KvDMR1 locus appears to influence imprinting, other regulatory elements are likely to act synergistically with KvDMR1 to control the allele-specific expression of the genes of this region.
Figure 3.
Southern blot analysis of the methylation status at the KvDMR1 locus. DNAs from six randomly selected HCCs and matched normal livers were double-digested with BamHI and HpaII, a CpG methylation-sensitive restriction enzyme. EST AA155639 was used as a probe to detect the KvDMR1 locus, as described by Mitsuya et al. (10). The maternal locus, which is hypermethylated, is detectable as a 6.0-kb fragment, whereas the demethylated paternal locus is detectable as a 2.6-kb fragment. Lanes with DNAs digested with BamHI or MspI (methylation-insensitive) were internal controls. In three samples, the maternal-specific methylation was lost specifically in the HCC DNA. Li, normal liver; HCC, hepatocarcinoma; Mat, maternal methylation; Pat, paternal methylation.
Table 2.
Relationship between KvDMR1 methylation and imprinting in HCC samples
| Gene | Ratio* | HCCKvDMR1 methylation†
|
Fisher exact test probability‡ | |
|---|---|---|---|---|
| P = M | P > M | |||
| SLC22A1L | Abnormal | 2 | 2 | 0.51 |
| Normal | 6 | 3 | ||
| CDKN1C | Abnormal | 0 | 6 | 0.035 |
| Normal | 2 | 0 | ||
| IGF2 | Abnormal | 1 | 4 | 0.039 |
| Normal | 4 | 0 | ||
Abnormal ratio indicates the detection of GOI, LOI or INV. Normal ratio indicates the presence of the same allelic expression pattern in normal and matched HCC.
†P, paternal pattern of methylation; M, maternal pattern of methylation.
‡ Probability <0.05 is statistically significant.
Discussion
The present study provides a demonstration of somatic GOI at 11p15 maternally expressed genes such that normally active genes are repressed. GOI was detected in a large fraction of HCCs and might represent a pathogenetic mechanism, possibly occurring in other neoplasms or during development. In principle, loss of 11p15 maternal alleles in pediatric tumors have the same consequence as GOI in HCCs of inactivating imprinted, maternally expressed genes. Furthermore, this mechanism recalls BWS cases with paternal uniparental disomy (31, 32), and suggests a unifying common picture, in which loss of function of 11p15 maternal alleles, through various different mechanisms, may be the critical event associating this chromosomal region with tumorigenesis and BWS. The loss of maternal specific methylation of KvDMR1, not only in HCCs but also in BWS (9, 10), supports this hypothesis.
In addition to GOI, we detected other abnormal allelic expression, including LOI and inversion of alleles. For example, we detected LOI in CDKN1C in one hepatocarcinoma (HCC-1435), but never at the IGF2 locus, which is instead frequently affected in Wilms' tumors (33, 34). These results suggest a heterogeneity in the mechanisms affecting imprinted genes in different tumors, and the possible involvement of independent fetal and adult imprinting centers as well as tissue-specific control elements, whose existence at chromosome 11p15 is supported by studies on SLC22A1L and KvLQT1 (23, 35, 36).
Partial or complete extinction of expression of various 11p15 genes as a consequence of abnormal control of imprinting may be relevant for tumorigenesis in general and not only for hepatocarcinogenesis. Just as mutations in CDKN1C have indicated a role of this gene in BWS (16), GOI leading to extinction of CDKN1C expression in HCC raises the possibility that this gene, especially in light of the cyclin-dependent cell cycle inhibitor it encodes, is involved in tumorigenesis. However, studies in mouse models have proven the role of CDKN1C in BWS (37) but not in tumor development. Similarly, the direct role of SLC22A1L in tumorigenesis is unclear, although the detection of mutations in this gene in some tumor samples (7, 15) and the present finding of down-regulated expression of the most active allele in some HCC samples support its role in tumorigenesis. Because IGF2 is believed to be an important autocrine growth factor in childhood and adult tumors (13, 38), its loss or reduction of expression is unlikely to have a role in the pathogenesis of HCCs; however, its detection may represent the evidence of a mechanism that shuts off the expression of imprinted loci present over a large region of the maternal chromosome. The demonstration that GOI can affect expression of various genes raises the possibility that not a single, but the combined loss of expression of various 11p15 imprinted genes may contribute to the tumorigenic process. We do not exclude the possibility that other 11p15 imprinted genes not investigated in the present study, such as H19 or BWR1C, might also be affected and have relevance for tumorigenesis and BWS.
The mechanism that generates abnormal imprinting in HCCs remains elusive. Our results indicate that somatic cells can, in some cases, activate or inactivate the machinery responsible for genomic imprinting. Insight into how this occurs in a somatic cell may help to understand genomic imprinting switch in germinal cells.
Acknowledgments
We thank Augusto Bevilacqua and Pietro Zucchini for their excellent technical assistance. This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro, from Telethon, and from Ministero dell'Universitá e della Ricerca Scientifica e Tecnologica ex-60%.
Abbreviations
- BWS
Beckwith–Wiedemann syndrome
- SSCP
single-strand conformation polymorphisms
- LOI
loss of imprinting
- GOI
gain of imprinting
- HCC
hepatocarcinoma
- LOH
loss of heterozygosity
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090087497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090087497
References
- 1.Sutcliffe J S, Nakao M, Christian S, Orstavik K H, Tommerup N, Ledbetter D H, Beaudet A L. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- 2.Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls R D, Horsthemke B. Nat Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- 3.Reik W, Maher E R. Trends Genet. 1997;13:330–334. doi: 10.1016/s0168-9525(97)01200-6. [DOI] [PubMed] [Google Scholar]
- 4.Junien C. Curr Opin Genet Dev. 1992;2:431–438. doi: 10.1016/s0959-437x(05)80154-6. [DOI] [PubMed] [Google Scholar]
- 5.Schroeder W T, Chao L Y, Dao D T, Strong L C, Pathak S, Riccardi V M, Lewis W K, Saunders G F. Am J Hum Genet. 1987;40:413–420. [PMC free article] [PubMed] [Google Scholar]
- 6.Williams J, Brown K, Mott M, Maitland N. Lancet. 1990;1:283–284. doi: 10.1016/s0140-6736(89)91300-7. [DOI] [PubMed] [Google Scholar]
- 7.Schwienbacher C, Sabbioni S, Campi M, Veronese A, Bernardi G, Menegatti A, Hatada I, Mukai T, Ohashi H, Barbanti-Brodano G, et al. Proc Natl Acad Sci USA. 1998;95:3873–3878. doi: 10.1073/pnas.95.7.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu R J, Lee M P, Connors T D, Johnson L A, Burn T B, Su K, Landes G M, Feinberg A P. Genomics. 1997;46:9–17. doi: 10.1006/geno.1997.4981. [DOI] [PubMed] [Google Scholar]
- 9.Smilinich N J, Day C D, Fitzpatrick G V, Caldwell G M, Lossie A C, Cooper P R, Smallwood A C, Joyce J A, Schofield P N, Reik W, et al. Proc Natl Acad Sci USA. 1999;96:8064–8069. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsuya K, Meguro M, Lee M P, Katoh M, Schulz T C, Kugoh H, Yoshida M A, Niikawa N, Feinberg A P, Oshimura M. Hum Mol Genet. 1999;8:1209–1217. doi: 10.1093/hmg/8.7.1209. [DOI] [PubMed] [Google Scholar]
- 11.Matsuoka S, Edwards M C, Bai C, Parker S, Zhang P, Baldini A, Harper J W, Elledge S J. Genes Dev. 1995;9:650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 12.Lee M-H, Reynisdottir I, Massague J. Genes Dev. 1995;9:639–649. doi: 10.1101/gad.9.6.639. [DOI] [PubMed] [Google Scholar]
- 13.Scott J, Cowell J, Robertson M E, Priestley L M, Wadey R, Hopkins B, Pritchard J, Bell G I, Rall L B, Graham C F, Knott T J. Nature (London) 1985;317:260–262. doi: 10.1038/317260a0. [DOI] [PubMed] [Google Scholar]
- 14.Qian N, Franck D, O'Keefe D, Dao D, Zhao L, Yuan L, Wang Q, Keating M, Walsh C, Tycko B. Hum Mol Genet. 1997;6:2021–2029. doi: 10.1093/hmg/6.12.2021. [DOI] [PubMed] [Google Scholar]
- 15.Lee M P, Reeves C, Schmitt A, Su K, Connors T D, Hu R-J, Brandenburg S, Lee M J, Miller G, Feinberg A P. Cancer Res. 1998;58:4155–4159. [PubMed] [Google Scholar]
- 16.Hatada I, Ohashi H, Fukushima Y, Kaneko Y, Inoue M, Komoto Y, Okada A, Ohishi S, Nabetani A, Morisaki H, et al. Nat Genet. 1996;14:171–173. doi: 10.1038/ng1096-171. [DOI] [PubMed] [Google Scholar]
- 17.O'Keefe D, Dao D, Zhao L, Sanderson R, Warburton D, Weiss L, Anyane-Yeboa K, Tycko B. Am J Hum Genet. 1997;61:295–303. doi: 10.1086/514854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogawa O, Eccles M R, Szeto J, McNoe L A, Yun K, Maw M A, Smith P J, Reeve A E. Nature (London) 1993;362:749–751. doi: 10.1038/362749a0. [DOI] [PubMed] [Google Scholar]
- 19.Rainier S, Johnson L, Dobry C J, Ping A J, Grundy P E, Feinberg A P. Nature (London) 1993;362:747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 20.Brown K W, Villar A J, Bickmore W, Clayton-Smith J, Catchpoole D, Maher E R, Reik W. Hum Mol Genet. 1996;5:2027–2032. doi: 10.1093/hmg/5.12.2027. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki H, Veda R, Takahashi T, Takahashi T. Nat Genet. 1994;6:332–333. doi: 10.1038/ng0494-332. [DOI] [PubMed] [Google Scholar]
- 22.Randhawa G S, Cui H, Barletta J A, Strichman-Almashanu L Z, Talpaz M, Kantarjian H, Deisseroth A B, Champlin R C, Feinberg A P. Blood. 1998;91:3144–3147. [PubMed] [Google Scholar]
- 23.Cooper P R, Smilinich N J, Day C D, Nowak N J, Reid L H, Pearsall R S, Reece M, Prawitt D, Landers J, Housman D E, et al. Genomics. 1998;49:38–51. doi: 10.1006/geno.1998.5221. [DOI] [PubMed] [Google Scholar]
- 24.Matsuoka S, Thompson J S, Edwards M C, Barletta J M, Grundy P, Kalikin L M, Harper J W, Elledge S J, Feinberg A P. Proc Natl Acad Sci USA. 1996;93:3026–3030. doi: 10.1073/pnas.93.7.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takodoro K, Fujii H, Inoue T, Yamada M. Nucleic Acids Res. 1991;19:6967. doi: 10.1093/nar/19.24.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Negrini M, Rasio D, Hampton G M, Sabbioni S, Rattan S, Carter S L, Rosenberg A L, Schwartz G F, Shiloh Y, Cavenee W K, Croce C M. Cancer Res. 1995;55:3003–3007. [PubMed] [Google Scholar]
- 27.Cui H, Horon I L, Ohlsson R, Hamilton S R, Feinberg A P. Nat Med. 1998;4:1276–1280. doi: 10.1038/3260. [DOI] [PubMed] [Google Scholar]
- 28.Strauss W M, Dausman J, Beard C, Johnson C, Lawrence J B, Jaenisch R. Science. 1993;259:1904–1907. doi: 10.1126/science.8096090. [DOI] [PubMed] [Google Scholar]
- 29.Wutz A, Smrzka O W, Schweifer N, Schellander K, Wagner E F, Barlow D P. Nature (London) 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 30.Constancia M, Pickard B, Kelsey G, Reik W. Genome Res. 1998;8:881–900. doi: 10.1101/gr.8.9.881. [DOI] [PubMed] [Google Scholar]
- 31.Henry I, Puech A, Riesewijk A, Ahnine L, Mannens M, Beldjord C, Bitoun P, Tournade M F, Landrieu P, Junien C. Eur J Hum Genet. 1993;1:19–29. doi: 10.1159/000472384. [DOI] [PubMed] [Google Scholar]
- 32.Slatter R E, Elliott M, Welham K, Carrera M, Schofield P N, Barton D E, Maher E R. J Med Genet. 1994;31:749–753. doi: 10.1136/jmg.31.10.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniguchi T, Sullivan M J, Osamu O, Reeve A. Proc Natl Acad Sci USA. 1995;92:2159–2163. doi: 10.1073/pnas.92.6.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steenman M J C, Rainier S, Dobry C J, Grundy P, Horon I L, Feinberg A P. Nat Genet. 1994;7:433–439. doi: 10.1038/ng0794-433. [DOI] [PubMed] [Google Scholar]
- 35.Dao D, Frank D, Qian N, O'Keefe D, Vosatka R J, Walsh C P, Tycko B. Hum Mol Genet. 1998;7:597–608. doi: 10.1093/hmg/7.4.597. [DOI] [PubMed] [Google Scholar]
- 36.Lee M P, Hu R J, Johnson L A, Feinberg A P. Nat Genet. 1997;15:181–185. doi: 10.1038/ng0297-181. [DOI] [PubMed] [Google Scholar]
- 37.Zhang P, Liegeois N J, Wong C, Finegold M, Hou H, Thompson J C, Silverman A, Harper J W, dePinho R A, Elledge S J. Nature (London) 1997;387:151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- 38.Yee D, Cullen K J, Paik S, Perdue J F, Hampton B, Lippman M E, Rosen N. Cancer Res. 1988;48:6691–6696. [PubMed] [Google Scholar]