Abstract
A mathematical model is presented to describe the catalytic mechanism of mammalian NAD-linked isocitrate dehydrogenase (NAD-IDH), a highly regulated enzyme in the tricarboxylic acid cycle, a crucial pathway in energy metabolism and biosynthesis. The mechanism accounts for allosteric regulation by magnesium-bound isocitrate and EGTA and calcium-bound ATP and ADP. The developed model is used to analyze kinetic data for the cardiac enzyme and to estimate kinetic parameter values. Since the kinetic mechanism is expressed in terms of chemical species (rather than biochemical reactants), the model explicitly accounts for the effects of biochemical state (ionic strength, pH, temperature, and metal cation concentration) on the kinetics. Because the substrate isocitrate competes with allosteric activators (ATP and ADP) and an inhibitor (EGTA) for metal ion cofactors (Ca2+ and Mg2+), the observed kinetic relationships between reactants, activator and inhibitor concentrations, and catalytic flux are complex. Our analysis reveals that under physiological conditions, the ADP/ATP ratio plays a more significant role than Ca2+ concentration in regulating the enzyme's activity. In addition, the enzyme is highly sensitive to Mg2+ concentration in the physiological range, pointing to a potential regulatory role of [Mg2+] in mitochondrial energy metabolism.
Introduction
NAD-linked isocitrate dehydrogenase (NAD-IDH, EC 1.1.1.41) catalyzes the biochemical reaction
where abbreviations for biochemical reactants are listed in Table 1. This enzyme is located exclusively in the mitochondria in mammalian cells and is a key regulatory enzyme in the TCA cycle [1-3]. NAD-IDH from a variety of animal tissue has been shown to require Mg2+, Mn2+, or Co2+ for activity [4-7]. In fact, it is apparent that the active substrate of the enzyme is the divalent metal ion chelate [8]. It also has been observed that the activity of this enzyme is enhanced by ADP [9] and inhibited by NADH, NADPH, and ATP [10]. NADH competes with NAD for binding to the active site [11]; adenine nucleotide binding at a regulatory site modifies the apparent Km for Mg-isocitrate [11]. ATP and AMP compete with ADP for the nucleotide regulatory site while the free enzyme must bind ADP before it can bind the substrate Mg-isocitrate [12].
Table 1.
Thermodynamic Parameter Values for Isocitrate Dehydrogenase (298.15 K, 1 M reactants, I = 0.15 M, P = 1 atm)
| Reactant |
Reference Species |
Ion-bound species |
pK |
|---|---|---|---|
| NAD |
NAD+ |
|
|
| HICIT2- | 5.64 | ||
| ICIT | ICIT3− | H2ICIT- | 4.33 |
| MgICIT- | 2.72 | ||
| |
|
CaICIT- |
2.47 |
| AKG |
AKG2− |
- |
- |
| NADH |
NADH0 |
- |
- |
| CO2tot |
CO32− |
HCO3− |
9.75 |
| EGTA | EGTA4− | HEGTA3− | 9.46 |
| H2EGTA2− | 8.85 | ||
| H3EGTA− | 2.65 | ||
| H4EGTA | 2.00 | ||
| MgEGTA2− | 5.21 | ||
| CaEGTA2− | 11.00 | ||
| |
|
KEGTA3− |
1.22 |
| ADP | ADP3− | HADP2− | 6.42 |
| H2ADP− | 3.82 | ||
| H3ADP | 1.00 | ||
| MgADP− | 3.05 | ||
| MgHADP | 1.61 | ||
| Mg2ADP+ | 1.00 | ||
| KADP2− | 1.53 | ||
| |
|
CaADP− |
2.86 |
| ATP | ATP3− | HATP2− | 6.49 |
| H2ATP− | 3.94 | ||
| H3ATP | 1.90 | ||
| MgATP− | 4.36 | ||
| MgHATP | 2.30 | ||
| Mg2ATP+ | 1.70 | ||
| KATP2− | 0.96 | ||
| CaATP− | 3.86 | ||
| |
|
MgKATP− |
1.31 |
|
P EDTA |
EDTA4− |
HEDTA3− |
10.26 |
| H2EDTA2− | 6.16 | ||
| H3EDTA− | 2.67 | ||
| H4EDTA | 1.99 | ||
| MgEDTA2− | 8.69 | ||
| CaEDTA2− | 10.07 | ||
| |
|
KEDTA3− |
1.22 |
| HPO4 | HPO42− | H2PO4− | 6.70 |
| H3PO4 | 1.96 | ||
| MgHPO4 | 1.95 | ||
| KHPO4− | 1.22 | ||
| |
|
KH2PO4 |
0.2 |
| SO4 | SO42− | HSO4− | 1.54 |
| MgSO4− | 1.48 | ||
| |
|
CaSO4− |
1.53 |
| * HEPES | HEPES2− | H2EPES− | 7.52 |
NAD-IDH is one of the three dehydrogenases within mammalian mitochondrial that have been shown to be regulated by Ca2+ ions. NAD-IDH and 2-oxoglutarate dehydrogenase (OGDH) are activated by lowering the apparent Km values for substrate (in the presence of an adenine nucleotide), with little effect of Ca2+ at saturating concentrations of substrates [13-15]. The pyruvate dehydrogenase (PDH) complex, on the other hand, is activated through increases in the active dephosphorylated form of the enzyme, largely through stimulation of PDHP phosphatase [16]. Each of these three enzymes thus represents a site at which electrophysiological stimulation, endocrine signals, and other stimuli potentially influence mitochondrial oxidative metabolism through cytosolic Ca2+ concentration [1, 17-19]. However the details remain unclear. Both activation and inhibition of NAD-IDH activity in extracts of animal tissues by added calcium have been reported. Newsholme and coworkers concluded that Ca2+ increases the apparent Km of the rat heart enzyme for isocitrate in both the presence and absence of ADP [20, 21]. Aogaichi et al. reported that purified isocitrate dehydrogenase from bovine heart was stimulated by free Ca2+ in the presence of ADP and sub-saturating levels of Mg-isocitrate, but not in absence of ADP [14]. They found that Ca2+ is not absolutely required for ADP activation. Similarly, it has been observed that calcium affects NAD-IDH from extracts of rat heart mitochondria by lowering the apparent Km for isocitrate without affecting Vmax in the presence of ADP while slightly increasing the apparent Km for isocitrate when ADP is absent [13]. Rutter and Denton [22] investigated the effects of adenine nucleotides, bivalent metal Mg2+ on the regulation of the enzyme by Ca2+. They concluded that the sensitivity of NAD-IDH to Ca2+ is shown to be critically dependent on the concentration of Mg2+.
Rutter and Denton reported the range over which changes in ADP/ATP ratio may affect the sensitivity of the enzyme to Ca2+ [23]. It is also shown that increases in ADP/ATP ratio result in a decrease in apparent Km values for the respective substrates [3]. Based on these observations, it is suggested that increases in the ADP/ATP ratio may stimulate the supply of NADH for the respiratory chain through the activation of NAD-IDH [23]. Another important regulation of flux through this enzyme is the ration of NADH/NAD+ [2, 10]. Inhibition by NADH or NADPH of NAD isocitrate dehydrogenase has been reported for the soluble enzyme from heart [12] and liver [6]. Simulations predict that this ratio is a more important regulator of TCA cycle flux than the ADP/ATP ratio [24]. Inhibition of enzyme activity by NADH cannot be reversed by ADP regardless of the Ca2+ concentration [15].
Experimental studies of NAD-IDH typically use EGTA and/or EDTA to buffer Ca2+ and Mg2+ ions in reaction solutions. However, EGTA, EDTA, and other nitrogen-containing polycarboxylate Ca2+ chelators may inhibit the activity of NAD-specific isocitrate dehydrogenase in the presence and absence of ADP by a mechanism that cannot be attributed solely to the sequestration free Ca2+ [13, 15]. Therefore it is necessary to properly account for these inhibition mechanisms to analyze the available in vitro data. However it remains uncertain which chelate forms of EGTA and EDTA are responsible for inhibition and by what mechanism.
Although models have been proposed [9], to date no mechanistic model consistently explains the available kinetic data sets for this enzyme. In this work, a simple model is proposed to resolve observations from a number of studies. A systematic analysis provides clarification for several apparently contradictory results reported previously.
Methods and Results
Kinetic equations for ordered bi-bi enzyme mechanism
Here we model the kinetics of NAD-IDH based on the compulsory ordered bi-bi mechanism (or ordered-addition bi bi). This basic mechanism for the reaction A+B ⇌ P+Q involves four enzyme state transitions:
| (1) |
where each state transition is assumed to proceed by mass action [25-27]. Here E1 represents free (unbound) enzyme; E2 represents the complex formed between enzyme and the species A; E3 represents the enzyme complex bound to both substrates; and E4 represents the enzyme complex bound to product Q. Although the reaction of NAD-IDH involves three biochemical products, there are not enough data to resolve a catalytic mechanism accounting for the carbon dioxide-dependent kinetics. Therefore the developed mechanism does not explicitly account for CO2.
The quasi-steady flux for this mechanism may be expressed using the method of King and Altman, [28, 29]:
| (2) |
where
| (3) |
and
| (4) |
| (5) |
where the kinetic constants are defined as:
| (6) |
Other than Vmf and Vmr, all of these parameters have units of concentration (mass per unit volume); Vmf and Vmr have units of mass per unit time per unit volume.
Note that for a given value of the equilibrium constant for the biochemical reaction, the eleven kinetic parameters cannot vary independently. The eleven parameters are related to Keq via the equation:
| (7) |
Here we apply this general form to the analysis of data from isocitrate dehydrogenase to determine kinetic parameters for this enzyme and to elucidate the mechanisms of this enzyme.
Model of Isocitrate Dehydrogenase
Isocitrate dehydrogenase involves the fourth step in the oxidation of acetyl-CoA in the citric acid cycle. The reference chemical reaction is
| (8) |
where the abbreviations for the biochemical species are listed in Table 1. The corresponding biochemical reaction is
| (9) |
In Equation (9), biochemical reactants represent the sums of species. For example, isocitrate is made up of several rapidly interconverting species: [ICIT] = [ICIT3−] + [HICIT2−] + [H2ICIT−] + [MgICIT−] + [CaICIT−]. The reactant CO2tot represents the sum of aqueous carbon dioxide and biocarbonate species (, and H2CO3). All reactants and associated chemical species are listed in Table 1. Also listed in the table are species that may act as allosteric regulators and/or contribute to ion buffering in the experiments analyzed below.
The standard Gibbs free energy of the reference reaction is computed [30, 31]:
| (10) |
where the basic thermodynamic data are listed in Table 1. The equilibrium constant for the reference reaction () is calculated:
| (11) |
The species concentrations in the reference reaction can be computed using the binding polynomials defined as [31]:
Here the K's represent dissociation constants for the binding/unbinding between biochemical species and cations. For example, KHICIT is the dissociation constant for the reaction HICIT2− ⇌ ICIT3− + H+. Where no significant cation-bound states occur, the binding polynomial is set to equal to one. Each term in a binding polynomial represents the relative fraction of a given reactant in a particular species [32]. For example, the term [H+]/KHICIT in PICIT represents the relative fraction of isocitrate with one hydrogen ion bound to the ICIT3− reference state. The first term (equal to one) is the relative fraction in the reference state. Therefore the binding polynomials can be used to convert between reactant (sum of species) concentration and reference species concentration. For example, for the reactants and products of the NAD-IDH reaction,
| (12) |
The apparent equilibrium constant for the biochemical reaction is computed [31]
| (13) |
Here we hypothesize the following simple mechanism may explain the observed kinetics of the NAD-IDH [10, 12]:
This mechanism assumes ordered binding of the two substrates NAD+ and MgICIT−, with an irreversible step that includes the formation of the reaction products AKG2− and CO32− , and a final step that produces NADH. Since this mechanism is irreversible, it may be used only to model the forward flux through the catalytic cycle. The expression for the forward flux can be obtained by removing the P-dependent terms in the numerator and denominator of the flux expression for the ordered bi-bi mechanism. The resulting expression for forward flux is
| (14) |
where [A] = [NAD+], [B] = [MgICIT−] and [Q]=[NADH].
Mammalian NAD-IDH is composed of four subunits in the stoichiometry 2α:1β:1γ[33-35]. The α subunits contain the primary catalytic sites, while the other subunits are thought to play regulatory roles [33]. These structural features are likely responsible for the observed cooperative behavior in terms of overall flux versus substrate (MgICIT−) concentration. Although it is has not been determined whether or not the βand γsubunits significantly bind MgICIT− [35], catalytic flux is strongly cooperative with MgICIT− [9, 13]. To account for the observed cooperativity, we make the assumption that the binding of MgICIT− proceeds with the pseudo-first order binding constant , where n is the Hill coefficient:
| (15) |
If n = 3, then binding to two regulatory sites and a single active site are lumped into a single step. Thus it is assumed that binding to one catalytic site does not influence binding affinity to the other. Although this assumed mechanism is rather simplified in that individual biding and unbinding steps for MgICIT− are not considered, it provides the appropriate level of detail to explain the available data and to effectively simulate reaction flux as a function of reactant concentrations.
Accounting for cooperativity, the flux expression becomes
| (16) |
To account for additional allosteric activators and inhibitors, the flux expression must be further modified. As demonstrated below, the observed kinetics are explained by assuming that MgEGTA acts as a linear mixed-type noncompetitive inhibitor for unbound enzyme state E1; and CaADP, CaATP and ADP act as non-essential activators that competitively bind to E2. Accounting for these mechanisms, the forward flux expression becomes:
| (17) |
where the parameters KiMgEGTA, KaCaADP, KaCaATP, KaADP, a1, a2, a3, and a4 must be estimated. Using the relationship J−/J+ = exp(ΔG/RT) to determine the reverse flux, the full flux expression is [36]
| (18) |
To account for additional pH effects on the catalytic activity of the enzyme, it is assumed that the active sites acts as a monobasic acid [37] with dissociation constant KiH. The maximal flux Vmf in Equation (17) is expressed:
| (19) |
where the is a constant.
Computational Methods
The developed model has 14 adjustable parameters. Parameter values were estimated by finding the optimized values that minimize the difference between model prediction and experimental data in Figures 1–10, as detailed below. An error analysis was performed to estimate the sensitivity of error to small change of parameter values. The sensitivity was computed using [24]:
| (20) |
where E* is the minimum mean squared difference between model simulations and experimental data, and xi is the optimal value of the ith parameter. We used the FMINCON algorithm in Matlab to solve this non-linear optimization problem. The results of the sensitivity analysis are listed in Tables 2 and 3.
Figure 1.
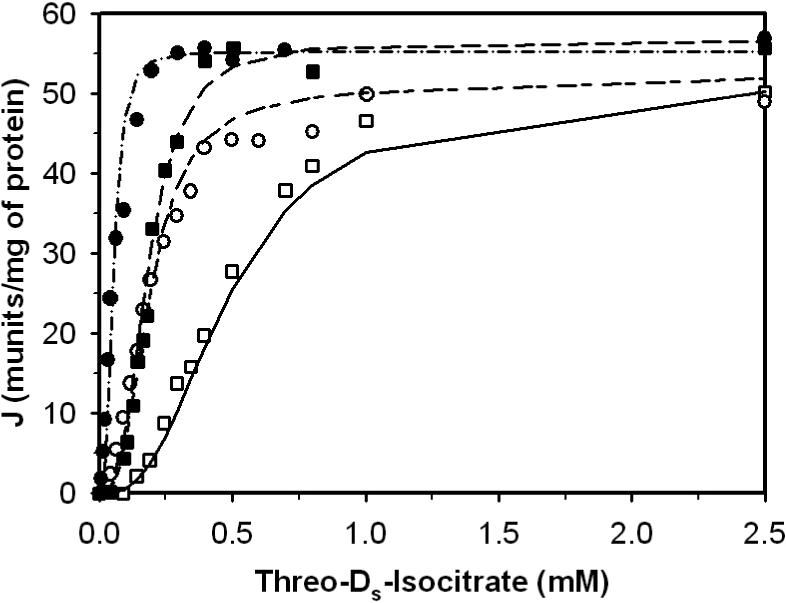
Fits to kinetic data from Figure 2a (left) of [23] on the forward operation of toluene-permeabilized enzyme. Open symbols (□, ○) represent data obtained without significant Ca2+ in the assay. Closed symbols (■, •) represent the presence of 0.1 mM free [Ca2+]. Circles and squares correspond to data obtained in the presence of 1.5 mM ADP and ATP, respectively; All data were obtained from Figure 2a of [23]. Solid line, long-short line, long dash line and dash-dot line are fits to the data points represented by symbols □, ○, ■ , and •, respectively.
Figure 10.
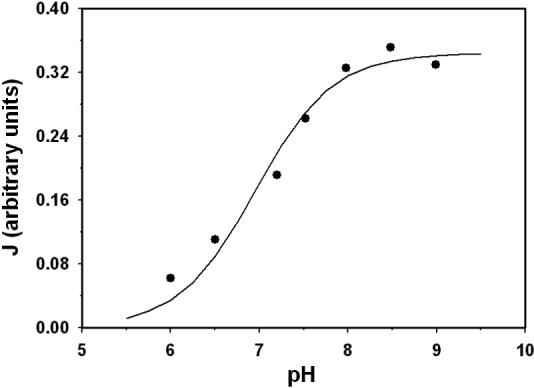
The maximum enzyme activity as a function of pH. Data points represent the measured maximum flux (relative) obtained by extrapolation of double-reciprocal plots [40]. Model fit is plotted as solid line.
Table 2.
Kinetic Parameter Values for Isocitrate Dehydrogenase from Rat Heart Data ([23])
| Parameter |
Parameter |
Sensitivity |
|---|---|---|
| Vmf (μmol·min−1·μg−1) | 69.2 | 67.0 |
| KmA (μM) | 503.3 | 6.94 |
| KmB (μM) | 148.9 | 103.3 |
| Kia (μM) | 77.6 | 0.0148 |
| KaCaADP (μM) | 13.3 | 0.483 |
| KaCaATP (μM) | 288.6 | 2.96 |
| KaADP (mM) | 61.3 | 14.1 |
| KiMgEGTA (μM) | 84.1 | 0.207 |
| Kiq (μM) | - | - |
| KiH (μM) | 0.11a | 1.13 |
| a1 | 0.0012 | 4.06 |
| a2 | 0.0097 | 2.07 |
| a3 | 0.0004 | 7.70 |
| a4 | 2.21 | 0.0026 |
| n | 3b | - |
Estimated from ox-brain mitochondria [40]
not an adjustable parameter
Table 3.
| Parameter |
Bovine heart data |
Sensitivity |
|---|---|---|
| Vmf (nmol/ml/min) | 8.62 | 19.86 |
| KmA (μM) | 909.3 | 7.29 |
| KmB (μM) | 168.4 | 194.0 |
| Kia (μM) | 52.7 | 0.0103 |
| KaCaADP (μM) | 52.8 | 4.43 |
| KaCaATP (μM) | - | |
| KaADP (mM) | 187 | 16.3 |
| KiMgEGTA (μM) | 7.05 | 0.0023 |
| Kiq (μM) | 4.75 | 0.0826 |
| KiH (μM) | 0.11a | 1.13 |
| a1 | 0.0006 | 16.2 |
| a2 | - | |
| a3 | 0.001 | 4.92 |
| a4 | 2.4 | 0.0005 |
| n | 3b | - |
Estimated from ox-brain mitochondria [40]
not an adjustable parameter
Analysis of data from toluene-permeabilized rat heart mitochondria
Rutter and Denton conducted a number of experiments to study the kinetics of NAD-linked isocitrate dehydrogenase and 2-oxoglutarate dehydrogenase within toluene-permeabilized rat heart mitochondria [23]. Their data are used here to identify the kinetic parameters for NAD-linked IDH in rat heart mitochondria.
Figures 1, 2, 3 and 4 illustrate model fits to data obtained from Figure 2(a) left, Figure 2(a) right, 3 and 4(a) in Rutter and Denton [23]: Figures 1 and 2 plot the flux versus threo-Ds-isocitrate at different Ca2+, ADP and ATP concentrations; Figure 3 plots the relative enzyme activity as a function of Ca2+ in the presence of ADP or ATP; the effect of ADP/ATP ratio on the relative enzyme activity at difference Ca2+ levels is plotted in Figure 4. All experimental data in Figures 1, 2, 3 and 4 were obtained at pH 7.2 and 30°C in a reaction medium detailed in [23]. Measured enzyme activity is expressed in munit/mg protein, where 1 munit is defined as the amount of enzyme catalyzing the c rate per min at 30°C. Based on the components of the reaction medium the overall ionic strength is estimated to be 0.15 M for these experiments.
Figure 2.
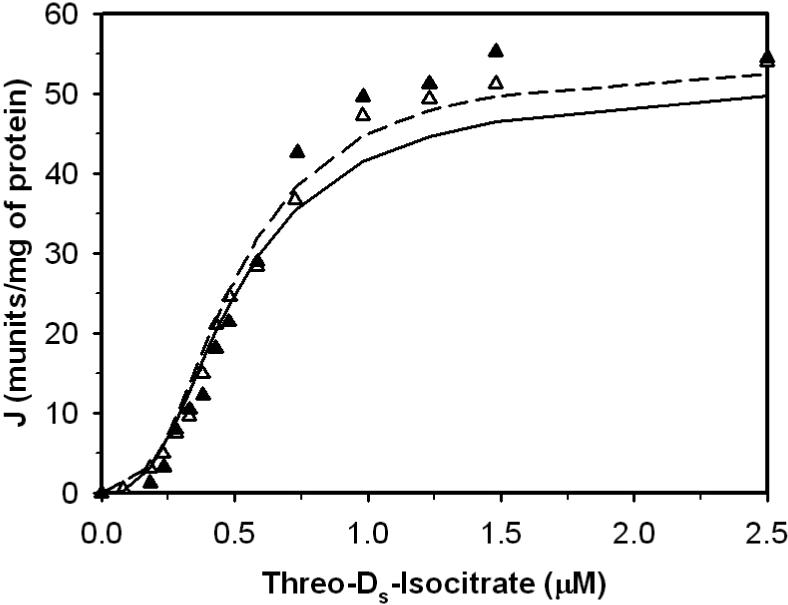
Fits to kinetic data from Figure 2a (right panel) of [23] on the forward operation of toluene-permeabilized enzyme. Symbols ▲ and Δ represents the pre sence and the absence of calcium in the buffer, respectively. Solid and dashed lines are fits to the data with and without calcium, respectively. No ATP or ADP was present for these experiments. Free [Ca2+] was 0.1 mM for the ▲ data set.
Figure 3.
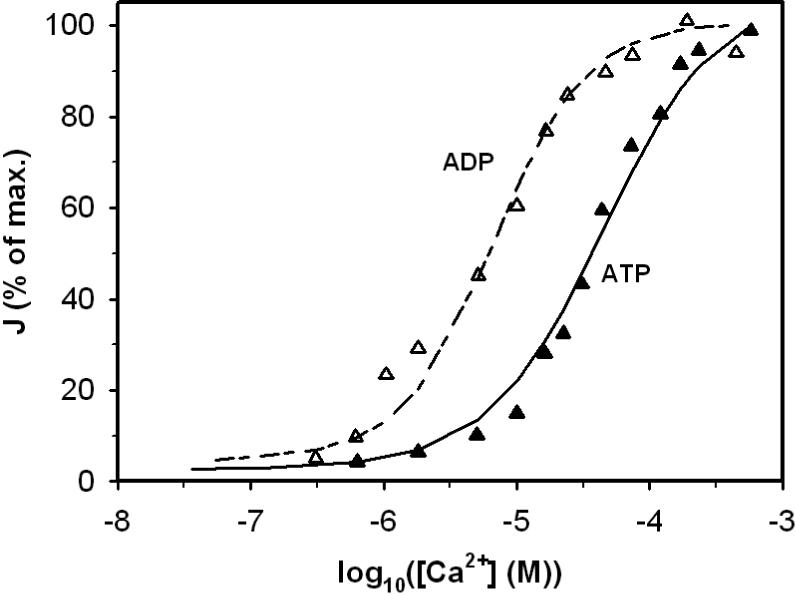
The effect of ATP and ADP on the activation effect of calcium to the enzyme activity. Symbols ▲ and Δ represent data obtained from Figure 3 of [23], obtained in the presence of either 1.5 mM [ATP] or [ADP]. Solid line and long-short line are fits to the data.
Figure 4.
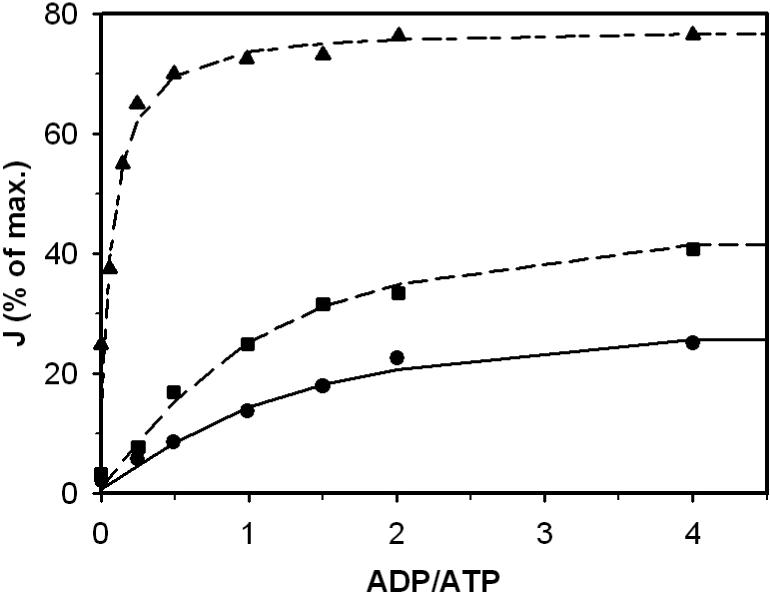
The effect of ADP/ATP ratio on the enzyme activity at different calcium concentrations. Solid line, long-dash line and long-short line are fits to the kinetic data obtained from Figure 4 of [23]. The sum of ATP and ADP concentration was held fixed 1.5 mM. Symbols •, ■ and ○ represent free calcium concentrations of 1 nM, 1 μM and 0.1 mM, respectively.
The data shown in Figure 1 and 2 are obtained from Rutter and Denton, where, NAD-IDH activity was measured by isocitrate (racemic mixture of D- and L-isocitrate) in the presence of 2 mM NAD+ in their ”basic incubation medium” [23]. The medium contained Mops (50 mM), triethanolamine (35.5 mM), KCl (75 mM), sucrose (60 mM), KH2PO4 (2 mM), EGTA (1 mM), HEDTA (1 mM), and rotenone (1 μg/ml). MgCl2 was added to give 1.0 mM free [Mg2+] before the addition of DL-isocitrate. Open symbols represent data obtained without significant Ca2+ in the assay. Closed symbols represent the presence of 0.1 mM free [Ca2+]. Circles and squares correspond to data obtained in the presence of 1.5 mM ADP and ATP, respectively; and triangles indicate neither ADP nor ATP was present.
To analyze these experiments, we calculated species concentrations (free ions and metal-ion bound species of ATP, ADP, isocitrate, EGTA, EDTA) in the complex media based on the reactant concentrations used in the experiments. Our calculations assume that both stereoisomers of isocitrate have the same dissociation constants, as indicated in Table 1. Total MgCl2 and CaCl2 were calculated to give [Mg2+] = 1.0 mM at zero isocitrate concentration and [Ca2+] = 0.0 or 1.0 mM, depending on the protocol.
Rutter and Denton indicated that the free Mg2+ concentration was 1 mM at the beginning of all experiments and free Ca2+ concentration was maintained at 0.1 mM in experiments to obtain the closed symbol data in Figure 1. These total concentrations and total DL-isocitric acid concentration were used to compute the concentrations of all ions of interest in the complex reaction media based on the dissociation constants listed in Table. Our calculations reveal that the free [Mg2+] varied between 0.35 mM and 0.42 mM at the highest concentration of DL-isocitrate for experiments in Figures 1 and 2. These estimates are similar to those reported by Rutter and Denton, who estimated the free [Mg2+] to be 0.5 mM at the maximal isocitrate concentration employed. Differences between our estimates and that of Rutter and Denton are due to the fact we account more bound states (all binding states indicated in Table 1 are included in our calculation) and we have adjusted the values of the dissociation constants to account for ionic strength and temperature.
Figure 3 shows enzyme activity measured while increasing free Ca2+ concentration [23]. The open and close triangles denote the presence of 1.5 mM ADP and ATP, respectively. Figure 4 plots enzyme activity measured at different values of the ADP/ATP ratio [23]. Here the sum of ATP and ADP concentrations was held fixed at 1.5 mM. Triangle, square, and circle data points represent free calcium concentrations of 0.1 mM, 1.0 μM, and 1.0 nM. In analyzing these data it was assumed that free [Mg2+] was 1.0 mM in the assay before experiments began. The free ion concentrations were again used to determine total concentrations and compute the distribution of ATP, ADP, isocitrate, EGTA, EDTA species.
The complete data from Figure 1, 2, 3 and 4 were used to estimate 12 of the 13 adjustable parameter values in our proposed model for NAD-IDH by determining values for which the model best fits the data. Since NADH was not present in the reaction media used for any of these experiments, we are not able to estimate Kiq from these data. The parameter values associated with the best fits to the data are listed in Table 2 and the corresponding model predictions are plotted as solid lines in the figures.
Rutter and Denton calculated kinetic constants using the equation
| (21) |
where Vmax is the maximum enzyme activity, K0.5 is the apparent Michaelis-Menten constant for the substrate, and n is the Hill coefficient [23]. Their estimated Vmax value was 51.0 ± 5.7 munits/mg of mitochondrial protein, and n varied between 1.6 and 3.2 for permeabilized mitochondria. Our model predicted a slightly higher maximum activity (Vmf = 69.2 munits/mg of mitochondrial protein), using a fixed value (n = 3) of the Hill coefficient.
The value n = 3 is in the range reported by Denton [13] and other [38]. To test the validity of our use of n = 3, we allowed n to vary as one of the parameters and re-performed the parameter estimation. In this case the Hill coefficient is estimated to be 2.91 and other re-estimated parameters remain nearly unchanged. Model predictions for fluxes using n = 3 and n = 2.91 are not significantly different.
Our estimate of the KmA value—the apparent Michaelis-Menten constant for NAD+—is within the range that was previously reported. We estimate KmA to be approximately 500 μM, while previous estimates are in the range of 74−900 μM [38, 39]. However, since our estimate is based on the experiments in which the NAD+ concentration was saturating ([NAD+] = 2 mM), the estimate of 500 μM for KmA may not be accurate.
Analysis of data for the bovine enzyme
The data set of Aogaichi et al. [14], which reported on the effects of calcium and lanthanide ions on the activity of NAD-IDH purified from bovine heart, was used to parameterize and validate the model. Specifically, data from Figure 1, Figure 2, and Figure 7 of Aogaichi et al. [14] were used to estimate 10 of the 13 kinetic parameters for NAD-IDH. As for the analysis data for the rat heart enzyme, the data of Aogaichi et al. do not provide a basis to estimate Kiq. This parameter is estimated from another data set, as detailed below. In addition, these data do not provide a basis for estimating KaCaATP or a2 because ATP is not included in the reaction media in any of the experiments. The parameter values associated with the best fits to the Aogaichi et al. data are listed in Table 3.
Figure 7.
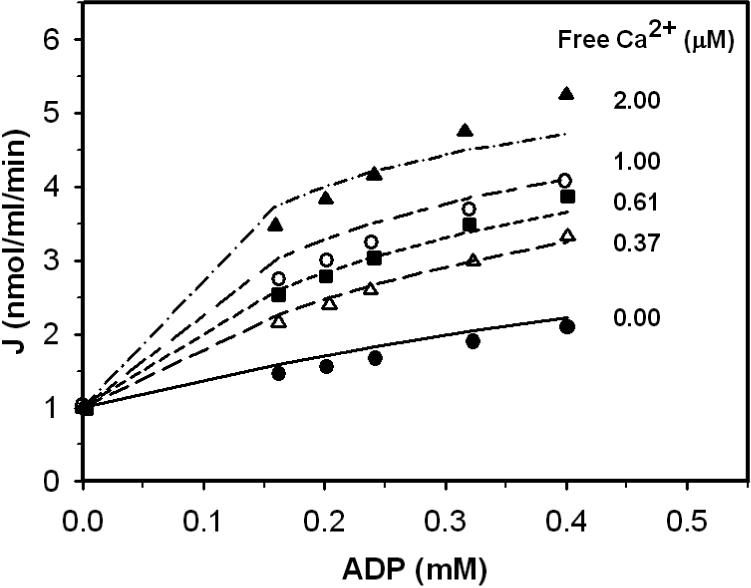
Activation effect of ADP to bovine heart enzyme in the presence of calcium. Measured flux in units of nmol/ml/min was obtained from Figures 7 of [14]. Flux is plotted as a function of activator ADP concentration for free [Ca2+] = 0 μM (•), 0.37 μM (Δ), 0.61 μM (■), 1.00 μM (○) and 2.00 μM (▲). Solid line, long-short line, long dash line and dash-dot line are model fits to •, Δ, ■, ○ , and ▲, respectively.
These experiments were conducted in solution containing 167 mM Na-Hepes at pH = 7.4, with 0.5 mM NAD+. The ionic strength is not available in Aogaichi et al.; and we assumed overall ion strength of 0.15 M. Reaction fluxes were measured at 25°C, and assumed to be the forward reaction flux with no product present in the assays.
Figures 5, 6, and 7 plot data obtained from Figure 1, Figure 2, and Figure 7 of Aogaichi et al. [14]. Figure 5 illustrates the inhibition effect of EGTA in the presence and absence of ADP on enzyme activity. Reaction mixtures contain 1.0 mM MgSO4, 0.2 mM DL-isocitrate and 2.3 μM free Ca2+. Triangles represent data obtained in the presence of 0.5 mM ADP, and circles represent obtained the absence of ADP. Figure 6 shows how EGTA affects ADP activation of the enzyme. Reaction mixtures for this experiment contained 1.0 mM MgSO4, 0.2 mM DL-isocitrate and 3.0 μM free Ca2+. Triangles represent data obtained in the absence of EGTA; circles represent data obtained in the presence of 100 μM EGTA. Figure 7 shows the enzyme behavior as a function of ADP concentration at various calcium levels. Reaction mixtures contained 0.18 mM MgSO4 and 52 mM DL-isocitrate. CaCl2 was added into buffer to adjust free Ca2+ concentration from 0.37 to 2.0 μM. Near z ero free Ca2+ in the buffer was obtained by adding 100 mM EGTA to reaction mixtures.
Figure 5.
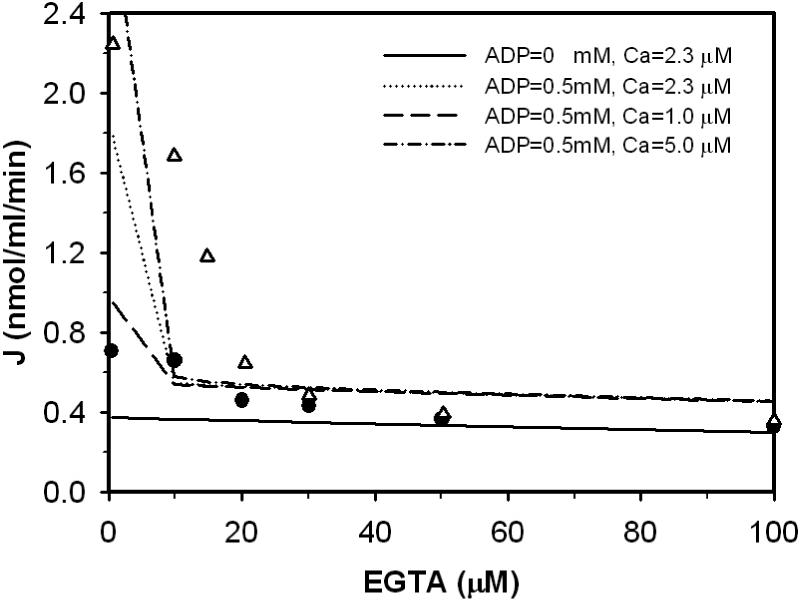
Fits to kinetic data on the forward operation of purified NAD-IDH from bovine heart. Measured flux as a function of concentrations of ADP and EGTA was obtained from Figures 2 of [14]. Data symbols are as follows: Δ data obtained at 0.5 mM ADP; • data obtained in the absence of ADP. Dot-dot line and solid line represent model fits to the data with free [Ca2+] set to 2.3 μM. Dash-dash line and dash-dot line represent model predictions at 1.0 and 5.0 μM [Ca2+].
Figure 6.
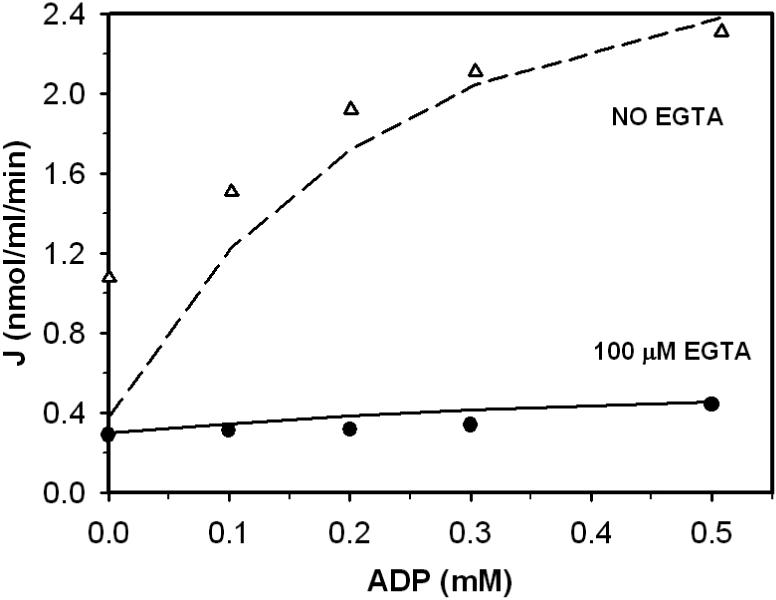
Impact of [EGTA2−] and [ADP] on bovine heart enzyme. Measured flux in units of nmol/ml/min was obtained from Figure 3 of [14]. The buffer contained 1.0 mM MgSO4, 0.2 mM DL-isocitrate and 3.0μM endogenous Ca2+. Symbols • and Δ represents fluxes measured as a function of ADP in the presence and the absence of EGTA in the buffer, respectively. Solid and dashed lines represent model fits to data with and without EGTA.
As for the rat enzyme experiments, species concentrations in the complex media were computed to satisfy equilibria of metal ion-organic ion binding reactions. The reported free Ca2+ concentrations were used to calculate the total calcium used in these calculations. The total concentration were used to compute the concentrations of all ions of interest in the complex reaction media based on the dissociation constants listed in Table 1.
Model fits to data are plotted as lines in Figures 5, 6, and 7. Here, the model is able to effectively explain the effects of EGTA and ADP in Figure 6 and 7. However the model predicts lower activities than observed experimentally at low EGTA for the data shown in Figure 5 and at low level of ADP with no EGTA present in Figure 6. In particular, at the indicated free [Ca2+] of 2.3 μM the model underpredicts the observed flux at low EGTA in Figure 5. A possible explanation is that the model does not accurately capture the effects of EGTA inhibition of the enzyme. Another possible explanation is that Aogaichi et al. underestimated the free Ca2+ in their experiments. They did not specify how they estimated this value in their experiments. In fact, these results are expected to by highly sensitive to Ca2+ concentration. Model predictions (with the estimated parameter set) are indicted for Ca2+ concentrations of 1.0 and 5.0 μM in Figure 5. Indeed, the predicted activity increases sharply when Ca2+ concentration is increased.
Analysis of NADH inhibition of NAD-IDH
Gabriel and Plaut studied the inhibition effect of NADH on NAD-IDH from bovine heart mitochondria [10]. Data from Figure 1A and Figure 4B of their study were used here to validate the product inhibition mechanism proposed in our model.
Figure 8 plots data obtained from experiments conducted to show the effect of ADP on inhibition by NADH [10]. The reaction medium contained 0.167 M NaHepes, 0.25 mM NAD+, 0.2 Mm Mg-DL-isocitrate and 0.45 mM isocitrate. Open and closed circles represent data obtained in the presence and absence of ADP. Enzyme activities were measured at pH 7.4 and different NADH concentrations. Percent inhibition is computed 100(J0-J)/J0, where J0 and J are the initial velocities in the absence and presence of inhibitor.
Figure 8.
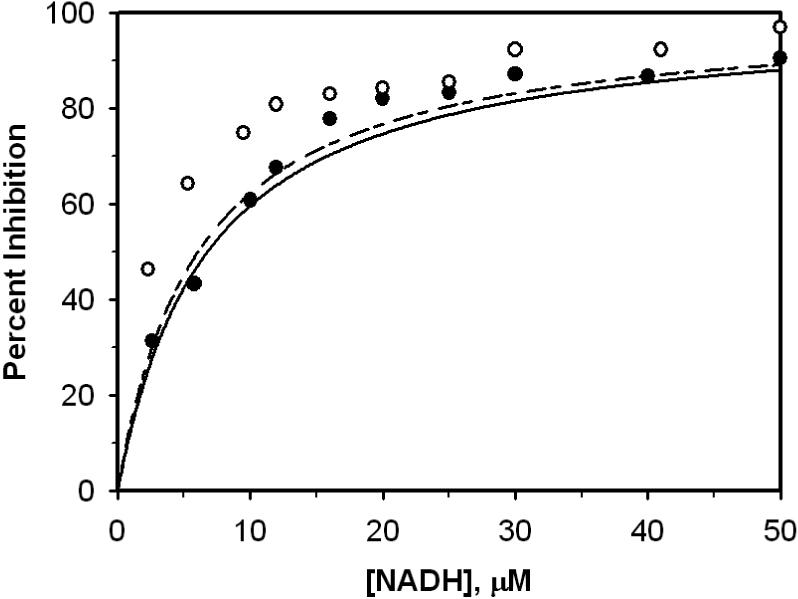
Inhibition effect of NADH on the enzyme activity in the absence and presence ([ADP3−]=0.2 mM] of ADP. Percent inhibition was computed from 100(v0-v)/v0, whre v0 and v were the initial velocities in the absence and presence of inhibitor; Open circle and closed circle represent data obtained in the presence and absence of ADP. Dash line and solid line are model predictions to ○ and •.
Figure 9 plots data obtained from experiments conducted to compare enzyme activity in the presence (closed circle) with the absence (open circle) of NADH [10]. The reaction medium contained 0.167 M NaHepes, 0.2 mM Mg DL-isocitrate and 0.45 mM free DL-isocitrate. Enzyme activity was measured at pH 7.4 and various NAD+ concentrations. Based on matching model predictions to these data, the value of Kiq was estimated while holding the other parameter values from Table 3 constant. The values of the fixed parameters were chosen to be the same as that obtained from the analysis of bovine data [14]. The estimated Kiq value is 4.75μM, agre eing with the range (2.9 − 6.3μM) estimated by Garbriel and Plaut [10].
Figure 9.
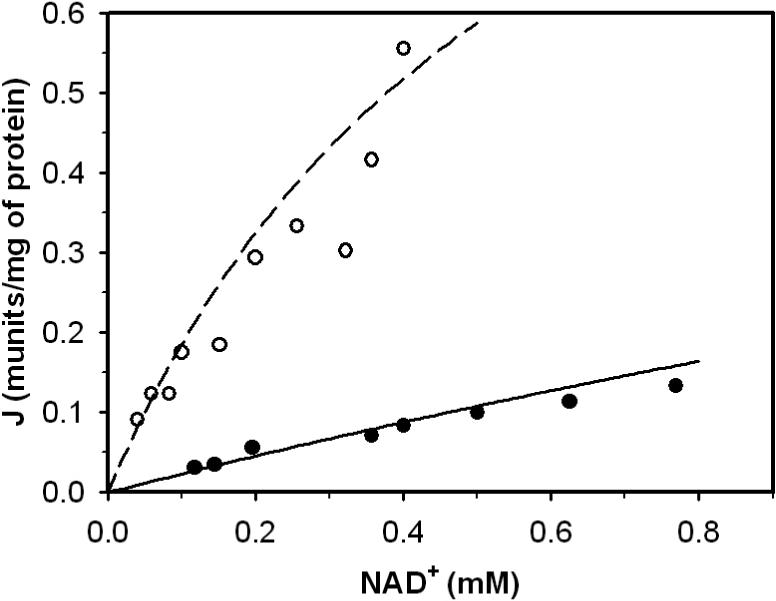
Enzyme activity as a function of NAD+ in the absence of presence of NADH. Closed circle and open circle represent data obtained in the presence ([NADH] = 0.039 mM) and absence of NADH. Dash line and solid line are model predictions to ○ and •.
pH dependency of NAD-IDH
In computing detailed species distributions, our model explicitly accounts for the effect of pH on the apparent thermodynamics of the IDH reaction. In addition to these effects, the catalytic kinetics are expected to be pH-dependent if the chemical properties of the regulatory and catalytic biding sites are pH-dependent [26].
Willson and Tipton [40] report that the sigmoid dependence of the initial reaction rate on the concentration of isocitrate is greatly reduced as the pH is decreased, for NAD-IDH derived from ox brain. Figure 10 plots the data points (from Figure 1 of [40]) that show the effect of pH on the maximum velocity of the enzyme turnover [41]. These data are used here to estimate the value of KiH in Equation (19). For these studies reaction medium contained 1mM, 1 mM NAD+, 10 mM MgSO4, 100 mM imidazole-HCl (pH 6.0 and 6.5), and 100 triethanolamine chloride (pH 7.2−9.0). The fit illustrated in Figure 10 is obtained with KiH = 0.11 μM.
Willson and Tipton also report that the apparent Hill coefficient describing the shape of the curve for flux versus Mg-isocitrate concentration increases from 1 to 2.5 as pH is increased from 6 to 8. Nichols et al. [42] report a similar but less dramatic effect in NAD-IDH purified from rat heart. However it is not clear how the Hill coefficient was estimated in these studies. Willson and Tipton [40] provide data on measured flux verses total D-isocitrate concentration at two extreme pH values: 6.0 and 8.5. Our model (for which the apparent Hill coefficient remains nearly constant over this pH range) effectively matches these data.
Predicted regulation of NAD-IDH in vivo
To explore how NAD-IDH activity is controlled in vivo, relative activity was computed at physiologically reasonable reactant concentrations while varying concentrations of allosteric regulators.
The effects of divalent cation concentration were studied by varying free Mg2+ between 0 and 2.5 mM and free Ca2+ between 0 and 2.5 μM. Figure 11 shows relative activity over these ranges for two different sets of reactant concentrations, corresponding to “rest” and “work” states. Reactant concentrations were obtained using the striated muscle cell energetics model of Wu et al. [24]. Kinetic constants in Table 2 were used for the model predictions.
Figure 11.
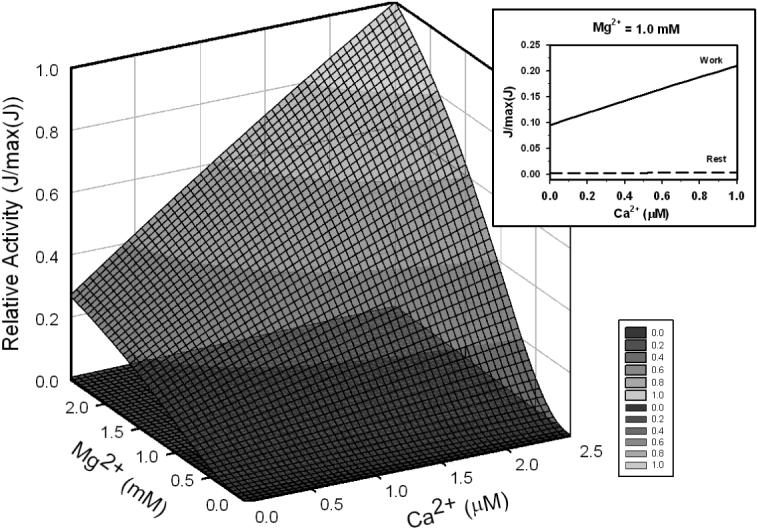
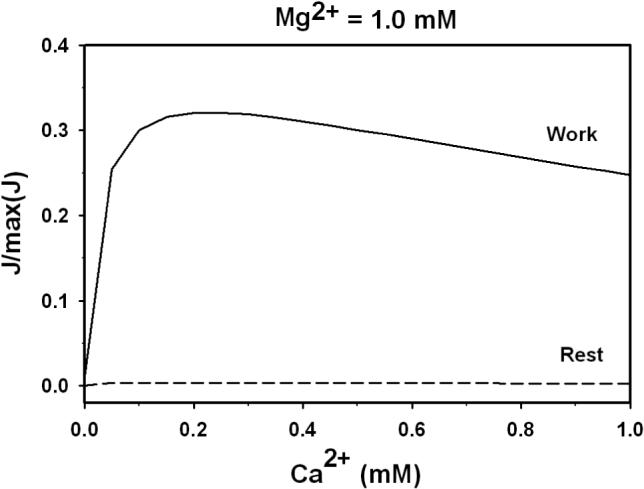
Regulation effect of Mg2+ and Ca2+on the enzyme behavior in rest (ATPase = 0 and work conditions (ATPase = 0.25 mmol sec−1 (liter cytoplasm)−1). Reactant concentrations are computed for steady states using the cell energetics model of Wu et al. [24]; concentrations are listed in Table 4. Insert: activation effect of Ca2+ on the enzyme at free [Mg2+] = 1.0 mM; solid line represents work condition and long-dash line represents rest condition.
For the rest condition, cellular ATP hydrolysis rate was set to 0; for the work condition cellular ATP hydrolysis rate was set to 0.25 mmol sec−1 (liter cytoplasm)−1. The resulting substrate concentrations are listed in Table 4. In both the rest and work states, the activity is sensitive to both Mg2+ and Ca2+. Over a large range of mitochondrial calcium (0 to 1 μM, shown in the insert of Figure 11), the activity varies over an approximately two-fold range. (However, an increase in Ca2+ to extremely high levels results in a decrease of enzyme activity, as discussed below.) Here the free NADH concentration has been estimated to account for reports that mitochondrial NADH is predominantly protein bound, where NAD+ is largely unbound [43-45]. Tischler et al. [44] estimated that the ratio of mitochondria free NAD/NADH in hepatocytes is in the range 8.5−22.5% based on the lactate dehydrogenase redox couple at pH 7.0 and b-hydroxybutyrate dehydrogenase redox couple at pH 7.4. Choosing a reasonable value within this range, we assumed that mitochondrial free NADH is 15% of total NADH.
Table 4.
Reactant concentrations used in Figures 10, 11, and 12
| |
Rest state, ATPase = 0 mmol sec−1 (l cyto)−1 |
High work state, ATPase = 0.25 mmol sec−1 (l cyto)−1 |
|---|---|---|
| ATPx | 6.54 mM | 9.1 mM |
| ADPx | 3.45 mM | 0.83 mM |
| ICITx | 13 | 27 |
| AKGx | 0.342 | 4.5 |
| NADHx | 2.50 mM | 0.52 mM |
| NADx | 0.47 mM | 2.45 mM |
| CO2,tot | 21.4 M | 21.4 M |
Figure 12 compares the roles of that mitochondrial ADP/ATP ratio and Ca2+ in regulating enzyme activity. The relative activity is plotted for the ADP/ATP ratio over the range 0 to 0.5 and free Ca2+ over the range 0 to 2.5 μM. Here the sum of ATP and ADP was held fixed at 10 mM. Other reactant concentrations were set at the values indicated in Table 4 for the 0.25 mmol sec−1 (liter cytoplasm)−1 ATP hydrolysis level. Here it is apparent that the ADP/ATP ratio has a significant impact on enzyme activity and the relationship between ADP/ATP and activity is modulated by calcium.
Figure 12.
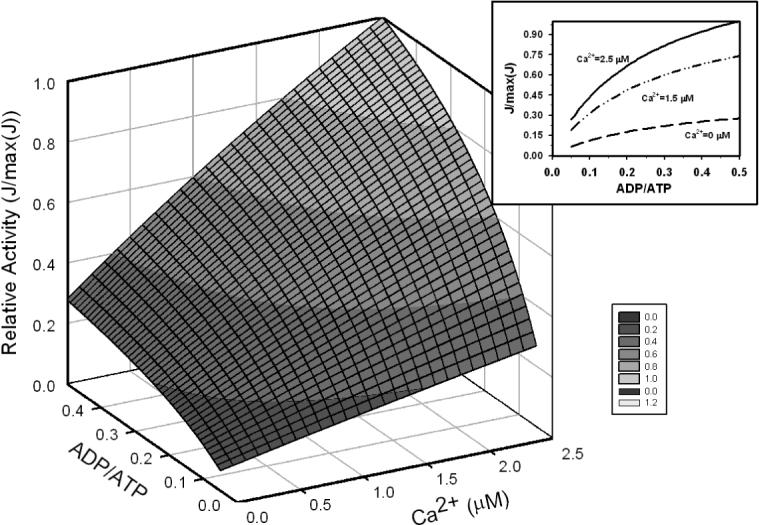
Enzyme activity as a function of ADP/ATP ratio and Ca2+. Insert: activation effect of ADP/ATP ratio on the enzyme at Ca2+=0 μM (long-dash), 1.5 μM (long-dot-dot) and 2.5 μM (solid). Total [ATP] + [ADP] is fixed at 10 mM; other reactant concentrations are set to values corresponding to the work state in Table 4. All calculations were done at free [Mg2+] = 1.0 mM.
Discussion
Here we have developed a mathematical model to describe the catalytic mechanisms of NAD-linked ICD. The model assumes that the enzyme catalyzes the conversion of isocitrate to α-ketoglutarate through a four-state process. The flux expression was derived based on general form of the ordered bi-bi enzyme kinetic mechanism, and modified to include appropriate activators and inhibitors.
Calculation of ion concentrations in complex media
To analyze kinetic data for this enzyme obtained from complex reaction media, it was necessary to compute the equilibrium distribution of all metal ion and proton binding states of reactants and other species in the media. The ligand and free metal ion concentrations in many previous reports were calculated based on the method of Feldman et al. using apparent stability constants [13, 14, 23, 46], which ignores the complex binding in reaction mixtures, including proton-chelate binding. This approximation may result in the inaccurate determination of reactant and ion concentrations. Here we performed detailed analysis of ionic species concentrations in all experimental samples. The analysis explicitly considered the binding among different species, protons, K+, Mg2+, phosphate, etc. In addition, the dissociation constants were appropriately adjusted for overall ionic strength in the reaction media. These calculations accurately determine reactant concentrations in terms of species. Small differences between our calculations and previous estimates may have quantitatively significant impacts our analyses. For example, in the reaction mixtures in Figure 2 of Rutter and Denton [23], the free [Mg2+] was determined to vary from 0.35 mM to 0.42 mM at the highest concentration of DL-isocitrate, while Rutter and Denton estimated that the free concentration did not drop below 0.5 mM.
Computer code we used to calculate the free species concentrations in different experimental buffers is available to the public on our website (bbc.mcw.edu/computation).
Alternative models of NAD-IDH
A number of previous models of NAD-linked isocitrate dehydrogenase kinetics have been reported. The simple phenomenological expression of Equation (2) used by Rutter and Denton [22, 23] describes the cooperative relationship between flux and isocitrate concentration, but does not mechanistically describe the kinetics in terms of additional reactants. Kohn and Garfinkel [11] proposed in their model MgICIT and ADP as activators, and ATP and NADH as inhibitors [39]. Cortassa et al. [38] assumed the enzyme activity is a function of pH, and ADP and Ca2+ concentrations, as well as substrate concentrations. Wu et al. [24] modified the Kohn and Garfinkel model for use in simulating mitochondrial energy metabolism. In all of these models, the kinetic constants are obtained from the values reported in the literature. Here we have developed a mechanistic model in which CaADP, CaATP, and ADP act as non-essential activators in reaction step 2 and MgEGTA acts as non-competitive inhibitor acting on step 1. Kinetic constants are obtained by reanalyzing primary data on the kinetics of the enzyme.
While all of these models phenomenologically capture the allosteric activation mechanisms elucidated here, none are able to quantitatively predict the data. In particular, the models of Kohn and Garfinkel [11] and Wu et al. [24] can match the impacts of the ADP/ATP ratio and Mg2+ on flux, but does not capture Ca2+-mediate effects. The model of Cortassa et al. [38] treats ADP and Ca2+ as independent activators, rather than treating the mechanistic effects of Ca2+-bound ATP and ADP, and does not capture Mg2+-dependent effects.
In addition, we tested the ability of the random bi-bi mechanism to fit the data sets analyzed here (results not shown). While we could not reproduce the observed data using a random-binding model, our analysis does not rule out all possible alternatives or modifications to the current model. The current model should be thought of as a quantitative representation of the catalytic kinetics of this enzyme that is consistent with the available data and therefore useful as a component for integrated modeling of biochemical systems and a template model for investigating other dehydrogenase enzyme with similar regulatory mechanisms.
Inhibition by EGTA
EGTA, which is used in many studies of NAD-linked IDH behavior to control Ca2+ concentration [13, 14, 22, 23], has been shown to introduce inhibition effect on the enzyme's activity [13]. One mechanism for this inhibition is through reducing free Ca2+ concentration, decreasing the CaADP induced activation to the enzyme. However it is also apparent that EGTA inhibits the enzyme in the absence of ADP [13, 14]. Gabriel and Plaut suggested that EGTA may interact directly with the enzyme [15], although they did not suggest a specific inhibition mechanism.
To determine which species of EGTA is responsible for the inhibition, we computed the free EGTA concentration in the studies of Aogaichi et al. [14] illustrated in Figure 5 of this paper, and found it remains almost unchanged in the presence and absence of ADP. This indicates that free EGTA is not a likely candidate to act as an important inhibitor. Denton also studied the enzyme activity from rat heart with the presence and absence of Ca2+ and observed no significant difference in the enzyme activity [13]. Therefore CaEGTA is not expected to act as a significant inhibitor at the concentrations obtained in these studies. Here we are able to explain the observed data using a model where MgEGTA that acts as a linear mixed noncompetitive inhibitor on reaction step 1. This assumption is consistent with the observed data and the inhibition constant is estimated at KiMgEGTA = 84.1 μM for rat enzyme and 7.05 μM for bovine enzyme.
Activation by magnesium, calcium, ADP and ATP
Many studies have reported that Ca2+, ADP and ATP cause the activation of NAD-linked IDH. We hypothesized that CaADP, CaATP and ADP act as non-essential activators in reaction step 2 and showed that the observed data are consistent with this hypothesis. Parameter estimation shows that CaATP, CaADP and ADP activate the enzyme through decreasing the apparent Km for Mg-isocitrate in both isoforms studied here. We also found that CaADP has much stronger activation effect than CaATP, in general agreement with previous reports [15, 23]. Rutter and Denton reported 2−10 fold higher K0.5 values for Ca2+ in the presence of ATP than ADP. We found our KaCaATP value is about 20 times higher than KaCaATP. In addition we found that ADP3− can bind to the same activation site as CaADP− and CaATP2−, but with KaADP values higher than the physiological range of ADP3− concentration.
It has been suggested that calcium acts as a feed-forward signal stimulating mitochondrial dehydrogenases in parallel to activating contraction in muscle [19, 47] In addition it is apparent that Mg2+ concentration can have a significant effect on enzyme activity. Magnesium ions are highly buffered by binding to ATP, ADP, and other anionic biochemical species. As a result [Mg2+] concentrations can vary significantly as the energetic state changes. In addition, the presence of a Mg2+-selective channel in the mitochondrial inner membrane suggests a role of Mg2+ as a regulator of mitochondrial metabolism [48].
Inhibition by high calcium concentration
It has been reported that Ca2+ may act as an inhibitor by increasing the apparent Km of the rat heart enzyme for isocitrate [20, 21]. In Figure 13 enzyme activity is plotted as a function of free [Ca2+] for un-physiologically high [Ca2+]. It is found that the enzyme activity decreases for [Ca2+] > 200 μM, agreeing with the observations of Denton et al. [13]. Our analysis reveals that this inhibition effect is the result of the competitive of Mg2+ and Ca2+ for isocitrate in the presence of high Ca2+ concentration.
Figure 13.
Inhibition effect of high Ca2+ concentration. This figure plots the enzyme activity under the conditions in the insert of Figure 11 at higher Ca2+ concentration. The solid line represents work condition (ATPase = 0.25 mmol sec−1 (liter cytoplasm)−1) and dashed line represents rest condition (ATPase = 0). All calculations were done at free [Mg2+] = 1.0 mM.
Acknowledgements
The authors thank Fan Wu, Ranjan Dash, and Kalyan Vinnakota for advice and illuminating discussion. This work was supported by NIH grant HL072011.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hansford RG. Relation between mitochondrial calcium transport and control of energy metabolism. Rev Physiol Biochem Pharmacol. 1985;102:1–72. doi: 10.1007/BFb0034084. [DOI] [PubMed] [Google Scholar]
- 2.Williamson JR, Cooper RH. Regulation of the citric acid cycle in mammalian systems. FEBS Lett. 1980;117(Suppl):K73–85. doi: 10.1016/0014-5793(80)80572-2. [DOI] [PubMed] [Google Scholar]
- 3.Gabriel JL, Zervos PR, Plaut GW. Activity of purified NAD-specific isocitrate dehydrogenase at modulator and substrate concentrations approximating conditions in mitochondria. Metabolism. 1986;35(7):661–667. doi: 10.1016/0026-0495(86)90175-7. [DOI] [PubMed] [Google Scholar]
- 4.Plaut GW, Sung SC. Diphosphopyridine nucleotide isocitric dehydrogenase from animal tissues. J Biol Chem. 1954;207(1):305–314. [PubMed] [Google Scholar]
- 5.Goebell H, Klingenberg M. [Dpn-Specific Isocitrate Dehydrogenase of Mitochondria. I. Kinetic Properties, Occurrence and Function of Dpn-Specific Isocitrate Dehydrogenase.]. Biochem Z. 1964;340:441–464. [PubMed] [Google Scholar]
- 6.Plaut GW, Aogaichi T. Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver. J Biol Chem. 1968;243(21):5572–5583. [PubMed] [Google Scholar]
- 7.Stein AM, Kirkman SK, Stein JH. Diphosphopyridine nucleotide specific isocitric dehydrogenase of mammalian mitochondria. II. Kinetic properties of the enzyme of the Ehrlich ascites carcinoma. Biochemistry. 1967;6(10):3197–3203. doi: 10.1021/bi00862a030. [DOI] [PubMed] [Google Scholar]
- 8.Colman RF. Mechanisms for the oxidative decarboxylation of isocitrate: implications for control. Adv Enzyme Regul. 1975;13:413–433. doi: 10.1016/0065-2571(75)90028-x. [DOI] [PubMed] [Google Scholar]
- 9.Plaut GW, Schramm VL, Aogaichi T. Action of magnesium ion on diphosphopyridine nucleotide-linked isocitrate dehydrogenase from bovine heart. Characterization of the forms of the substrate and the modifier of the reaction. J Biol Chem. 1974;249(6):1848–1856. [PubMed] [Google Scholar]
- 10.Gabriel JL, Plaut GW. Inhibition of bovine heart NAD-specific isocitrate dehydrogenase by reduced pyridine nucleotides: modulation of inhibition by ADP, NAD+, Ca2+, citrate, and isocitrate. Biochemistry. 1984;23(12):2773–2778. doi: 10.1021/bi00307a037. [DOI] [PubMed] [Google Scholar]
- 11.Kohn MC, Garfinkel D. Computer simulation of metabolism in palmitate-perfused rat heart. II. Behavior of complete model. Ann Biomed Eng. 1983;11(6):511–531. doi: 10.1007/BF02364082. [DOI] [PubMed] [Google Scholar]
- 12.Chen RF, Plaut GW. Activation and Inhibition of Dpn-Linked Isocitrate Dehydrogenase of Heart by Certain Nucleotides. Biochemistry. 1963;2:1023–1032. doi: 10.1021/bi00905a020. [DOI] [PubMed] [Google Scholar]
- 13.Denton RM, Richards DA, Chin JG. Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and other tissues. Biochem J. 1978;176(3):899–906. doi: 10.1042/bj1760899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aogaichi T, Evans J, Gabriel J, Plaut GW. The effects of calcium and lanthanide ions on the activity of bovine heart nicotinamide adenine dinucleotide-specific isocitrate dehydrogenase. Arch Biochem Biophys. 1980;204(1):350–356. doi: 10.1016/0003-9861(80)90043-0. [DOI] [PubMed] [Google Scholar]
- 15.Gabriel JL, Plaut GW. NAD-specific isocitrate dehydrogenase from bovine heart. Interaction with Ca2+ chelators. Biochem J. 1985;229(3):817–822. doi: 10.1042/bj2290817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denton RM, Randle PJ, Martin BR. Stimulation by calcium ions of pyruvate dehydrogenase phosphate phosphatase. Biochem J. 1972;128(1):161–163. doi: 10.1042/bj1280161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denton RM, McCormack JG. Ca2+ transport by mammalian mitochondria and its role in hormone action. Am J Physiol. 1985;249(6 Pt 1):E543–554. doi: 10.1152/ajpendo.1985.249.6.E543. [DOI] [PubMed] [Google Scholar]
- 18.Denton RM, McCormack JG, Midgley PJ, Rutter GA. Hormonal regulation of fluxes through pyruvate dehydrogenase and the citric acid cycle in mammalian tissues. Biochem Soc Symp. 1987;54:127–143. [PubMed] [Google Scholar]
- 19.McCormack JG, Denton RM. Influence of calcium ions on mammalian intramitochondrial dehydrogenases. Methods Enzymol. 1989;174:95–118. doi: 10.1016/0076-6879(89)74013-1. [DOI] [PubMed] [Google Scholar]
- 20.Vaughan H, Newsholme EA. The effects of Ca(2+) and ADP on the activity of NAD-linked isocitrate dehydrogenase of muscle. FEBS Lett. 1969;5(2):124–126. doi: 10.1016/0014-5793(69)80311-x. [DOI] [PubMed] [Google Scholar]
- 21.Zammit VA, Newsholme EA. Effects of calcium ions and adenosine diphosphate on the activities of NAD+-linked isocitrate dehydrogenase from the radular muscles of the whelk and flight muscles of insects. Biochem J. 1976;154(3):677–687. doi: 10.1042/bj1540677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutter GA, Denton RM. Rapid purification of pig heart NAD+-isocitrate dehydrogenase. Studies on the regulation of activity by Ca2+, adenine nucleotides, Mg2+ and other metal ions. Biochem J. 1989;263(2):445–452. doi: 10.1042/bj2630445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutter GA, Denton RM. Regulation of NAD+-linked isocitrate dehydrogenase and 2-oxoglutarate dehydrogenase by Ca2+ ions within toluene-permeabilized rat heart mitochondria. Interactions with regulation by adenine nucleotides and NADH/NAD+ ratios. Biochem J. 1988;252(1):181–189. doi: 10.1042/bj2520181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu F, Yang F, Vinnakota KC, Beard DA. Computer modeling of mitochondrial tricarboxylic acid cycle, oxidative phosphorylation, metabolite transport, and electrophysiology. J Biol Chem. 2007;282(34):24525–24537. doi: 10.1074/jbc.M701024200. [DOI] [PubMed] [Google Scholar]
- 25.Segel IH. Enzyme kinetics : behavior and analysis of rapid equilibrium and steady-state enzyme systems. Wiley; New York: 1975. [Google Scholar]
- 26.Cornish-Bowden A. Fundamentals of enzyme kinetics. Third edn. Portland Press; London: 2004. [Google Scholar]
- 27.Segel IH. Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems. Wiley; New York: 1975. [Google Scholar]
- 28.King EL, Altman C. A schematic method of deriving the rate laws for enzyme-catalyzed reactions. J Phys Chem. 1956;60:1375–1378. [Google Scholar]
- 29.King EL, Altman C. A schematic method of deriving the rate laws for enzyme catalyzed reactions. The Journal of Physical Chemistry. 1956;60:1375–1378. [Google Scholar]
- 30.Alberty RA. Thermodynamics of Biochemical Reactions. Wiley-Interscience; Hoboken, N.J.: 2003. [Google Scholar]
- 31.Beard DA, Qian H. Chemical Biophysics: Quantitative Analysis of Cellular Systems. Cambridge University Press; Cambridge, UK: 2008. Conventions and calculations for biochemical systems. pp. 24–40. [Google Scholar]
- 32.Beard DA, Qian H. Chemical Biophysics: Quantitative Analysis of Cellular Processes. Cambridge University Press; Cambridge, UK: 2008. Enyzme-catalyzed reactions: cycles, transients, and non-equilibrium steady state. pp. 69–104. [Google Scholar]
- 33.Soundar S, Park JH, Huh TL, Colman RF. Evaluation by mutagenesis of the importance of 3 arginines in alpha, beta, and gamma subunits of human NAD-dependent isocitrate dehydrogenase. J Biol Chem. 2003;278(52):52146–52153. doi: 10.1074/jbc.M306178200. [DOI] [PubMed] [Google Scholar]
- 34.Ehrlich RS, Colman RF. Interrelationships among nucleotide binding sites of pig heart NAD-dependent isocitrate dehydrogenase. J Biol Chem. 1982;257(9):4769–4774. [PubMed] [Google Scholar]
- 35.Ehrlich RS, Colman RF. Binding of ligands to half of subunits of NAD-dependent isocitrate dehydrogenase from pig heart. Binding of manganous ion, isocitrate, ADP and NAD. J Biol Chem. 1981;256(3):1276–1282. [PubMed] [Google Scholar]
- 36.Beard DA, Qian H. Relationship between thermodynamic driving force and one-way fluxes in reversible processes. PLoS ONE. 2007;2(1):e144. doi: 10.1371/journal.pone.0000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beard DA, Vinnakota KC, Wu F. Detailed enzyme kinetics in terms of biochemical species: study of citrate synthase. PLoS ONE. 2008;3(3):e1825. doi: 10.1371/journal.pone.0001825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cortassa S, Aon MA, Marban E, Winslow RL, O'Rourke B. An integrated model of cardiac mitochondrial energy metabolism and calcium dynamics. Biophys J. 2003;84(4):2734–2755. doi: 10.1016/S0006-3495(03)75079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohn MC, Achs MJ, Garfinkel D. Computer simulation of metabolism in pyruvate-perfused rat heart. II. Krebs cycle. Am J Physiol. 1979;237(3):R159–166. doi: 10.1152/ajpregu.1979.237.3.R159. [DOI] [PubMed] [Google Scholar]
- 40.Willson VJ, Tipton KF. The effect of pH on the allosteric behaviour of ox-brain NAD+-dependent isocitrate dehydrogenase. Eur J Biochem. 1980;109(2):411–416. doi: 10.1111/j.1432-1033.1980.tb04809.x. [DOI] [PubMed] [Google Scholar]
- 41.Willson VJC, Tipton KF. The Effect of pH on the Allosteric Behaviour of Ox-Brain NAD+-Dependent Isocitrate Dehydrogenase. 1980;109:411–416. doi: 10.1111/j.1432-1033.1980.tb04809.x. [DOI] [PubMed] [Google Scholar]
- 42.Nichols BJ, Rigoulet M, Denton RM. Comparison of the effects of Ca2+, adenine nucleotides and pH on the kinetic properties of mitochondrial NAD(+)-isocitrate dehydrogenase and oxoglutarate dehydrogenase from the yeast Saccharomyces cerevisiae and rat heart. Biochem J. 1994;303(Pt 2):461–465. doi: 10.1042/bj3030461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chance B, Baltscheffsky H. Respiratory enzymes in oxidative phosphorylation. VII. Binding of intramitochondrial reduced pyridine nucleotide. J Biol Chem. 1958;233(3):736–739. [PubMed] [Google Scholar]
- 44.Tischler ME, Hecht P, Williamson JR. Effect of ammonia on mitochondrial and cytosolic NADH and NADPH systems in isolated rat liver cells. FEBS Lett. 1977;76(1):99–104. doi: 10.1016/0014-5793(77)80129-4. [DOI] [PubMed] [Google Scholar]
- 45.Bucher T, Brauser B, Conze A, Klein F, Langguth O, Sies H. State of oxidation-reduction and state of binding in the cytosolic NADH-system as disclosed by equilibration with extracellular lactate-pyruvate in hemoglobin-free perfused rat liver. Eur J Biochem. 1972;27(2):301–317. doi: 10.1111/j.1432-1033.1972.tb01840.x. [DOI] [PubMed] [Google Scholar]
- 46.Feldman H, Rodbard D, Slevine D. Mathematical theory of cross-reactive radioimmunoassay and ligand-binding systems of equilibrium. Anal Biochem. 1972;45(2):530–556. doi: 10.1016/0003-2697(72)90216-3. [DOI] [PubMed] [Google Scholar]
- 47.McCormack JG, Denton RM. The role of Ca2+ ions in the regulation of intramitochondrial metabolism and energy production in rat heart. Mol Cell Biochem. 1989;89(2):121–125. [PubMed] [Google Scholar]
- 48.Schindl R, Weghuber J, Romanin C, Schweyen RJ. Mrs2p forms a high conductance Mg2+ selective channel in mitochondria. Biophys J. 2007;93(11):3872–3883. doi: 10.1529/biophysj.107.112318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Portzehl H, Caldwell PC, Rueegg JC. The Dependence of Contraction and Relaxation of Muscle Fibres from the Crab Maia Squinado on the Internal Concentration of Free Calcium Ions. Biochim Biophys Acta. 1964;79:581–591. doi: 10.1016/0926-6577(64)90224-4. [DOI] [PubMed] [Google Scholar]


