Abstract
Background and purpose:
Several P2X7 receptor antagonists are allosteric inhibitors and exhibit species difference in potency. Furthermore, N2-(3,4-difluorophenyl)-N1-(2-methyl-5-(1-piperazinylmethyl)phenyl)glycinamide dihydrochloride (GW791343) exhibits negative allosteric effects at the human P2X7 receptor but is a positive allosteric modulator of the rat P2X7 receptor. In this study we have identified several regions of the P2X7 receptor that contribute to the species differences in antagonist effects.
Experimental approach:
Chimeric human-rat P2X7 receptors were constructed with regions of the rat receptor being inserted into the human receptor. Antagonist effects at these receptors were measured in ethidium accumulation and radioligand binding studies.
Key results:
Exchanging regions of the P2X7 receptor close to transmembrane domain 1 modified the effects of KN62, 4-(4-fluorophenyl)-2-(4-methylsulphinylphenyl)-5-(4-pyridyl)1H-imidazole (SB203580) and GW791343. Further studies, in which single amino acids were exchanged, identified amino acid 95 as being primarily responsible for the differential allosteric effects of GW791343 and, to varying degrees, the species differences in potency of SB203580 and KN62. The species selectivity of pyridoxalphosphate-6-azophenyl-2′,4′-disulphonic acid was affected by multiple regions of the receptor, with potency being particularly affected by the amino acid 126 but not by amino acid 95. A further region of the rat receptor (amino acids 154–183) was identified that, when inserted into the corresponding position in the human receptor, increased ATP potency 10-fold.
Conclusions:
This study has identified several key residues responsible for the species differences in antagonist effects at the P2X7 receptor and also identified a further region of the P2X7 receptor that can significantly affect agonist potency at the P2X7 receptor.
Keywords: P2X7 receptor; BzATP; KN62, SB203580; GW791343; PPADS
Introduction
The P2X7 receptor is a non-selective cation channel receptor gated by extracellular ATP (North, 2002). The receptor plays a key role in the release of the inflammatory cytokine, interleukin-1β (Ferrari et al., 2006), and in recent years there has been considerable interest in the development of selective P2X7 receptor antagonists due to the potential utility of such compounds in the treatment of inflammatory disorders and pain (Donnelly-Roberts and Jarvis, 2007). This has led to the development of a new generation of P2X7 receptor antagonists to complement the earlier tool compounds used to define this receptor such as oxidized ATP (Murgia et al., 1993), KN62 (Gargett and Wiley, 1997) and brilliant blue G (Jiang et al., 2000). The new generation of compounds includes compounds described in patents or publications from pharmaceutical companies and academic groups (Baraldi et al., 2004; Romagnoli et al., 2005; Donnelly-Roberts and Jarvis 2007) and more recently specific exemplars have been described including AZ11645373 (Stokes et al., 2006), A-740003 (Honore et al., 2006), compound-17 (Michel et al., 2007) and N2-(3,4-difluorophenyl)-N1-(2-methyl-5-(1-piperazinylmethyl)phenyl)glycinamide dihydrochloride (GW791343) (Michel et al., 2008).
The new generation of P2X7 antagonists, similar to earlier antagonists such as KN62, often displays marked selectivity for human as opposed to rodent P2X7 receptors, which clearly hampers preclinical studies to evaluate these compounds (Donnelly-Roberts and Jarvis, 2007). These compounds are also highly selective and specific P2X7 receptor antagonists (Honore et al., 2006; Stokes et al., 2006). This would seem unexpected if the compounds were simple competitive antagonists of ATP given the ubiquity of ATP and nucleotide-binding proteins. However, recent studies have shown that several P2X7 receptor antagonists are allosteric inhibitors, which may explain their specificity (Michel et al., 2007, 2008). One of these compounds, GW791343, exhibits quite unusual properties in that it inhibits ATP responses at the human receptor but increases ATP responses at the rat P2X7 receptor (Michel et al., 2008).
There are also species differences in agonist potency between the species orthologues, and recent studies have started to identify regions of the P2X7 receptor that contribute to the differences in agonist potency. In particular, residues at positions 127 and 284 have been shown to greatly affect agonist potency at rat, human and mouse receptors (Thompson, 2001; Young et al., 2007). However, there is little structural information regarding the residues responsible for the species differences in antagonist potency. A previous study identified residues in the extracellular domain (ECD) of the P2X7 receptor as being responsible for the species selectivity of KN62 (Humphreys et al., 1998), while we evaluated antagonists in chimeric human–rat receptors and found that undefined residues within the first 255 amino acids of the human P2X7 receptor were responsible for the higher potency of KN62 (Thompson, 2001) and 4-(4-fluorophenyl)-2-(4-methylsulphinylphenyl)-5-(4-pyridyl)1H-imidazole (SB203580) (Michel et al., 2006a) at the human than at the rat P2X7 receptor.
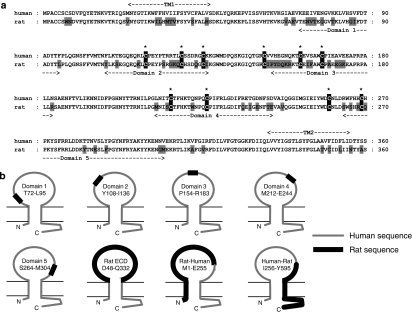
In this study, we have extended our earlier work to identify the regions of the human and rat P2X7 receptor that contribute to the species differences in antagonist effects. For this study, we focused on identifying regions of the receptor that contributed to the selectivity of GW791343, as this compound produced such contrasting effects at human and rat receptors (Michel et al., 2008). We have constructed chimeric human–rat receptors comprising the human P2X7 receptor into which specific residues or short domains of the rat sequence have been introduced. The domains were selected to contain regions of the species orthologues that exhibited the lowest levels of homology (Figure 1a). In addition, one of the domains contained amino acids 79–86 of the P2X7 receptor, which are in the ECD and are not present in other members of the P2X receptor family (North, 2002).
Figure 1.
Comparison of rat and human P2X7 receptors and illustration of receptor constructs used for these studies. (a) Sequence alignment of the first 360 amino acids of the human and rat P2X7 receptors with residues that differ between the receptors shaded in grey. The location of the five domains of the receptor that were exchanged between rat and human receptors is shown. These domains were in the extracellular portion of the receptor between the two putative transmembrane domains (TM1 and TM2). The ten conserved cysteine residues of the P2X7 receptor are shaded black and indicated by an asterix. (b) Representation of the P2X7 receptor to illustrate the approximate location of the various constructs.
From these studies, we have identified a key amino acid at position 95 that seems to be largely responsible for the species difference in effect of GW791343 and that also contributes to the species selectivity in antagonist effect of SB203580 and KN62. In addition, we have identified other regions of the P2X7 receptor that affect agonist and antagonist potency.
Materials and methods
Plasmid construction, chimeric receptor generation and site-directed mutagensis
The nomenclature used for describing the P2X7 receptor conforms to Br J Pharmacol's Guide to Receptors and Channels (Alexander et al., 2008). The generation of the chimeric rat–human P2X7 and human–rat P2X7 receptors have been described previously (Michel et al., 2006a). BacMam reagents for expression of human and rat P2X7 receptors have been described previously (Michel et al., 2007; Fonfria et al., 2008).
A human–rat chimeric receptor containing the entire ECD of the rat receptor (amino acids 48–332) was prepared in the background of an otherwise wild-type human receptor (rat ECD receptor, Figure 1). The DNA sequence of the rat ECD was based on the rat cDNA clone described previously (Fonfria et al., 2008) and was synthesized as a Pml1-to-Xcm1 fragment in pUC19 (Midland Certified Reagent Co. Inc., Midland, TX, USA, 79701). The rat ECD Pml1/Xcm1 fragment was then subcloned into the unique Pml1- and Xcm1-digested pFBM1-humP2X7 plasmid used for generating BacMam reagents (Fonfria et al., 2008).
The five mini-domain swap ECD mutants (Figure 1) were obtained using a modification of the mutagenesis method described previously (Geiser et al., 2001). For each of the domain swap mutants, a small region of the DNA sequence of the human receptor was replaced with that of the rat DNA sequence as indicated in Figure 1. The plasmid constructs were obtained using the following oligonucleotide pairs: (5′3′) Domain1=AAGGTGAAGGGGATAGCAGAGGTGACAGAGAATGTCACGGAGGGC and AAAGAGTTCCCCTGCAAAGGGAGGGTGTAGTCGGCCGTGTC; Domain 2=AGGGGAACTCTTTCTTCGTGATGACAAATTATCTCAAGTCAGAAGGC and GCTCTGGGGGTCCATCCATCCCTTTATACAACCCTGGTCAGAATG; Domain 3=ATTCAGACCGGAAGGTGTATACCTTACGACCAGAAGAGG and CACAGTGAAGTTTTCGGCGCTCCTCAAGAGTGCAGGCCGTGG; Domain 4=ACCACGAGAAACATCCTGCCAGGTATGAACATCTCTTGTACC and CATTATTCCGCCCTGAATTGCCACCTCTGTAAAGTTCTCTCC; and Domain 5=TACTGGTACTGCAACCTAGACAGCTGGTCCCATCGCTGTCAAC, and GACTTTTATCAGAGTCCGTTTCTCCATGCCATTTTCCTTATAGTAC.
P2X7 receptors containing single amino-acid point mutations (rat amino acid introduced into wild-type human receptor) were obtained using a Quick Change II site-directed mutagensis kit following the manufacturer's protocol. The following single point mutants were generated using the plasmid pFBM1-humP2X7 previously described: lysine 72 to threonine (K72T), aspartate 74 to asparagine (E74N), asparagine 78 to glycine (N78G), lysine 81 to threonine (K81 T), serine 86 to glycine (S86G), phenylalanine 95 to leucine (F95L), arginine 126 to glycine (R126G), lysine 128 to glutamate (L128Q), arginine 133 to glutamate (R133Q). In addition, a histidine 155 to tyrosine human receptor mutant (H155Y) was generated, but this was in a human P2X7 receptor containing a naturally occurring single-nucleotide polymorphism (SNP) at amino acid 270. This receptor contained arginine at amino acid 270 instead of histidine (H270R) as was present in the wild-type receptor used for the other studies. Each of the plasmids created above were used to generate BacMam baculoviruses as described previously (Clay et al., 2003).
In addition to these single-point mutations in the human receptor, we also generated a single-point mutation in the rat receptor using the same procedure described above. This was the leucine 95 to phenylalanine (L95F) mutant. This was produced in rat pcDNA3.1, and HEK293 cells were transfected with this plasmid and a stable cell line generated for use in functional studies.
Cell culture and viral transductions
Studies on the human, rat, human–rat, rat–human, rat L95F and domain 3 receptors were performed using HEK293 cells stably transfected with the recombinant receptor. The rat ECD, other chimeric domain swaps and single-point mutant receptors were transiently expressed in human osteosarcoma U-2 OS cells using the BacMam viral transduction method (Fonfria et al., 2008). For the later studies, wild-type human and rat receptors were also expressed in U-2 OS cells for comparative purposes. The BacMam viral transductions for ethidium accumulation studies were conducted as described previously (Fonfria et al., 2008). Briefly, U-2 OS cells were maintained in adherent culture conditions in the presence of Dulbecco's modified Eagle's medium: nutrient mixture F-12 supplemented with Glutamax and 10% foetal bovine serum at 37 °C, 5% CO2. One day before assay, cells were harvested from the culture flasks using 0.05% trypsin/EDTA and suspended at a concentration of ∼750 × 103 cells mL−1 in culture media in the presence of the various BacMam viruses (1–2 × 108 plaque-forming units mL−1 of the BacMam virus stock in culture media). This concentration of BacMam virus produced maximal responses for all of the recombinant receptor (AD Michel, unpublished data). Cells (70–80 000) were plated into individual wells of poly-L-lysine-pretreated 96-well plates and the plates were incubated at 37 °C, 5% CO2 overnight. HEK293 cells stably expressing the human, rat, human–rat, rat–human, rat L95F or domain 3 recombinant P2X7 receptors were prepared in a similar manner except that the BacMam virus was omitted.
In studies to measure KN62 effects in radioligand-binding studies, U-2 OS cells were grown to confluence in T175 cm2 flasks and the media were replaced with fresh growth media containing BacMam virus (1–2 × 108 plaque-forming units mL−1). The cells were incubated overnight, harvested using Versene and membranes were prepared as described previously (Michel et al., 2007). Similar studies to determine the effects of agonists in radioligand-binding studies were performed using membranes prepared from HEK293 cells stably expressing the human, rat or domain 3 chimeric recombinant receptors using the same methods as described above for the U-2 OS cells.
Cellular ethidium accumulation measurements
Studies were performed as described previously (Michel et al., 2006a; Fonfria et al., 2008) using assay buffers comprising (in mM): HEPES 10, N-methyl-D-glucamine 5, KCl 5.6, D-glucose 10, CaCl2 0.5 (pH 7.4) and supplemented with either 280 mM sucrose (sucrose buffer) or 140 mM NaCl (NaCl buffer). Before use, growth medium was completely removed from the cells and they were rinsed with 350 μL of the appropriate assay buffer, which was also removed before assay additions were performed. All solutions were aspirated using 25-gauge bevelled syringe needles to provide complete solution removal. In all studies, the final assay volume was 100 μL and studies were performed at a room temperature (RT) of 19–21 °C.
Cells were incubated with antagonist for 40 min before addition of a mixture containing the agonists, ATP or 2′- and 3′-O-(4benzoylbenzoyl) ATP (BzATP), and ethidium bromide (100 μM final assay concentration). After agonist addition, incubations were continued until approximately 10–30% of maximal agonist-stimulated dye accumulation occurred. Reactions were rapidly terminated by addition of 25 μL of 1.3 M sucrose assay buffer containing 5 mM reactive black 5, and cellular accumulation of ethidium was determined by immediately measuring fluorescence (excitation wavelength of 530 nm and emission wavelength of 620 nm) from below the plate with a 96-well plate fluorescence reader (FlexStation, Molecular Devices, Wokingham, UK).
Radioligand-binding studies
The radioligand-binding studies using [3H]-compound-17 were performed as described previously (Michel et al., 2007). Briefly, studies with KN62 were performed using membranes prepared from U-2 OS cells transduced with human, rat or chimeric recombinant P2X7 receptors. Studies with agonists were performed using membranes prepared from HEK293 cells stably expressing human, rat or domain 3 chimeric recombinant P2X7 receptors. The radioligand, [3H]-compound-17, was used at a concentration of 2–3 nM. Incubations were carried out for 60 min at RT in a final assay volume of 200 μL of 50 mM Tris-HCl buffer containing 0.01% BSA (pH 7.4 at RT) and were terminated by vacuum filtration. Non-specific binding was defined using 10 μM compound-17.
Materials
The Quick Change II site-directed mutagensis kit was obtained from StrataGene (La Jolla, CA, USA); αβ-methylene-ATP, adenosine-5′-O-(3-thiotriphosphate) (ATPγS), 2-methylthio-ATP, ATP, BzATP, ethidium bromide, KN62, pyridoxalphosphate-6-azophenyl-2′,4′-disulphonic acid (PPADS) and reactive black 5 were obtained from Sigma (Poole, UK). SB203580 and GW791343 were synthesized in the Chemistry Department of GSK (Harlow, UK). All culture media, 0.05% trypsin/EDTA and Versene were obtained from Invitrogen (Paisley, Scotland), whereas other reagents were obtained from VWR (Loughborough, UK); the poly-L-lysine-pretreated 96-well plates were obtained from Costar (High Wycombe, UK); foetal bovine serum was obtained from PAA laboratories GmBH (Pasching, Austria). [3H]-compound-17 was from Tritec (Basel, Switzerland; specific activity was 2.1TBq mmol−1 and purity was >99% by HPLC). KN62 (1-(N,O-bis(5-isoquinolinesulphonyl)-N-methyl-1-tyrosyl)-4-phenylpiperazine), SB203580, compound-17 and GW791343 were dissolved in DMSO as a 10 mM solution and were stored as frozen aliquots at −20 °C until required.
Data analysis
Individual concentration–effect or inhibition curves from each experiment were fitted to a four-parameter logistic function to determine the maximum and minimum responses and to calculate the EC50 or IC50 values and the Hill slope. For graphical purposes, most concentration–effect and inhibition curves are presented as a percentage of the maximal response obtained in the control group.
As the compounds produced non-competitive antagonist effects in the Schild studies (for example, see Figure 3), the data from the Schild studies were also analysed to calculate antagonist pIC50 values at each agonist concentration, as this provided some quantitative estimate of antagonist potency. To represent the effect of agonist concentration on antagonist pIC50 graphically, the agonist concentration was expressed relative to its EC50 at the various receptors (logarithm [Agonist Concentration/Agonist EC50]). This enabled a simpler comparison of antagonist pIC50 values between the species orthologues and chimeric receptors. In addition, for the single-point mutants, the relationship between pIC50 and (logarithm [agonist concentration/agonist EC50]) was linear (Figure 3e). To provide a statistical comparison of antagonist potency between these mutants, the pIC50 data for each experiment were analysed by linear regression and the antagonist pIC50 at the agonist EC50 was calculated from the fit. In graphical terms, this corresponded to the pIC50 at 0 on the x axis (Figure 3e), and this value is referred to as the normalized pIC50. This approach was only followed for analyses where the slope of the best fit from the regression analysis did not differ between receptors being compared.
Statistical comparisons were made using Student's t-test or one-way ANOVA followed by Tukey's post hoc test. Differences were assessed as significant when P<0.05. In these studies, the data are the mean±s.e.mean of 3–7 experiments. All curve fitting and statistical analysis was performed using GraphPad Prism 3 (San Diego, CA, USA).
Drug/molecular target nomenclature
Drug/molecular target nomenclature conforms with Br J Pharmacol's Guide to Receptors and Channels (Alexander et al., 2008).
Results
Effect of GW791343 at chimeric receptors
GW791343 inhibits P2X7 receptor-mediated responses in cells expressing human P2X7 receptors, but in cells expressing the rat P2X7 receptor, the predominant effect of GW791343 is to increase P2X7 receptor-mediated responses (Michel et al., 2008). To identify the regions of the P2X7 receptor contributing to the species differences in effects of GW791343, we constructed chimeric receptors in which regions of the rat P2X7 receptor were inserted into the corresponding positions of the human P2X7 receptor.
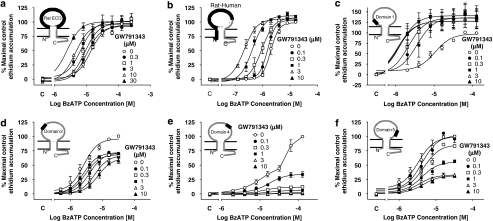
The rat ECD and rat–human chimeric receptors could only be studied in sucrose buffer, as there was no detectable agonist-stimulated ethidium accumulation in NaCl buffer (data not shown). At both of these chimeric receptors, GW791343 increased responses to BzATP (Figures 2a and b). In contrast, GW791343 inhibited responses at the human–rat chimeric receptor in both sucrose and NaCl buffer (data not shown). These data suggest that the positive allosteric effect of GW791343 is determined by residues in the ECD and within the N-terminal 255 amino acids of the receptor.
Figure 2.
The effect of GW791343 on BzATP-induced ethidium accumulation in cells expressing human–rat recombinant chimeric P2X7 receptors. All studies were performed at RT in sucrose buffer in cells pre-equilibrated for 40 min with the indicated concentrations of GW791343 before measuring BzATP responses. Studies were performed in HEK293 cells stably expressing the recombinant receptor (b) or in U-2 OS cells transiently transduced with recombinant P2X7 receptors using BacMam virus (a, c–f). The effect of GW791343 is shown at (a) rat ECD, (b) rat-human, (c) domain 1, (d) domain 2, (e) domain 4 or (f) domain 5 receptors. Basal ethidium accumulation in the absence or presence of GW791343 is indicated on the X-ordinate as C. The data are the mean±s.e.mean of 3–4 separate experiments.
The regions of the rat P2X7 receptor responsible for the ability of GW791343 to increase agonist responses were further localized by studying the effect of GW791343 at chimeric receptors in which smaller domains of the extracellular region of the rat receptor were inserted into the human P2X7 receptor (Figure 1). These studies were complicated by the fact that we could only study the domain 1, 2, 4 and 5 receptors in sucrose buffer, as responses in NaCl buffer were too small for quantitative analysis (data not shown). In contrast, the domain 3 receptor could only be studied in NaCl buffer (see below).
Despite these complications, it was evident that GW791343 inhibited responses at the domain 2, 4 and 5 receptors but increased responses at the domain 1 receptor when studied using BzATP in sucrose buffer (Figures 2c–f). In cells expressing the domain 2, 4 and 5 receptors, the inhibitory effects of GW791343 were all slightly different (Figure 2). At the domain 2 and 4 receptors, the lowest concentration of GW791343 tested (100 nM) significantly reduced the maximal effect of BzATP (Figures 2d and e, P<0.05, one-way ANOVA followed by Tukey's post hoc test). This contrasts with the lack of effect of 100 nM GW791343 at the human P2X7 receptor (Figure 3a) and the domain 5 receptor (Figure 2f). GW791343 inhibited responses at the domain 3 receptor when studied in NaCl buffer, and the effect was similar to that observed at the wild-type human receptor (data not shown).
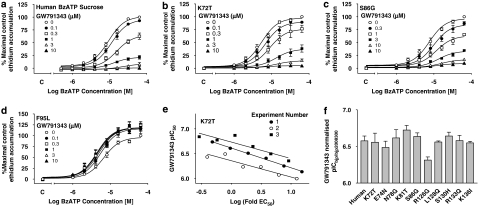
Figure 3.
The effect of GW791343 on BzATP-induced ethidium accumulation in cells expressing human–rat recombinant chimeric P2X7 receptors. All studies were performed at RT in sucrose buffer in U-2 OS cells transiently transduced with recombinant P2X7 receptors using BacMam virus and pre-equilibrated for 40 min with the indicated concentrations of GW791343 before measuring BzATP responses. The effect of GW791343 is shown at (a) human, (b) K72T, (c) S86G and (d) F95L receptors. Basal ethidium accumulation in the absence or presence of GW791343 is indicated on the x ordinate as C. (e) The data from (b) have been analysed to calculate the pIC50 of GW791343 at each concentration of agonist. Agonist concentration is expressed relative to agonist EC50 such that log (fold EC50) represents logarithm [agonist concentration/agonist EC50]. The data were fitted to a straight line by linear regression. From this analysis, the pIC50 at the agonist EC50 (the normalized pIC50) was determined as described in Materials and methods. (f) Normalized pIC50 values for GW791343 at human and mutant receptors. There were no significant differences in normalized pIC50 between any of the single-point mutants (P>0.05, one-way ANOVA followed by Tukey's post hoc test). The data are the mean±s.e.mean of three separate experiments.
Amino acid 95 is a key residue in determining the species difference in effects of GW791343
To further explore the role of residues within domain 1 in modulating the effects of GW791343, a series of P2X7 receptors were generated in which single residues of the human P2X7 receptor were exchanged with the corresponding residue from the rat P2X7 receptor. GW791343 was a potent antagonist at the K72T, E74N, N78G, K81T and S86G single-point mutant receptors (Figures 3b, c and f). Interestingly, in contrast to the studies using the larger domain swaps (Figure 2), the antagonist effect of GW791343, when observed, was similar to the wild-type human receptor for all of the single-point mutant receptors (Figures 3a–c). The relationship between pIC50 and (logarithm [agonist concentration/agonist EC50]) was linear for all of the single-point mutant receptors (Figure 3e, data for K72T and data not shown) and the slopes of the lines of best fit were not significantly different between the various receptors. To compare potency of GW791343 between the various mutant receptors, the data from each experiment were fitted using linear regression, and the pIC50 value at the agonist EC50 (normalized pIC50) was determined from the fit as described in Materials and methods. Graphically, the normalized pIC50 corresponds to the pIC50 value at the intercept of the line of best fit with 0 on the x axis (Figure 3e). For the K72T, E74N, N78G, K81T and S86G mutant receptors, the normalized pIC50 values were similar to that at the wild-type human receptor (Figure 3f, P>0.05, one-way ANOVA followed by Tukey's post hoc test).
In contrast, GW791343 increased responses to BzATP at the F95L mutant receptor (Figure 3d), although the magnitude of this effect was not as marked as at the rat ECD, rat–human or domain 1 receptor (Figures 2a–c).
The effects of GW791343 were also evaluated at five single-point mutant receptors in which residues from domain 2 of the human receptor were exchanged with the corresponding residues from the rat receptor (R126G, L128Q, S130H, R133Q, K136I). At all of these mutant receptors, GW791343 produced similar antagonist effects to those observed at the wild-type human receptor (data not shown). The normalized pIC50 values of GW791343 for the five single-point mutant receptors in domain 2 were similar to that observed at the wild-type receptor (Figure 3f, P>0.05, one-way ANOVA followed by Tukey's post-hoc test). The mean pIC50 at the R126G mutant receptor was slightly lower than at the human receptor, but this difference was not statistically significant (Figure 3f).
Amino acid 95 contributes to the species differences in the effects of SB203580
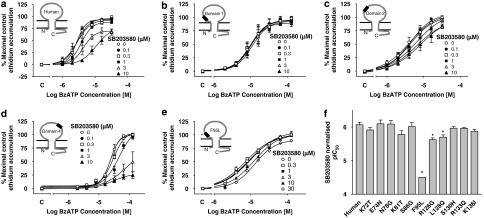
We have previously shown that SB203580 is a weak antagonist of the human P2X7 receptor (pIC50 5.5–6.5) but has no activity at the rat P2X7 receptor at concentrations up to 10 μM (Michel et al., 2006a). Furthermore, at a chimeric human–rat P2X7 receptor containing the first 255 amino acids of the human receptor, SB203580 was an antagonist, but it had no effect at a chimeric receptor containing the first 255 amino acids of the rat receptor (Michel et al., 2006a). Consistent with this, SB203580 blocked BzATP responses at the domain 5 receptor in which amino acids 264–304 were exchanged from the rat into the human receptor (data not shown) and the effect was similar to that observed at the wild-type human receptor (Figure 4a). SB203580 also blocked BzATP responses at the domain 2 and 4 receptors (Figures 4c and d), although its effects at the domain 2 receptor (Figure 4c) were less than that observed at the wild-type human receptor (Figure 4a). SB203580 did not produce any detectable antagonist effect at the domain 1 receptor at concentrations up to 10 μM (Figure 4b).
Figure 4.
The effect of SB203580 on BzATP-induced ethidium accumulation in cells expressing human–rat recombinant chimeric P2X7 receptors. All studies were performed at RT in sucrose buffer in U-2 OS cells transiently transduced with recombinant P2X7 receptors using BacMam virus and pre-equilibrated for 40 min with the indicated concentrations of SB203580 before measuring BzATP responses. The effects of SB203580 are shown at (a) human wild type, (b) domain 1, (c) domain 2, (d) domain 4 or (e) F95L receptors. Basal ethidium accumulation in the absence or presence of SB203580 is indicated on the x ordinate as C. (f) Normalized pIC50 values for SB203580 at the indicated single-point mutant receptors were determined as described in Materials and methods. *Significantly different from normalized pIC50 determined at human receptor, P<0.05, one-way ANOVA followed by Tukey's post hoc test. The data are the mean±s.e.mean of 3–7 separate experiments.
When the single-point mutant receptors from within domain 1 and 2 were examined, the effect of SB203580 was greatly reduced at the F95L mutant receptor, although there was some antagonist effect at both 10 and 30 μM (Figure 4e). In contrast, at the other single-point mutant receptors from within domain 1 and 2, the antagonist effects of SB203580 were similar to those observed at the wild-type human receptor (data not shown). Furthermore, the normalized pIC50 values were similar to that determined at the wild-type human receptor (P>0.05, one-way ANOVA followed by Tukey's post hoc test) with the exceptions of the R126G and L128Q mutant receptors (Figure 4f). However, the reduction in pIC50 at the latter receptors was minimal (pIC50 values at human, R126G and L128Q receptors were 6.07±0.08, 5.62±0.06 and 5.70±0.07, respectively).
The antagonist effects of SB203580 were attenuated at the domain 3 receptor such that the compound only produced antagonist effects at 10 and 30 μM (data not shown), and the normalized pIC50 of 5.06±0.12 was significantly lower than the normalized pIC50 value of 6.26±0.04 at the human wild-type receptor (P<0.05, one-way ANOVA followed by Tukey's post hoc test). We only constructed one single-point mutant receptor from within domain 3. This was the H155Y mutation, which also exists as a natural SNP in the human receptor (Cabrini et al., 2005). This mutant was constructed in a human receptor also containing an H270R single SNP, which complicates comparisons. However, SB203580 effects at the H155Y–H270R receptor were similar to those observed at the wild-type human receptor (data not shown), and the normalized pIC50 value of 5.99±0.04 was not significantly different to the normalized pIC50 value of 6.26±0.04 at the human wild-type receptor (P>0.05, one-way ANOVA followed by Tukey's post hoc test).
Effect of KN62 at chimeric receptors
KN62 is an effective antagonist of the human P2X7 receptor (Figure 5a) but had no effect at the rat P2X7 receptor at concentrations up to 10 μM (data not shown). In studies on the rat–human P2X7 receptor and the rat ECD P2X7 receptor, KN62 had no effect, but at the human-rat P2X7 receptor, KN62 was an antagonist (data not shown). This suggested that the residues responsible for the antagonist effects of KN62 were in the ECD and within the first 255 amino acids of the receptor.
Figure 5.
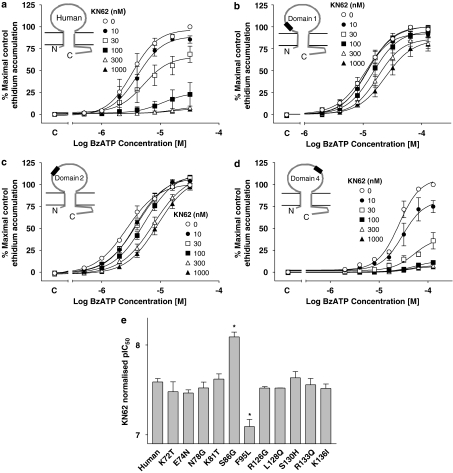
The effect of KN62 on BzATP-induced ethidium accumulation in cells expressing human–rat recombinant chimeric P2X7 receptors. Studies were performed at RT in sucrose buffer in U-2 OS cells transiently transduced with recombinant P2X7 receptors using BacMam virus and then pre-equilibrated for 40 min with the indicated concentrations of KN62 before measuring BzATP responses. The effect of KN62 is shown at (a) wild-type human, (b) domain 1, (c) domain 2 or (d) domain 4 receptors. Basal ethidium accumulation in the absence or presence of KN62 is indicated on the x ordinate as C. (e) Normalized pIC50 values for KN62 at the indicated single-point mutant receptors were determined as described in Materials and methods. *Significantly different from normalized pIC50 determined at the human receptor, P<0.05, one-way ANOVA followed by Tukey's post hoc test. The data are the mean±s.e.mean of 3–6 separate experiments.
KN62 was an antagonist at the domain 1, 2, 4 and 5 receptors (Figures 5b–d and data not shown), although its effects were reduced at the domain 1 and 2 receptors. In these receptors, KN62 produced more limited shifts in the BzATP concentration–effect curve and did not suppress maximal responses to the same extent as observed in studies on the wild-type human P2X7 receptor (Figures 5a–c). The effects of KN62 were similarly modified at the domain 3 receptor when studied in a NaCl buffer (data not shown), although the normalized pIC50 value at the domain 3 receptor (8.32±0.16) was not significantly different (P>0.05, Student's t-test) from that determined at the wild-type human receptor (7.92±0.02).
In studies on the single-point mutant receptors from within domains 1 and 2, KN62 antagonist effects were almost identical to those at the human receptor for the K72T, E74N, N78G, K81T, R126G, L128Q, S130H, R133Q and K136I mutants and there was no significant difference in normalized pIC50 when compared to the human receptor for any of these mutant receptors (Figure 5e, P>0.05, one-way ANOVA followed by Tukey's post hoc test). However, at the F95L receptor, the effect of GW791343 on BzATP responses (data not shown) was very similar to that observed at the domain 1 receptor (Figure 5b), and this was associated with a significant decrease in normalized pIC50 compared with the human receptor (Figure 5e). In contrast, the normalized pIC50 of KN62 at the S86G mutant was higher than at the human P2X7 receptor (Figure 5e).
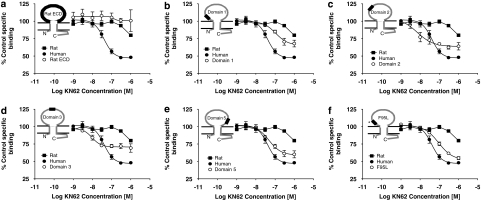
We did not evaluate the effects of GW791343 or SB203580 in binding studies as the potency of GW791343 is similar at rat and human P2X7 receptors, while SB203580 only produces a small inhibition of binding (Michel et al., 2007, 2008). However, KN62 discriminated between the human and rat P2X7 receptor in binding studies, as it had no effect at the rat P2X7 receptor but inhibited binding at the human P2X7 receptor (Figure 6). Consequently, we determined whether radioligand-binding studies could provide more quantitative information on the effect(s) of KN62 at the various chimeric receptors. However, the data were not easy to interpret. Thus, although KN62 had similar effects at the rat and rat ECD P2X7 receptors (Figure 6a), and its effects were reduced at the domain 1 and F95L receptors (Figures 6b and f), the effects of KN62 were also modified at the domain 2, domain 3 and domain 5 receptors (Figures 6c–e). Indeed, the only domain at which KN62 effects were not modified was the domain 4 receptor where the KN62 inhibition curve was identical to that at the human receptor (data not shown). Interestingly, KN62 potency was increased, but maximal inhibition decreased, at the domain 2 and 3 receptors (Figures 6c and d), although the maximal inhibition was slightly reduced at the domain 5 receptor (Figure 6e).
Figure 6.
The potency of KN62 to inhibit [3H]-compound-17 binding to membranes prepared from cells expressing human–rat recombinant chimeric P2X7 receptors. Membranes were prepared from U-2 OS cells transiently transduced with recombinant P2X7 receptors using BacMam virus. The radioligand concentration was 2 nM and specific binding was defined with 10 μM compound-17. The effect of KN62 at wild-type rat and human receptors is compared with effects at (a) rat-ECD, (b) domain 1, (c) domain 2, (d) domain 3, (e) domain 5 or (f) F95L receptors. The data are the mean±s.e.mean of four separate experiments.
Effect of PPADS at chimeric receptors
Pyridoxalphosphate-6-azophenyl-2′,4′-disulphonic acid is a slowly reversible antagonist of the P2X7 receptor (Chessell et al., 1998). However, it appears to interact at the ATP-binding site and there appears to be little interaction with the other antagonists such as SB203580 (AD Michel, unpublished data) or GW791343 (Michel et al., 2008). PPADS is also selective for human over rat receptors (Hibell et al., 2001) and so was an interesting compound to evaluate at the various chimeric P2X7 receptors.
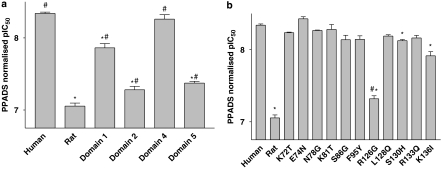
The potency of PPADS at the domain 4 receptor was similar to that measured at the human P2X7 receptor (Figure 7a, P>0.05, one-way ANOVA followed by Tukey's post hoc test). In contrast, the potency of PPADS was reduced at the domain 1, 2 and 5 receptors (Figure 5a, P<0.05, one-way ANOVA followed by Tukey's post hoc test), although the reduction in potency at the domain 1 receptor was modest. The decrease in potency of PPADS at the domain 2 and 5 receptors was more marked, although the normalized pIC50 values were significantly greater than at the rat receptor (P<0.05, one-way ANOVA followed by Tukey's post hoc test).
Figure 7.
The potency of PPADS to block BzATP-induced ethidium accumulation in cells expressing human–rat recombinant chimeric P2X7 receptors. (a) Data from studies using domain 1, 2, 4 and 5 receptors are shown, whereas panel b shows data obtained using single-point mutations from within domain 1 and 2. Studies were performed at RT in sucrose buffer in U-2 OS cells transiently transduced with recombinant P2X7 receptors using BacMam virus and then pre-equilibrated for 40 min with PPADS before measuring BzATP responses. The normalized pIC50 was calculated as described in Materials and methods for each receptor. *Significantly different from normalized pIC50 determined at human receptor, P<0.05, one-way ANOVA followed by Tukey's post hoc test. #Significantly different from normalized pIC50 determined at rat receptor, P<0.05, one-way ANOVA followed by Tukey's post hoc test.
Although PPADS potency was slightly reduced at the domain 1 receptor, there was no significant effect of any of the single-point mutations within this domain on PPADS potency (Figure 7b). In the case of the domain 2 single-point mutant receptors, the potency of PPADS was most markedly reduced at the R126G mutant, although the normalized pIC50 was significantly greater than at the rat receptor (P<0.05, one-way ANOVA followed by Tukey's post hoc test). PPADS potency at the S130H and K136I receptors was also significantly lower than at the human receptor, although the differences in potency were small (normalized pIC50 values at human, S130H and K136I receptors were 8.34±0.18, 8.13±0.02 and 7.92±0.06, respectively). We did not construct any single-point mutant receptors from within domain 5.
Studies on the domain 3 receptor were complicated by the need to perform the studies in NaCl buffer, by the use of different expression systems and by the use of a different form of the human receptor for some of the studies. Nevertheless, the potency of PPADS was slightly, but significantly, higher (P<0.05, Student's t-test) at the domain 3 receptor (normalized pIC50=7.75±0.02) than at the wild-type human receptor (normalized pIC50=7.42±0.03) when studied in HEK293 cells following stable expression. When studied in U-2 OS cells after transient expression, the normalized pIC50 values of PPADS at the wild-type, H270R and H155Y-H270R receptors were 7.18±0.03, 7.21±0.02 and 7.17±0.03, respectively, and were not significantly different from each other (P>0.05, one-way ANOVA followed by Tukey's post hoc test), suggesting that the residue at position 155 does not contribute to the slight differences in potency between the domain 3 and the wild-type human receptor. Note that in these studies, using NaCl buffer, PPADS potency is lower than in sucrose buffer. However, the human versus rat species difference in potency was maintained (data not shown).
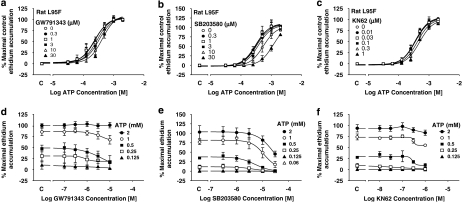
Effect of antagonists at the rat L95F receptor
The rat L95F receptor exhibited a high degree of tonic activity in sucrose buffer and so studies were performed in NaCl buffer. GW791343 increased responses at the wild-type rat receptor (Michel et al., 2008) but was a weak antagonist of the rat L95F receptor (Figures 8a and d). SB203580 has no effect at the rat receptor (Michel et al., 2006a) but was an antagonist of the rat L95F receptor (Figures 8b and e). For SB203580, the antagonist effects at the L95F receptor were not as marked as observed at the human receptor (cf. Figures 4a and 8b). This was also true for GW791343 (cf. Figures 3a and 8a). KN62 has no effect at the rat receptor (Hibell et al., 2001 and data not shown) and had little effect on the L95F receptor, although it did produce a modest inhibition of responses to the lower concentrations of ATP (Figures 8c and f).
Figure 8.
Antagonist effects on ATP-induced ethidium accumulation in cells expressing the rat L95F receptor. All studies were performed at RT in NaCl buffer in HEK293 cells stably expressing the recombinant receptor and pre-equilibrated for 40 min with the indicated concentrations of antagonist before measuring ATP responses. The effect on ATP responses is shown for (a) GW791343, (b) SB203580 or (c) KN62. (d) Transposition of the data shown in (a) to show effect of GW791343 on ATP responses. (e) Transposition of the data shown in (b) to show effect of SB203580 on ATP responses. (f) Transposition of the data shown in (c) to show effect of KN62 on ATP responses. Basal ethidium accumulation in the absence or presence of GW791343 is indicated on the x ordinate as C in (a–c). The data are the mean±s.e.mean of 3–4 separate experiments.
Increase in agonist potency at the domain 3 P2X7 receptor
There were differences in agonist potency or effect at several of the mutant receptors constructed. This was most pronounced at the domain 3 receptor where agonist potency was higher than at the other receptors studied. Indeed, this receptor could not be examined in sucrose buffer as it appeared to be constitutively activated, presumably by endogenously released ATP. Thus, there was a high level of basal ethidium uptake that could be blocked by apyrase (1 unit mL−1) or P2X7 receptor antagonists such as decavanadate (10 μM) or compound-17 (1 μM) (data not shown).
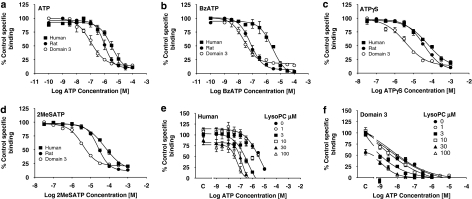
In NaCl buffer, BzATP also possessed high potency at the domain 3 receptor with a pIC50 of 5.94±0.09. The pEC50 value for BzATP at the domain 3 receptor was significantly higher than at the rat or human receptors (P<0.05, one-way ANOVA followed by Tukey's post hoc test) where pEC50 values were 5.30±0.08 and 4.43±0.05, respectively (Fonfria et al., 2008). The maximal response produced by BzATP at the domain 3 receptor was similar to that produced by ATP (97.3±1.07% of the ATP maximum; P>0.05, one-way ANOVA followed by Tukey's post hoc test). This contrasts with effects at the wild-type human receptor where BzATP produces a smaller maximal effect than ATP (Fonfria et al., 2008). In this study, the ATP pEC50 at the domain 3 receptor was 4.16±0.08, and this was significantly higher than at the rat or human receptors (P<0.05, one-way ANOVA followed by Tukey's post hoc test) where pEC50 values were 3.66±0.02 and 3.06±0.04, respectively (Fonfria et al., 2008).
The differences in agonist potency were also observed in binding studies where ADP and αβ-methylene-ATP (data not shown) as well as ATP, BzATP, adenosine-5′-O-(3-thiotriphosphate) and 2-methylthio-ATP were 3- to 10-fold more potent at the domain 3 receptor than at the rat P2X7 receptor and between 16- and 100-fold more potent than at the human P2X7 receptor (Figures 9a–d). Lysolipids can increase agonist potency at the P2X7 receptor by at least 100-fold (Michel and Fonfria, 2007), raising the possibility that the domain 3 residue was mimicking the effects of lipids in some way. However, palmitoyl L-α-lysophosphatidylcholine (lysoPC) increased ATP potency at the domain 3 receptor and the effect was comparable with its effects at the human receptor (Figures 9e and f). These studies were complicated by the reduction in binding produced by lysoPC. This probably reflects its ability to augment the inhibitory effects on radioligand binding of endogenous ATP present in the membranes, as the inhibitory effect of lysoPC was not observed in the presence of 1 unit mL−1 of the ATP degrading enzyme, apyrase (data not shown).
Figure 9.
Inhibition by agonists of [3H]-compound-17 binding to membranes prepared from cells expressing wild-type human, rat and domain 3 chimeric recombinant P2X7 receptors. Membranes were prepared from HEK293 cells stably expressing the recombinant P2X7 receptors. The radioligand concentration was 2 nM and specific binding was defined with 10 μM compound-17. The effect of agonists at the domain 3 receptor is compared with effects at the rat and human receptors for (a) ATP, (b) BzATP, (c) adenosine-5′-O-(3-thiotriphosphate) or (d) 2-methylthio-ATP. The effect of lysoPC on ATP inhibition of binding is shown in (e) the human wild-type receptor and (f) the domain 3 receptor. The effect of lysoPC on radioligand binding in the absence of ATP is indicated on the x ordinate as C. The data are the mean±s.e.mean of three separate experiments.
Discussion
The main finding of this study is that the nature of the amino acid in position 95 of the P2X7 receptor has a marked effect on the potency, or effects, of several P2X7 receptor antagonists and so may be a key determinant of the species differences in effects of these antagonists.
GW791343 is an allosteric modulator of the P2X7 receptor, being a negative allosteric modulator at human and a positive allosteric modulator at rat P2X7 receptors (Michel et al., 2008). Studies on the human chimeric and mutant receptors sequentially localized the amino acids responsible for this species difference in effect to the ECD, the first 255 amino acids, amino acids 72–95 and finally to amino acid 95 of the receptor. This residue was phenylalanine in the human P2X7 receptor, at which GW791343 inhibited responses, but was leucine in the rat P2X7 receptor where GW791343 increased responses to ATP. This amino acid was also important in the species selectivity of SB203580 and partially contributed to the species difference in the potency of KN62. We also generated complimentary data using the rat receptor where the corresponding L95F mutation appreciably modified the effects of both GW791343 and SB203580 and also produced a subtle change in the effects of KN62. Although there was compelling evidence that the residue at position 95 could greatly affect the species selectivity of the antagonists, it should be noted that exchanging this residue did not result in a complete species interconversion of antagonist effects even for GW791343. Thus, the increase in ATP responses produced by GW791343 at the human F95L receptor was not as marked as observed at the rat receptor, whereas the antagonist effect of GW791343 at the rat L95F receptor was less than observed at the human receptor.
It is not clear from this study exactly how the residue at amino acid 95 influenced antagonist effects or potency. Phenylalanine has been implicated in the binding of ATP to some P2X receptors but, on the basis of previous site-directed mutagenesis studies, it seems unlikely that amino acid 95 is part of the ATP-binding site (Vial et al., 2004). Furthermore, PPADS interacts at the ATP-binding site (Michel et al., 2006b), and its potency was not influenced by the amino acid at position 95. Similarly, the potency of ATP and BzATP differ between rat and human P2X7 receptors, although only 3- to 10-fold, yet their potency estimates at the F95L human P2X7 chimeric receptor were not significantly different from those at the human receptor (AD Michel, unpublished data).
It seems unlikely that the residue at amino acid 95 exerts any direct effect on the binding of GW791343, as the affinity of GW791343, assessed using [3H]-compound-17, did not differ greatly between human and rat receptors (Michel et al., 2008). Furthermore, GW791343 is an allosteric modulator of the P2X7 receptor, and the main difference in GW791343 effect when exchanging phenylalanine with leucine was to change the compound from a negative to a positive allosteric modulator. Consequently, it seems more likely that amino acid 95 can modulate putative conformational changes that occur as a consequence of GW791343 binding. In the case of the human receptor, the residue is phenylalanine and this may result in conformational changes following GW791343 binding that lead to inhibition of channel opening, either by direct steric hindrance from the large phenylalanine residue or by an indirect effect on receptor flexibility. In the case of the rat receptor, where the residue is a smaller and potentially more flexible leucine, this may lead to the conformational changes induced by GW791343 facilitating, rather than inhibiting, channel opening. In the case of KN62 and SB203580, where decreases in potency were observed in the chimeric F95L human P2X7 receptor, it may be that the large aromatic group of phenylalanine coordinates ligand binding better than leucine, although it seems more plausible that the efficacy, or extent of effect, of these allosteric antagonists is affected by the residue at position 95.
Interestingly, amino acid 95 is close to an amino-acid sequence (79–86) that is unique to the P2X7 receptor (North, 2002). Although we did not observe any marked change in GW791343 or SB203580 effects at the two mutants from within this region, K81T or S86G, binding of compounds at such a region may be a plausible explanation for the considerable selectivity of most P2X7 receptor antagonists over other P2X receptor types (see Introduction).
It seems likely that other residues in the P2X7 receptor apart from those in domain 1 can affect the functional effects of GW791343 and SB203580. Thus, their inhibitory potency was also changed in several of the other domains studied. This may not be unexpected when exchanging large tracts of receptor between orthologues, as this may lead to quite large changes in tertiary structure of the receptor, and further studies would be required to determine if these effects can be localized to a single residue or result from a general change in receptor conformation. SB203580 effects were also reduced in the domain 2 and 3 receptors, but we could not identify a single residue responsible for this.
The situation with KN62 was more complicated. Its potency was reduced in domain 1 and also at the F95L chimera but its potency was also slightly increased in the S86G mutant. The significance of this increase in potency at the S86G receptor is not known, but it provides further evidence that this region of the receptor is important for the effects of allosteric antagonists. The effects of KN62 were also modified at the domain 2 receptor, although we could not identify a specific residue responsible for this change. It should also be noted that we have evaluated several other structurally diverse P2X7 receptor antagonists and have found several compounds that differentiate between human and rat P2X7 receptors, but their potency is not affected in either the F95L or R126G (see below) mutants, suggesting that there may be multiple regions of the receptor responsible for the species selectivity of antagonists (AD Michel, unpublished data).
The results with PPADS provided further information on the sites of the receptor responsible for antagonist effects. PPADS was an interesting compound to study, as mechanistic studies suggested that it interacts with the P2X7 receptor in a distinct manner to KN62, GW791343 and SB203580 (Michel et al., 2006a, 2008), yet it also displays higher potency at human than at rat receptors. PPADS potency was modified in several of the chimeric receptors. There was a small reduction in potency at the domain 1 receptor, but this was not detected in any of the single-point mutant receptors within domain 1, including F95L. In the domain 2 receptor, PPADS potency was markedly reduced and this may be due to the amino acid at position 126. Interestingly, this residue is adjacent to a key amino acid at position 127, which is thought to contribute to the mouse and rat species differences in the potency of BzATP (Young et al., 2007). In the domain 5 receptor, PPADS potency was also reduced, although we did not construct single-point mutations within this region. Interestingly, residue 284 is localized within domain 5 and this is thought to be a further key residue responsible for the species differences in ATP and BzATP effects at mouse and rat receptors (Young et al., 2007). However, this residue is asparagine at both human and rat receptors, suggesting it is unlikely to be the amino acid responsible for the species differences in PPADS effects. Given that PPADS appears to interact at the ATP-binding site (Michel et al., 2006b), it may be that residues close to amino acid 284 affect PPADS species selectivity. Certainly adjacent amino acids at positions 282, 285 and 288 differ between the human and the rat receptors.
There are limitations to the approach of exchanging large tracts of receptors between orthologues as illustrated by the data obtained with the domain 3 chimera. In this receptor, agonist potency was greatly increased. Indeed, the increase in ATP potency was so great that the receptor became tonically activated when studied in the sucrose assay buffer. This was, presumably, due to receptor activation by endogenously released ATP and was observed only in studies in sucrose buffer, as ATP has much higher potency in sucrose than in NaCl buffer (Michel et al., 1999). The increase in potency at the domain 3 receptor was not restricted to ATP and was also observed with other ATP analogues. The increased potency at the domain 3 receptor was also evident in ligand-binding studies and did not appear to reflect a simple switch in the pharmacological properties of the human receptor to those of the rat receptor, as agonist potency at the domain 3 receptor was even higher than at the rat receptor for all of the agonists studied. Interestingly, PPADS is thought to bind to the ATP-binding site, and PPADS potency was also increased, although only threefold, at the domain 3 receptor.
The reason(s) for the high agonist potency at the domain 3 receptor is not known and it would be interesting to determine if the change reflects the large exchange of protein between orthologues or can be localized to a single residue. In this respect, H155Y is a reported gain-of-function SNP in the human P2X7 receptor (Cabrini et al., 2005), and the residue is tyrosine in the rat receptor. However, it did not appear to be responsible for any of the changes in agonist or antagonist potency, although this comparison was complicated by the use of a human receptor containing an additional single SNP (H270R) in these studies. The domain 3 receptor also includes amino acid 173, and a previous study demonstrated that a single-point mutation of the rat P2X7 receptor, in which the human glycine residue at position 173 was inserted into the rat receptor, resulted in a chimeric receptor at which BzATP was 10- to 30-fold higher than at the rat receptor (Thompson, 2001). Although this change in potency is in the opposite direction to that observed in this study, where exchange of rat sequence into the human receptor increased potency, it is consistent with this region of the receptor being important for defining agonist potency at the P2X7 receptor.
In summary, we have identified a key residue at position 95, which seems to determine the nature of the allosteric effects of GW791343 and which is also partly responsible for the species difference in effects of SB203580 and KN62 but is not involved in the species selectivity of PPADS or in the species difference in agonist potency. A further residue at position 126 appeared to be important for the species difference in potency of PPADS, although residues in domain 5 also seemed to contribute to its species selectivity, perhaps implicating multiple regions of the receptor in the binding site for PPADS. Finally, we identified a further region of the P2X7 receptor that affected agonist and PPADS potency at the P2X7 receptor. This region is distinct from other areas of the receptor previously identified as being important for agonist binding and further study of residues within this domain may provide additional information on the interaction of agonists with the receptor.
Abbreviations
- BzATP
2′- and 3′-O-(4benzoylbenzoyl) ATP
- ECD
extracellular domain
- GW791343
N2-(3, 4-difluorophenyl)-N1-(2-methyl-5-(1-piperazinylmethyl)phenyl)glycinamide dihydrochloride
- LysoPC
palmitoyl L-α-lysophosphatidylcholine
- PPADS
pyridoxalphosphate-6-azophenyl-2′,4′-disulphonic acid
- RT
room temperature
- SB203580
4-(4-fluorophenyl)-2-(4-methylsulphinylphenyl)-5-(4-pyridyl)1H-imidazole
- SNP
single-nucleotide polymorphism
Conflict of interest
The authors are employed by GlaxoSmithKiline.
References
- Alexander SPH, Mathie A, Peters JA.Guide to receptors and Channels (GRAC) Br J Pharmacol 2008153Suppl 2S1–S209.3rd edn. (2008 revision) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraldi PG, Di Virgilio F, Romagnoli R. Agonists and antagonists acting at P2X7 receptor. Curr Top Med Chem. 2004;4:1707–1717. doi: 10.2174/1568026043387223. [DOI] [PubMed] [Google Scholar]
- Cabrini G, Falzoni S, Forchap SL, Pellegatti P, Balboni A, Agostini P, et al. A His-155 to Tyr polymorphism confers gain-of-function to the human P2X7 receptor of human leukemic lymphocytes. J Immunol. 2005;175:82–89. doi: 10.4049/jimmunol.175.1.82. [DOI] [PubMed] [Google Scholar]
- Chessell IP, Michel AD, Humphrey PPA. Effects of antagonists at the human recombinant P2X7 receptor. Br J Pharmacol. 1998;124:1314–1320. doi: 10.1038/sj.bjp.0701958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay WC, Condreay JP, Moore LB, Weaver SL, Watson MA, Kost TA, et al. Recombinant baculoviruses used to study estrogen receptor function in human osteosarcoma cells. Assay Drug Dev Technol. 2003;1:801–810. doi: 10.1089/154065803772613435. [DOI] [PubMed] [Google Scholar]
- Donnelly-Roberts DL, Jarvis MF. Discovery of P2X7 receptor-selective antagonists offers new insights into P2X7 receptor function and indicates a role in chronic pain states. Br J Pharmacol. 2007;151:571–579. doi: 10.1038/sj.bjp.0707265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari D, Pizzirani C, Adinolfi E, Lemoli RM, Curti A, Idzko M, et al. The P2X7 receptor: a key player in IL-1β processing and release. J Immunol. 2006;176:3877–3883. doi: 10.4049/jimmunol.176.7.3877. [DOI] [PubMed] [Google Scholar]
- Fonfria E, Clay WC, Levy DS, Goodwin JA, Roman S, Smith GD, et al. Cloning and pharmacological characterisation of the guinea-pig P2X7 receptor orthologue. Br J Pharmacol. 2008;153:544–556. doi: 10.1038/sj.bjp.0707596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargett CE, Wiley JS. The isoquinoline derivative KN-62 a potent antagonist of the P2Z-receptor of human lymphocytes. Br J Pharmacol. 1997;120:1483–1490. doi: 10.1038/sj.bjp.0701081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser M, Cebe R, Drewello D, Schmitz R. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques. 2001;31:88–92. doi: 10.2144/01311st05. [DOI] [PubMed] [Google Scholar]
- Hibell AD, Thompson KM, Xing M, Humphrey PP, Michel AD. Complexities of measuring antagonist potency at P2X7 receptor orthologs. J Pharmacol Exp Ther. 2001;296:947–957. [PubMed] [Google Scholar]
- Honore P, Donnelly-Roberts D, Namovic MT, Hsieh G, Zhu CZ, Mikusa JP, et al. A-740003 [N-(1-{[(cyanoimino)(5-quinolinylamino) methyl]amino}-2,2-dimethylpropyl)-2-(3,4-dimethoxyphenyl)acetamide], a novel and selective P2X7 receptor antagonist, dose-dependently reduces neuropathic pain in the rat. J Pharmacol Exp Ther. 2006;319:1376–1385. doi: 10.1124/jpet.106.111559. [DOI] [PubMed] [Google Scholar]
- Humphreys BD, Virginio C, Surprenant A, Rice J, Dubyak GR. Isoquinolines as antagonists of the P2X7 nucleotide receptor: high selectivity for the human versus rat receptor homologues. Mol Pharmacol. 1998;54:22–32. doi: 10.1124/mol.54.1.22. [DOI] [PubMed] [Google Scholar]
- Jiang LH, Mackenzie AB, North RA, Surprenant A. Brilliant blue G selectively blocks ATP-gated rat P2X7 receptors. Mol Pharmacol. 2000;58:82–88. [PubMed] [Google Scholar]
- Michel AD, Chambers LJ, Clay WC, Condreay JP, Walter DS, Chessell IP. Direct labelling of the human P2X7 receptor and identification of positive and negative cooperativity of binding. Br J Pharmacol. 2007;151:103–114. doi: 10.1038/sj.bjp.0707196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AD, Chambers LJ, Walter DS. Negative and positive allosteruic regulators of the P2X7 receptor. Br J Pharmacol. 2008;153:737–750. doi: 10.1038/sj.bjp.0707625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AD, Chessell IP, Humphrey PP. Ionic effects on human recombinant P2X7 receptor function. Naunyn Schmiedebergs Arch Pharmacol. 1999;359:102–109. doi: 10.1007/pl00005328. [DOI] [PubMed] [Google Scholar]
- Michel AD, Fonfria E. Agonist potency at P2X7 receptors is modulated by structurally diverse lipids. Br J Pharmacol. 2007;152:523–537. doi: 10.1038/sj.bjp.0707417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AD, Thompson KM, Simon J, Boyfield I, Fonfria E, Humphrey PP. Species and response dependent differences in the effects of MAPK inhibitors on P2X7 receptor function. Br J Pharmacol. 2006a;149:948–957. doi: 10.1038/sj.bjp.0706938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AD, Xing M, Thompson KM, Jones CA, Humphrey PP. Decavanadate, a P2X receptor antagonist, and its use to study ligand interactions with P2X7 receptors. Eur J Pharmacol. 2006b;534:19–29. doi: 10.1016/j.ejphar.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Murgia M, Hanau S, Pizzo P, Rippa M, Di Virgilio F. Oxidized ATP. An irreversible inhibitor of the macrophage purinergic P2Z receptor. J Biol Chem. 1993;268:8199–8203. [PubMed] [Google Scholar]
- North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- Romagnoli R, Baraldi PG, Di Virgilio F. Recent progress in the discovery of antagonists acting at P2X7 receptor. Expert Opin Ther Patents. 2005;15:271–287. [Google Scholar]
- Stokes L, Jiang LH, Alcaraz L, Bent J, Bowers K, Fagura M, et al. Characterization of a selective and potent antagonist of human P2X7 receptors, AZ11645373. Br J Pharmacol. 2006;149:880–887. doi: 10.1038/sj.bjp.0706933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KM.Structure function studies on P2X7 receptors 2001. PHD Thesis, University of Cambridge, UK
- Vial C, Roberts JA, Evans RJ. Molecular properties of ATP-gated P2X receptor ion channels. Trends Pharmacol Sci. 2004;25:487–493. doi: 10.1016/j.tips.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Young MT, Pelegrin P, Surprenant A. Amino acid residues in the P2X7 receptor that mediate differential sensitivity to ATP and BzATP. Mol Pharmacol. 2007;71:92–100. doi: 10.1124/mol.106.030163. [DOI] [PubMed] [Google Scholar]