Abstract
A surface immobilized optical protein sensor has been utilized to detect Interleukin-8 (IL-8) protein, an oral cancer marker, and can reach limit of detection (LOD) at 1.1 pM in buffer without using enzymatic amplification. Only after applying enzymatic amplification to increase the signal level by a few orders of magnitude, ELISA can reach the LOD of 1pm level. We then develop the confocal optics based sensor for further reducing the optical noise and can extend the LOD of the surface immobilized optical protein sensor two orders in magnitude. These improvements have allowed us to detect IL-8 protein at 4.0 fM in buffer. In addition, these sensitive LODs were achieved without the use of enzymatic signal amplification, such that the simplified protocol can further facilitate the development of point-of-care devices.
The ultra sensitive optical protein sensor presented in this paper has a wide number of applications in disease diagnoses. Measurements for detecting biomarkers in clinical sample are much more challenging than the measurements in buffer, due to high background noise contributed by large collections of non-target molecules. We used clinical salvia samples to validate the functionality of the optical protein sensor. Clinical detection of disease-specific biomarkers in saliva offers a non-invasive, alternative approach to using blood or urine. Currently, the main challenge of using saliva as a diagnostic fluid is its inherently low concentration of biomarkers. We compare the measurements of 40 saliva samples; half from oral cancer patients and half from a control group. The data measured by the optical protein sensor is compared with the traditional Enzyme-Linked Immunosorbant Assay (ELISA) values to validate the accuracy of our system. These positive results enable us to proceed to using confocal optical protein sensor to detect other biomarkers, which have much lower concentrations.
Keywords: Optical biosensor, Diagnostics, IL-8 protein, Oral cancer, Surface immobilization
1. Introduction
ELISA is a standard technique to quantify the amount of protein in a solution. Protein targets are immobilized on a surface and labeled with an enzyme that can continually produce a colored product which accumulates in the solution for detection. If we track the information pathway, we find that the signal originates in the fluid sample volume, moves to the surface and returns to the volume to be finally detected. Although the signal is amplified by an enzymatic reaction, the method for taking light absorbance measurements (fluid opacity) is a relatively insensitive principle compared to optical fluorescence methods (Wells, 2006; Janasek et al., 2006).
At low target concentrations, surface immobilization (Britland et al., 1992; Mooney, Hunt et al. 1996; Lahiri et al., 1999; MacBeath and Schreiber, 2000) is an efficient detection principle, especially when the surface-to-volume ratio is increased (Khandurina and Guttman, 2002; Luo et al., 2005). In this work, we examine the effects of eliminating the enzymatic signal amplification and instead, directly labeling the immobilized proteins with fluorescent probes (Espina et al., 2004; Bashir 2004). We increase the sensitivity of detection by using fluorescence optics to measure the intensity signal (Craighead, 2006) and reducing the sample volume for increased surface-to-volume ratio. The low sample volume requirement (tens of microliters) is advantageous for clinical situations in which only a small amount of the biological fluid is available for assay.
Detecting biomarkers from a 3D volume to a 2D surface needs smaller sample volumes and can improve LOD. The challenge of this approach is the non-specific binding of non-target molecules or fluorescent probes to the surface, both of which will affect the signal integrity. Our previous development of an ultra-sensitive electrochemical RNA/DNA biosensor (Gau et al., 2001) demonstrated that a well-controlled sensor surface could reduce the background noise due to non-specific molecular binding. With this optimized surface modification protocol, detection sensitivity much less than fM concentrations was achieved. This achievement effectively lead to the elimination of PCR amplification of DNA/RNA targets which is usually needed to enable detection (Wang et al., 2005). Adopting a similar surface modification approach, we have shown in this paper that IL-8 can be detected at 1.1 pM level by using a surface immobilized sandwich assay technique. In comparison, the commercial IL-8 ELISA assay has a similar LOD at 1 pM; thus, we are able to achieve the same sensitivity but without the use of enzymatic signal amplification.
IL-8 has a molecular weight of 8.5 kDa and has clinical significance for oral cancer diagnosis (St John et al., 2004). Oral cancer, the fifth most common cancer in the United States, makes up for the largest number of cancers in the head and neck. According to the American Cancer Society, approximately 34,000 people in the United States will be diagnosed with oral cancer in 2007. The survival rate of oral cancer is 60–80% when detected during its early stages; however, this number drops to 30–40% when the cancer is diagnosed during the advanced stages (Franzmann et al., 2005). Identifying molecular markers of early disease can aid in early diagnosis, which can improve patient prognosis (Sidransky et al., 2002). IL-8, as a salivary biomarker for early-stage oral squamous cell carcinoma (OSCC), was discovered through a previous tissue-based expression profiling effort (St John, 2004). Among all of the body fluids that can be tested in the clinic, saliva is the easiest to access and its collection is the least invasive to the patient. In this effort, we first use a commercial ELISA kit to execute endpoint detection of the OSCC protein biomarker from a patient’s saliva; it was found that IL-8 was significantly elevated in the saliva of oral cancer patients. It was highly discriminatory in detecting oral cancer by saliva analysis. The average level of IL-8 in cancer and control patients depends on several variables, such as the number of the subjects and their physical condition. Regardless of these variables, there is an overall increase in the average measurements of IL-8 in cancer patients (Wong, 2006; Rhodus et al., 2004).
Being a filtrate of the serum, saliva has been found to have an abundance of biomarkers with major clinical significance (Herath, 2003; Sonmezoglu et al., 2005). The biomarker concentration in whole saliva can be lower than that of the serum, as in the case of TNF-α, which is another protein biomarker for oral cancer. The salivary TNF-α concentration is approximately 30 pg/ml (2 pM) in oral cancer patients and 3 pg/ml (176 fM) in healthy individuals. For detecting TNF-α in saliva, the LOD, a measure of the signal-to-noise ratio, must be several orders of magnitude lower than the baseline concentration. A need of more sensitive protein sensor is obvious. At low target concentrations, the signal is usually dominated by the background noise, which are caused by non-specific binding and optical noise. We have minimized the non-specific binding by surface modification, In optical system, the optical noise can be reduced through the utilization of confocal optics (Shotton, 1989). The use of confocal optics allows us to confine the detection volume by allowing the signal to pass through a pinhole, thereby rejecting signals that are not from the focal plane of the microscope. Indeed, a LOD as low as 4.0 fM was achieved. This value is approximately 200 times lower than the LOD for surface immobilized techniques, which lack the confocal arrangement. Our confocal optics-based surface immobilized protein sensor is essentially an ultra-sensitive protein sensor, which has potential for a wide array of applications in disease diagnostics.
In this study, three advances in optical protein biosensor are presented: 1) without enzymatic signal amplification, the surface immobilized optical protein sensor can reach a LOD of detecting IL-8 at 1.1 pM in buffer, 2) with the utilization of confocal optics, the LOD of this sensor can be further reduced by two orders of magnitude (Schweitzer et al., 2002, Zajac et al., 2007), detecting IL-8 at 4.0 fM in buffer and 3) the optical protein sensor is validated through detecting IL-8 protein in clinical saliva samples by comparing with traditional ELISA measurements. Our results show that the patients with oral cancer can be clearly distinguished from the control subjects.
2. Materials and Methods
2.1. Bio-chemicals and reagents for the optical protein micro-sensor
The same monoclonal (MAb) and polyclonal (PAb) antibodies that make up the sandwich assay pair in the human IL-8 ELISA kits were used in the detection scheme for the optical protein sensor. ELISA kits for human IL-8 protein were purchased from Pierce Endogen, Rockford, IL. The Mouse anti-human IL-8 MAb, biotin-labeled (M802B), recombinant human IL-8 (RIL810), and rabbit anti-human IL-8 PAb (P801) were purchased from Pierce Biotechnology (Rockford, IL, USA). Alexa Fluor 488 labeled goat anti-rabbit IgG F(ab’)2 (A11078) was purchased from Molecular Probes, Invitrogen (Carlsbad, CA, USA). Tween20 and Bovine Serum Albumin (BSA) were obtained from Sigma Chemical and Tris Hydrochloride was purchased from Gibco (Gaithersburg, MD, USA).
3. IL-8 detection protocol by the optical protein micro-sensor
Streptavidin-coated glass cover slips were purchased from Xenopore (Hawthorne, NJ, USA). Individual sensor areas were created by adhering plastic stickers, which had pre-defined wells of approximately 60 µlit capacities (Grace Bio Labs). The adhesion of the stickers to the cover slips was water-tight, preventing any cross-contamination between wells. To prime the micro-sensors for experiments, each well was incubated with 50 µl of IL-8 specific capture probe (M802B at 6 µg/ml (40 nM)) for 60 minutes, followed by incubation with a 3% BSA blocking solution for 30 minutes. These micro-sensors were then incubated with solutions containing the IL-8 target, which were either protein standards prepared with purified hIL-8 or raw saliva from human subjects. Standard samples were incubated for 30 minutes and saliva samples were incubated for 60 minutes. The longer incubation time for saliva is because of higher solution viscosity. Next, the detection probe (P801 at 20 µg/ml (133.3 nM)) was added to the micro-sensors for 30 minutes, followed by the fluorescent reporter probe (A11078 at 20 µg/ml (133.3 nM)) for 15 minutes. Micro-sensors were individually washed with a buffer solution (50 mM Tris, 0.2% Tween20) in between incubation steps to remove unbound or non-specifically bound bio-molecules. The entire protocol was executed at room temperature.
4. Fluorescence intensity measurement and analysis
4.1. Fluorescence Microscopy
An epi-fluorescence microscope (Leica DMIRB) equipped with a 100W mercury lamp and 63X, NA 0.70, dry objective was used for experiments. Fluorescence intensities from the optical protein micro-sensor surface was observed in a dark box with a Chroma filter set (Ex 480 nm, bandwidth 40 nm; Em 535 nm, bandwidth 50 nm). A 12-bit cooled monochrome CCD camera (Cool SNAP HQ, Photometrics) captured fluorescence images. The optimum exposure time was 5.0 seconds. ImageJ software was used for image analysis and to obtain the mean pixel value of the entire image. Five areas per micro-sensor (105 × 145 µm2) were averaged for a single sensor measurement and the effective detection area was determined by the size of the captured image (1392 × 1040 pixels, 0.105 µm pixel−1 at 63x magnification).
4.2. Confocal Microscopy
A confocal microscope (Leica DM IRBE) with an Ar ion laser and 63X 1.3 oil immersion objective was used to measure fluorescence intensity. The illumination of the sample in the confocal microscope has double cone profile, with the point spread function in the z direction at the plane r = 0 given by (Webb, 1996):
| (1) |
where
In the expression above, NA is the numerical aperture of the objective, which is equal to n sin α , α is half the angle of the light cone accepted by the objective, and λ is the wavelength of the excitation light. The pinhole in the confocal microscope rejects the light rays originating from planes other than the focal plane (Andrews et al., 2002) and allows focused light from the center of the illumination to be recorded on the photomultiplier tube (PMT) (R6357 Hamamatsu). With a pinhole in front of the optical protein micro-sensor, the depth of view is reduced to about one micron. This reduction in the depth of view leads to a significant reduction in the optical noise. The quantum efficiency of the PMT for the emission of AlexaFluor 488, with an emission wavelength of 519 nm, is slightly less than 30%.
For measurements, four different 238 × 238 µm2 spots were imaged on the micro-sensor surface. At each spot, an image stack was generated where each frame was an image slice through the z-axis, perpendicular to the sensor plane. The distance between each frame is 0.1µm. The mean intensity of each image slice was determined and plotted against its order in the stack. An example of this plot is shown in Figure 5 (inset). The maximum intensity values from these plots are chosen for each of the 4 points and then averaged to obtain a data point for a particular IL-8 sample. A Matlab code was written to batch process the image stacks.
Figure 5.

A calibration curve of the optical surface-based IL-8 protein sensor using confocal optics. The inset is a typical scan in which the peak represents a signal of the fluorescent dye. The peak intensity drops with lowering concentration.
5. Saliva sample collection and ELISA assay
Saliva samples were obtained using the same protocol as (St John, 2004). ELISA kits for human IL-8 (Pierce Endogen, Rockford, IL) were used according to the manufacturer’s protocol. After development of the colorimetric reaction, the absorbance at 450 nm was determined by a spectrophotometer and the absorbance readings were converted to concentration based upon standard curves obtained with recombinant cytokine in each assay. Each sample was tested in at least two ELISA experiments and the data was calculated as the mean of the multiple tests on each sample.
6. Statistical Analysis
6.1. T-test: statistical significance between oral cancer and healthy groups
We conducted two-tailed t-tests on the mean data of the cancer group and the control group, for both the optical protein sensor and ELISA. A two-tailed t-test (Glantz, 2002) is conducted to ensure that the difference between the two groups is not coincidental. The hypothesis that the difference is by chance, is called null hypothesis. Essentially, this test assesses whether or not the means of two groups are statistically different from each other. The groups were of moderate sample size (n = 20) and exhibited symmetrical distribution. The t-test formula is the ratio of the difference of means to the variability of the groups (Glantz, 2002):
| (2) |
where X̅a ,X̅b are the sample means, reflect the sample variances and na,nb are the number of subjects in the cancer group and the control group respectively In order to determine the range of values for t to prove that the data is wrong, the calculated t values were evaluated on a significance table. From this table, P-values, which are the probabilities that the null hypothesis is true, were obtained. P-values less than 0.05 were considered to be statistically significant.
6.2. Statistical comparison of diagnostic accuracy by comparison of AUC values from 2 ROC plots
The distributions of data from cancer patients and control group are usually very close to normal distributions and they overlap. On this plot, different cut-off values can be chosen to delineate the two groups. In other words, values above the cut-off points are considered the oral cancer group or testing positive and values below are considered healthy or testing negative. The diagnostic capability for each detection method was determined by calculating the diagnostic sensitivity (the fraction of positive tests that actually have disease) and the diagnostic specificity (the fraction of negative tests that are indeed healthy) (Altman and Bland, 1994), based on different cut-off values. The diagnostic specificity and sensitivity are dependent and their values are dependant on the cutoff value. Increasing sensitivity decreases specificity and vice versa. We determined the capabilities of both the optical protein sensor and ELISA to correctly identify oral cancer patients from control subjects based on IL-8 protein detection from the same saliva samples by comparison of Receiver Operator Characteristics (ROC) (Akobeng, 2007) curve analyses. ROC curves is the plot of sensitivity versus 1-specificity, and they allow visual analyses of trade-offs between sensitivity and specificity when the cutoff is varied. Based on these plots, the area under the curves (AUCs) can be computed. AUC is a single number that represents how good the sensor is in distinguishing the patient group from the control group. An AUC value of one represents the most accurate sensor, while AUC values less than 0.5 indicate poor reliability and results that are by chance. We chose to perform a statistical comparison of the AUC of the optical protein sensor ROC and the AUC of ELISA’s ROC. The method and equations to compare two AUCs is described in Hanley and McNeil (Hanley and Mcneil, 1983). If there are significant differences between the AUC’s, then the conclusion of the analysis result in p-values less than 0.05.
7. Results and Discussion
7.1. Characterization of the optical protein sensor
The limit of detection (LOD) is a figure of merit that describes the ability of a biosensor to discriminate the true signals from the noise level. In a surface-immobilized target detection scheme, each step in the protocol allows for competitive binding between the desired molecules and other non-specific molecules on the surface. Biological fluids are heterogeneous solutions and in addition to our target molecules, there are many other non-target molecules present in disproportionate numbers. This non-specific binding is the main contributor to the background noise and becomes the limiting factor in obtaining a low LOD. Due to the variety of different molecular players in an antibody sandwich assay, and the complexity of molecular interactions, an optimized protocol needs to be custom developed for the specific target molecules as well as the medium, buffer or body fluids to achieve a low LOD.
Optimizing the detection protocol begins with purified target molecules in a buffer to determine the performance baseline in a “clean” sample. After a reliable value is obtained, this procedure is repeated using clinical samples. Detection in clinical samples presents more challenges, such as increased heterogeneity or fluid viscosity and generally requires additional steps for optimization. In the current work, our task is to develop a surface-based optical protein sensing technique that can distinguish the signal generated by low concentrations of IL-8 protein from the noise produced by other native molecules using a small sample volume. The LOD is first established using purified IL-8 in buffer. The substrate is a streptavidin-coated glass surface upon which a biotinylated monoclonal antibody, specific to hIL-8 protein, was added. Although the streptavidin-biotin and antibody-(IL-8) interactions have high binding affinity, they are large biomolecules and thus increase the possibility if non-specific binding. To address this issue, the concentration, volume, and incubation times for each molecular addition was characterized by secondary fluorescence labels. The fluorescence intensity was compared in the presence and absence of the molecule of interest and we used the signal-to-noise (S/N) ratio as a figure of merit. While the introduction of secondary fluorescence labels created potentials for additional molecular interactions, we found in this approach, of choosing the parameters that provided the best (S/N) ratios for the finalized protocol, did indeed improve the overall LOD of the surface-based optical detection technique.
Despite extensive parametric optimization, the background noise could not be completely eliminated; however, it was significantly reduced. We also were able to reduce non-specific binding events to a range that is comparably low, relative to the signal after executing the entire detection protocol. Figure 1 compares the S/N ratio of IL-8 with that of the negative control (3% BSA) and six other proteins that are found in saliva. The signals from the competing proteins are comparable to that of the negative control; thus, the optimized sensor demonstrates excellent specificity for detecting IL-8 protein in a heterogeneous solution.
Figure 1.

Cross reactivity of the optical surface-based IL-8 protein sensor with other common salivary proteins. The sensor exhibits high specificity for IL-8 protein.
To determine the LOD, we followed similar methods and definitions, as established by ELISA. First, a standard curve relating known IL-8 concentrations to intensity output was generated. IL-8 standards were prepared in a buffer solution ranging from 100 pM to less than 1pM. The negative control for this study was a solution without any IL-8 protein. The standard curve in Figure 2 shows that IL-8 protein could be detected at concentrations as low as 1.1 pM (resolution: 1.80 AU pM−1, R2 = 0.9951, n = 8). The LOD is determined from the intensity value, which is two standard deviations above the mean intensity of the negative control. The concentration that this intensity corresponds to is calculated from the equation of the standard curve line, and this theoretical concentration is defined as the LOD. Our calculated result is comparable to the LOD of the ELISA assay for IL-8 protein (1 pM).
Figure 2.
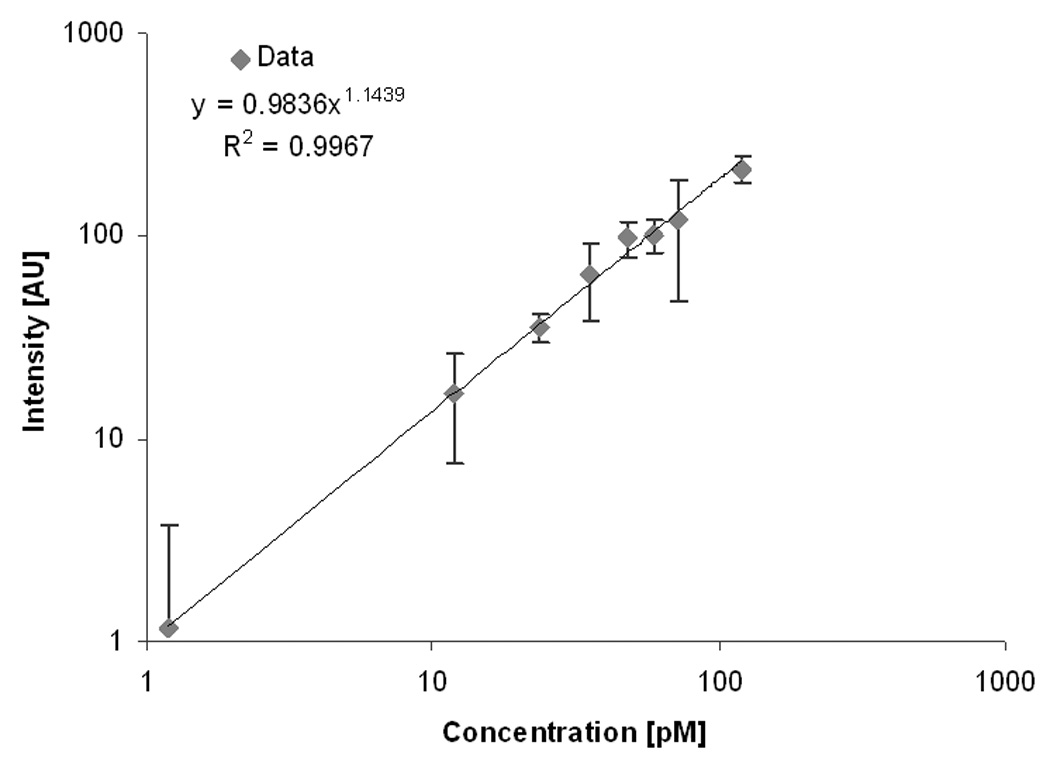
A calibration curve of the optical surface-based IL-8 protein sensor using a standard fluorescent microscope. Each data point represents the mean intensity of five imaged areas on the sensor surface. . The biosensor exhibits excellent linearity (on a log-log graph) at low concentrations with an LOD of 1.19 pM.
7.2. IL-8 measurement in clinical saliva samples
To demonstrate the clinical application of our sensor in biological fluid, we tested for IL-8 protein in saliva samples collected from 20 control subjects and 20 individuals with oral cancer. We compared our results with those of a traditional ELISA assay executed on identical saliva samples. The comparison was performed using statistical methods described earlier in this paper.
The results of the optical protein sensor are shown in Figure 3(a), in which the fluorescence intensity (AU) output is shown for each saliva sample. Using the optical protein sensor, the means were 439 AU (standard deviation = 125 AU) and 320 AU (standard deviation = 105 AU) for the cancer and control groups respectively. The t-test analysis confirms that the oral cancer and control groups are statistically significant, using the optical protein sensor with p < 0.05 (actual p = 0.007). The means of the both groups were calculated using the converted concentration values, based on a standard curve. The extrapolated concentration values are linearly related to the optical density values (signal output) from the ELISA assay. The cancer mean using ELISA was 1252 pg/ml (standard deviation = 456 pg/ml) and the control mean was 577 pg/gml (standard deviation = 355 pg/ml). The t-test analysis also confirmed that the oral cancer and control group were statistically different using ELISA (p = 1e-5).
Figure 3.

(a) Intensity measurements for each individual using the optical protein sensor (b) Comparison of ELISA and mircosensor technique on clinical samples. Assay measurements are normalized to test group average.
While the sensitivity and specificity of a diagnostic test vary with the choice of cut-off value, the ROC plots for the optical sensor and ELISA display all of the possible cut-off values and the resulting diagnostic sensitivity and specificity. And the ROC plot does not change with the change of the cut-off value. The AUC for the optical sensor and ELISA were 0.837 and 0.863 respectively. We use a statistical analysis method to compare the AUC from the two different ROC plots and found that they did not demonstrate any statistical difference (p = 0.33) (Hanley and Mcneil, 1983). Using AUC as an indicator of predictive ability, we can conclude that the optical sensor and ELISA have similar performances as diagnostic tests.
7.3. Characterization of the Confocal Optical Protein Sensor
The use of multiple biomarkers in a diagnostic test greatly improves the accuracy of disease diagnosis (Liu, 2005). In the case of oral cancer, it has been shown that the predictability for disease using IL-8 protein as a biomarker increases dramatically with the addition of another cytocine, IL-6 protein (St John, 2004). While there is a need to detect other proteins in saliva for improved diagnostics, the concentration of these other salivary proteins are very low. For instance, the concentration of TNF-α, another important oral cancer biomarker, is approximately 30 pg/ml (1.76 pM) in oral cancer patients and 3 pg/ml (176 fM) in healthy subjects (Rhodus et al., 2004). Unfortunately, these values are below the LOD of our optical protein sensor and ELISA can detect. To significantly improve upon salivary diagnostics, an even lower LOD needs to be achieved. Utilizing confocal optics on the sensor surface prepared with the previously described protocol, we have improved the LOD by two orders of magnitude.
A standard curve from different IL-8 concentrations generated after implementing confocal optics, is presented in Figure 5. The integration of the confocal image stack to generate a signal intensity value is described in the Materials and Methods section. The LOD, as determined from the equation of the line in Figure 5, is 4.0 fM. Excellent linearity (R2 = 0.988, n = 6) was obsreved over seven decades of IL-8 concentrations (6 fM–12 pM).
Stable and consistent output from a sensor is useful for quantitative measurements. The optical intensities at different protein concentrations were produced from multiple data sets, obtained at various times over the span of a several days. If the functional dependence is repeatable, data from these different experiments should fall into a single curve; however, this was not the case. By adjusting the intensity values at each IL-8 concentration by dividing the number of IL-8 molecules in the sample volume of 50 µl and use this normalized value instead, the set of curves do collapse into a single curve, as illustrated in Figure 6. The horizontal axis represents the number of molecules in the measured volume. These finding provides a calibration curve which doesn’t vary with time, thus greatly simplifying the measurement of absolute protein levels. Using this method, we don’t need to generate a standard curve for each experiment for protein quantification since the signal output from the optical protein detection technique can be manipulated in a way that is consistently comparable.
Figure 6.

Comparison between four experiments of different concentrations on different days. These results show that repeatable results are achievable and allows for simplified calibration. The sample volume for these measurements is 50 µl.
The original optical protein sensor has an LOD which is able to detect the IL-8 baseline concentrations (589.25 pg/ml (70 pM)) in the control group and the raised IL-8 levels in the cancer patients (1165 pg/ml (138.5 pM)). The confocal optical protein sensor has a LOD in the fM range and exceeds the minimum requirements for salivary IL-8 detection. However, this sensor can be applied to a variety of biomarkers, especially those that are clinically significant, but are present in very low concentrations.
Conclusion
We have developed a platform technology that can significantly increase the sensitivity and simplify the assay preparation for protein detection. The target protein is immobilized on the surface with capture probe. The emission light from fluorophore conjugated with the reporter probe is used as the detection signal. An advantage of the presented sensor is that it can be developed with attainable materials and instrumentation for straightforward integration for point-of care applications. For example, the functionalized sensor surface can be fabricated on various substrates (silicon, glass or polymers) that have exposed –OH groups. The OH group can be used for immobilization of different antibody on surface. If needed, the surface can be patterned with photolithography for higher density arrays (Wong et al., 2004). Another advantage of this sensor is the use of saliva, sampling saliva is the easiest and least invasive. Without applying an enzymatic amplification step, the optical protein sensor can detect 1.1 pM, which is very close to results obtained from traditional ELISA techniques with enzymatic amplification. In other words, we have improved the signals by orders of magnitude with a simplified protocol. Performing experiments on a group of 40 clinical saliva samples to measure the IL-8 protein show close agreements between data from the optical protein sensor and ELISA. In addition, the optical protein sensor has a slightly better capability of detecting true positives in cancer patients, whereas ELISA has a slight edge over to differentiate true negatives.
In order to minimize false positive and false negative diagnosis, the concentration of several biomarkers needs to be measured in parallel. Statistical analysis of the panel readouts can significantly improve the accuracy of molecular diagnosis. There are many biomarkers in body fluids that have very low concentrations; and ELISA is not sensitive enough for their detections. Using confocal optics to reduce the optical noise, we can further extend the LOD of the optical protein sensor by two orders of magnitude to 4.0 fM which is the lowest reported sensitivity for IL-8 detection. With femtomolar sensitivity and without enzymatic amplification, we can further extend the range of detection of this molecular diagnostic technique. More accurate diagnostics with higher sensitivity and specificity become possible to detect very low concentration biomarkers of the disease under study.
Figure 4.

The resulting ROC plot for the optical protein sensor using IL-8 as a cancer biomarker tested on 40 saliva samples.
Acknowledgments
This work is supported by the UO1 Research Grant DE15018 from the National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD 20892.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Akobeng AK. Acta Paediatrica. 2007;96(5):644–647. doi: 10.1111/j.1651-2227.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- Altman DG, Bland JM. British Medical Journal. 1994;308(6943):1552–1552. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews PD, Harper IS, Swedlow JR. Traffic. 2002;3(1):29–36. doi: 10.1034/j.1600-0854.2002.30105.x. [DOI] [PubMed] [Google Scholar]
- Bashir R. Advanced Drug Delivery Review. 2004;56(11):1565–1586. doi: 10.1016/j.addr.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Britland S, Perezarnaud E, Clark P, Mcginn B, Connolly P, Moores G. Biotechnology Progress. 1992;8(2):155–160. doi: 10.1021/bp00014a010. [DOI] [PubMed] [Google Scholar]
- Franzmann EJ, Reategui EP, Carraway KL, Hamilton KL, Weed DT, Goodwin WJ. Cancer Epidemiology Biomarkers & Prevention. 2005;14(3):735–739. doi: 10.1158/1055-9965.EPI-04-0546. [DOI] [PubMed] [Google Scholar]
- Gau JJ, Lan EH, Dunn B, Ho CM, Woo JCS. Biosensors & Bioelectronics. 2001;16(9–12):745–755. doi: 10.1016/s0956-5663(01)00216-0. [DOI] [PubMed] [Google Scholar]
- Glantz SA. Primer of Biostatistics. 5th Ed. McGrowHill; 2002. [Google Scholar]
- Hanley JA, Mcneil BJ. Radiology. 1983;148(3):839–843. doi: 10.1148/radiology.148.3.6878708. [DOI] [PubMed] [Google Scholar]
- Herath HMTU. Journal of Clinical Pathology. 2003;56(9):694–698. doi: 10.1136/jcp.56.9.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janasek D, Franzke J, Manz A. Nature. 2006;442(7101):374–380. doi: 10.1038/nature05059. [DOI] [PubMed] [Google Scholar]
- Khandurina J, Guttman A. Journal of Chromatography A. 2002;943(2):159–183. doi: 10.1016/s0021-9673(01)01451-0. [DOI] [PubMed] [Google Scholar]
- Lahiri J, Ostuni E, Whitesides GM. Langmuir. 1999;15(6):2055–2060. [Google Scholar]
- Liu AY, Schisterman EF, Zhu Y. Statistics in Medicine. 2005;24(1):37–47. doi: 10.1002/sim.1922. [DOI] [PubMed] [Google Scholar]
- Luo CX, Fu Q, Li H, Xu LP, Sun MH, Ouyang Q, Chen Y, Ji H. Lab on a Chip. 2005;5(7):726–729. doi: 10.1039/b500221d. [DOI] [PubMed] [Google Scholar]
- MacBeath G, Schreiber SL. Science. 2000;289(5485):1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- Mooney JF, Hunt AJ, McIntosh JR, Liberko CA, Walba DM, Rogers CT. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(22):12287–12291. doi: 10.1073/pnas.93.22.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodus NL, Ho V, Miller CS, Myers S, Ondrey F. Cancer Detection and Prevention. 2004;29(1):42–45. doi: 10.1016/j.cdp.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Schweitzer B, Roberts S, Grimwade B, Shao WP, Wang MJ, Fu Q, Shu QP, Laroche I, Zhou ZM, Tchernev VT, Christiansen J, Velleca M, Kingsmore SF. Nature Biotechnology. 2002;20(4):359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shotton DM. Journal of Cell Science. 1989;94:175–206. [Google Scholar]
- Sidransky D. Nature Reviews Cancer. 2002;2(3):210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- Sonmezoglu M, Baysal B, Ergen A, Barut SG. International Journal of Clinical Practice. 2005;59(4):433–436. doi: 10.1111/j.1368-5031.2005.00495.x. [DOI] [PubMed] [Google Scholar]
- St John MAR, Li Y, Zhou XF, Denny P, Ho CM, Montemagno C, Shi WY, Qi FX, Wu B, Sinha U, Jordan R, Wolinsky L, Park NH, Liu HH, Abemayor E, Wong DTW. Archives of Otolaryngology-Head & Neck Surgery. 2004;130(8):929–935. doi: 10.1001/archotol.130.8.929. [DOI] [PubMed] [Google Scholar]
- Wang TH, Peng YH, Zhang CY, Wong PK, Ho CM. Journal of the American Chemical Society. 2005;127(15):5354–5359. doi: 10.1021/ja042642i. [DOI] [PubMed] [Google Scholar]
- Webb RH. Reports on Progress in Physics. 1996;59(3):427–471. [Google Scholar]
- Wells M. Current Opinion in Biotechnology. 2006;17(1):28–33. doi: 10.1016/j.copbio.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Wong DT. Expert Review of Molecular Diagnostics. 2006;6(3):267–272. doi: 10.1586/14737159.6.3.267. [DOI] [PubMed] [Google Scholar]
- Wong PK, Wang TH, Deval JH, Ho CM. Ieee-Asme Transactions on Mechatronics. 2004;9(2):366–376. [Google Scholar]
- Zajac A, Song DS, Qian W, Zhukov T. Colloids and Surfaces B-Biointerfaces. 2007;58(2):309–314. doi: 10.1016/j.colsurfb.2007.02.019. [DOI] [PubMed] [Google Scholar]


