Abstract
We investigated mechanisms underlying GATA-6-mediated transcriptional activation of the alveolar epithelial type I cell-enriched gene aquaporin-5 (AQP5). GATA-6 expression increases in alveolar epithelial cells in primary culture, concurrent with upregulation of AQP5 and transition to a type I cell-like phenotype. Cotransfections in MLE-15 and NIH 3T3 cells demonstrated trans-activation by GATA-6 of a rat 1,716-bp-AQP5-luciferase (−1716-AQP5-Luc) reporter. Electrophoretic mobility shift assay and chromatin immunoprecipitation identified an interaction between GATA-6 and putative binding sites in the AQP5 promoter. However, mutation of these sites did not reduce GATA-6-mediated activation, implicating mechanisms in addition to direct binding of GATA-6 to DNA. A 5′-deletion construct, −358-AQP5-Luc, that does not encompass GATA motifs was still activated by GATA-6 by as much as 50% relative to −1716-AQP5-Luc. Internal deletion of the −358/−173 GC-rich domain, which includes several putative Sp1 consensus sites, reduced trans-activation by ∼60%, suggesting importance of this region for GATA-mediated activity. −358-AQP5-Luc was similarly activated by both GATA-6 and a GATA DNA-binding defective mutant, whereas cotransfections in Schneider S2 cells demonstrated dose-dependent trans-activation of −358-AQP5-Luc by Sp1. Activation of −358-AQP5-Luc by GATA-6 was dramatically reduced by Sp1 small-interfering RNA, and −358-AQP5-Luc was activated synergistically by GATA-6 and Sp1 in NIH 3T3 cells. Furthermore, association between endogenous GATA-6 and Sp1 was demonstrated by coimmunoprecipitation. These results suggest that transcriptional activation of AQP5 by GATA-6 is mediated at least in part through cooperative interactions with Sp1 occurring at the proximal promoter.
Keywords: transcription, alveolar epithelium, gene regulation, transfection, protein-protein interaction
the gata family of zinc finger transcription factors (TF) plays an essential role in developmental processes and tissue- and cell-specific gene transcription (6, 41). Six GATA TF are present in vertebrates and can be divided into two subgroups based on sequence homology and tissue distribution (6). GATA-1/2/3 are expressed predominantly in cells of the hematopoietic lineage (37, 47). GATA-4/5/6 are strongly expressed in various mesoderm- and endoderm-derived tissues, including heart, lung, liver, gonad, and gut, where they play important roles in organogenesis and regulation of tissue-specific gene expression (27, 32). Each GATA family member contains a highly conserved DNA-binding domain consisting of two zinc fingers with the motif Cys-X2-Cys-X17-Cys-X2-Cys that mediate binding to a core nucleotide sequence element (A/T)GATA(A/G) (19, 25, 35). Specificity of individual GATA family members is mediated to a large extent through interactions with both ubiquitous and cell-specific transcriptional activators (27).
GATA-6 is expressed during embryogenesis as early as the blastocyst stage at 3.5 days. Targeted mutagenesis of the mouse GATA-6 gene is lethal at 5.5 days postcoitum due to a defect in extraembryonic tissue formation (21). Although the expression pattern for GATA-4/5/6 genes largely overlaps during early embryogenesis, GATA-6 is the only GATA factor that is expressed in the distal epithelium of the developing lung (28, 45). GATA-6 is expressed in respiratory epithelial cells of developing lung tubules by embryonic day 10 and is essential for normal branching morphogenesis and epithelial cell differentiation (14). In addition, GATA-6 has been shown to regulate the promoters of a number of lung-specific genes, including Nkx2.1 (36), surfactant protein A (5), and surfactant protein C (SP-C) (24). Interestingly, expression of a GATA-6 Engrailed dominant-negative fusion protein in the distal lung epithelium of transgenic mice using the human 3.7-kb SP-C promoter resulted in absence of type I (AT1) cells in the distal lung during late alveolar development and decreased expression of the AT1 cell-specific gene aquaporin-5 (AQP5), suggesting a critical role for GATA-6 in regulation of alveolar epithelial (especially AT1) cell differentiation.
AQP5 is a member of the large family of aquaporin water channel proteins (1). AQP5 is expressed abundantly in lung, salivary, and lacrimal tissues (18). In the adult lung, AQP5 is selectively expressed in the apical membranes of AT1, but not type II (AT2) cells (18, 29). Expression of AQP5 increases substantially late in gestation, consistent with expansion of the AT1 cell population at this period in development (18, 38). AQP5 is also upregulated as a function of time following culture of freshly isolated AT2 cells on inflexible substrata concurrent with transdifferentiation of these cells to an AT1 cell-like phenotype in vitro, suggesting its utility as a phenotypic marker with which to investigate transcriptional mechanisms regulating gene expression in AT1 cells (4).
GATA-6 has been shown to activate the mouse AQP5 promoter in transient transfection assays (45). Although the proximal −1716-bp AQP5 promoter encompasses putative GATA DNA-binding sites, substantial transcriptional activation by GATA-6 was observed with deletion of these sites, suggesting the possibility that, in addition to direct binding to DNA, GATA-6 mediates transcriptional activation of AQP5 indirectly through interactions with other TF that themselves bind to DNA (24, 45). In this study, we investigated the mechanisms underlying transcriptional regulation of AQP5 by GATA-6 to gain insight into the mechanisms regulating expression of the differentiated AT1 cell phenotype.
MATERIALS AND METHODS
Rat alveolar epithelial cell isolation and culture.
All animal use procedures used in these studies were approved by the Institutional Animal Care and Use Committee of University of Southern California. AT2 cells were isolated from adult male Sprague Dawley rats by elastase disaggregation (2.0–2.5 U/ml) and panning on IgG-coated bacteriological plates (2). Enriched AT2 cells were resuspended in a minimal defined serum-free medium consisting of Dulbecco's modified Eagle's medium (DMEM) and Ham's F-12 nutrient mixture in a 1:1 ratio, supplemented with 1.25 mg/ml BSA, 10 mM HEPES, 0.1 mM nonessential amino acids, 2.0 mM glutamine, 100 U/ml sodium penicillin G, and 100 μg/ml streptomycin. Cells were seeded into tissue culture-treated polycarbonate (Nuclepore) filter cups at a density of 106 cells/cm2 and grown to confluence. AT2 cell purity (>90%) was assessed by staining freshly isolated cells with an antibody to lamellar membrane protein p180 (Covance, Richmond, CA). Cell viability (>95%) was measured by trypan blue dye exclusion.
Maintenance of cell lines.
MLE-15 mouse lung epithelial cells (J. Whitsett, Cincinnati, OH) (44) were cultivated in RPMI 1640 supplemented with 10 nM hydrocortisone, 5 μg/ml insulin, 5 μg/ml human transferrin, 10 nM β-estradiol, 5 μg/ml selenium, 2 mM l-glutamine, 10 mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2% FBS. NIH 3T3 cells were grown in DMEM supplemented with 10% FBS, 2.5 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Schneider's Drosophila Line 2 (S2) cells (ATCC, Manassas, VA) were grown without carbon dioxide at 22°C in Schneider's Drosophila Medium with l-glutamine (Invitrogen, Carlsbad, CA) supplemented with antibiotic-antimycotic solution (Invitrogen) and 10% heat-inactivated FBS (Hyclone, Logan, UT).
Western analysis.
Protein was extracted from alveolar epithelial cells (AEC) on days 0, 1, and 8, and from MLE-15 and NIH 3T3 cells, in 2% SDS buffer (62.5 mM Tris·HCl, 2% SDS, and 10% glycerol). Samples were separated by SDS-PAGE and transferred to Immuno-Blot polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hercules, CA). Membranes were blocked with 5% donkey serum in 20 mM Tris, pH 7.5, 500 mM NaCl, and 0.1% Tween 20 overnight at 4°C and incubated with goat anti-GATA-6 (sc-7244; Santa Cruz Biotechnology, Santa Cruz, CA) or rabbit anti-AQP5 (Chemicon, Temecula, CA) polyclonal antibodies for 1 h at room temperature. Membranes were incubated with peroxidase-conjugated donkey anti-goat (Jackson ImmunoResearch Laboratories, West Grove PA) or anti-rabbit (Promega, Madison, WI) secondary antibodies for 45 min at room temperature. To detect Sp1 expression, following blocking with 5% nonfat milk for 1 h at room temperature, blots were incubated with rabbit Sp1 antibody (sc-59X; Santa Cruz Biotechnology) overnight at 4°C. After being washed with PBS (pH 7.2), membranes were incubated with goat anti-rabbit secondary antibody (Promega). Peroxidase activity was detected with Super Signal (Pierce, Rockford, IL), and images analyzed using a FluorChem imager (Alpha Innotech, San Leandro, CA). To ensure equal loading, protein was normalized to either eukaryotic initiation factor 2α (eIF2α) or lamin A/C using rabbit anti-eIF2α and anti-lamin A/C polyclonal antibodies (Santa Cruz Biotechnology).
Preparation of nuclear extracts.
NIH 3T3 cells grown on 100-mm plates were transiently transfected with pcDNA3/GATA-6 expression plasmid for preparation of nuclear extracts for electrophoretic mobility shift assays (EMSA). After 24 h, cells were washed and harvested in PBS. After centrifugation, pellets were resuspended in 1 ml hypotonic buffer [10 mM HEPES (pH 7.5), 10 mM KCl, 3 mM MgCl2, 0.05% Nonidet P-40, 1 mM EDTA (pH 8.0), 10 mM NaF, and 0.1 mM Na3VO4] supplemented with protein inhibitor cocktail set III (5 μl/ml, Calbiochem, San Diego, CA) and incubated on ice for 20 min, followed by vortexing for 10 s and centrifugation at 500 g for 10 min at 4°C. Nuclear pellets were washed two times with hypotonic buffer and lysed in 200 μl lysis buffer [100 mM HEPES (pH 7.5), 0.5 M KCl, 5 mM MgCl2, 28% glycerol, and 5 μl/ml protein inhibitor cocktail set III] for 30 min on ice with shaking. After centrifugation at 20,000 g for 30 min to remove debris, nuclear extracts were separated into aliquots and stored at −70°C. Nuclear extracts from MLE-15 cells for coimmunopreciptation were generated using the Active Motif Nuclear Extract Co-IP Complex Kit (Carlsbad, CA).
EMSA and electrophoretic mobility supershift assays.
Double-stranded oligonucleotides encompassing nine putative GATA-6-binding sites identified in the proximal 1,716-bp of the rat AQP5 promoter and designated as sites 1–9 [no. 1: 5′-GAAAGCCCTCCAATCTCCTTTCAC-3′ (−1,363 to −1,339); no. 2: 5′-AGGAATGGGAGATAGCCGGGCAGA-3′ (−1,197 to −1,174); no. 3: 5′-AAGAACAAGGATAAAAAGCAAAC-3′ (−979 to −755); no. 4: 5′-AACGCCTCCTAGAGCCCGCT-3′ (−801 to −782); no. 5: 5′-CCCGCTGCCTAGAGCCAACA-3′ (−787 to −768); no. 6: 5′-GCGTCGAGGATTGCCATTAG-3′ (−755 to −735); no. 7: 5′-GCCACGGGCGGATTGGGAG-3′ (−653 to −633); no. 8: 5′-TTGGGAGCTAAGAGGCGC-3′ (−624 to −641); and no. 9: 5′-CCTCAGAGAATCGCATCAT-3′ (−601 to −620)] were labeled with 33P-ATP. Nuclear extract (∼4 μg) was incubated with ∼2 × 105 counts/min of 33P-labeled oligonucleotide for 20 min at 4°C. Competition for binding was performed with 100-fold molar excess of unlabeled oligonucleotide. DNA-protein complexes were resolved by electrophoresis on 5% nondenaturing polyacrylamide gels. Gels were blotted onto Whatman No.3 MM paper, vacuum dried for 1 h at 80°C, and exposed to X-ray film (Kodak, Rochester, NY).
Chromatin immunoprecipitation.
MLE-15 cells were cross-linked with 1% formaldehyde for 10 min at room temperature, followed by addition of glycine to 0.125 M and continued incubation for 5 min. Cells were lysed in hypotonic buffer, and nuclear pellets were collected by centrifugation and resuspended in nuclear lysis buffer. Solubilized chromatin was sheared by sonication (VirTis, Gardiner, NY). Chromatin was incubated with goat anti-GATA-6 antibody (sc-7244; Santa Cruz Biotechnology) or goat IgG at 4°C overnight. Complexes were recovered by incubation with A/G Plus-agarose beads at 4°C for 2 h. Beads were collected by centrifugation and washed thoroughly. Complexes were eluted by incubation with 150 μl immunoprecipitation elution buffer (1% SDS and 0.1% NaHCO3). NaCl was then added at a final concentration of 0.3 M, and samples were heated at 65°C overnight to reverse cross-linking, followed by phenol-chloroform extraction and ethanol precipitation. DNA was resuspended in 30 μl TE, and PCR was performed using primers corresponding to the regions −1,391 to −1,373 (5′-GCCAGTCCAAGAGTGCAAG-3′) and −1,251 to −1,235 (5′-TTCTAGCTCCATCTAGTTCGC-3′), which span conserved GATA binding site no. 2 in the mouse AQP5 promoter at −1,313 to −1,310 relative to the initiator methionine at +1. For input DNA, soluble chromatin not incubated with antibodies was subjected to PCR. Albumin was amplified as a nontarget gene to monitor for nonspecific binding.
AQP5 promoter reporter constructs and other plasmids.
−1716-, −1097-, −894-, −173- and −139-AQP5-luciferase constructs are as previously described (3). −1716(Δ−173/−358)-Luc was generated by digestion of −1716-AQP5-Luc with SacII followed by religation (see construct g in Fig. 4). −173(−173/−358)F1 (also called −358-AQP5-Luc) and −173(−173/−358)R AQP5-luciferase constructs were generated by inserting a SacII fragment spanning from −173 to −358 of the AQP5 promoter into −173-AQP5-Luc in forward or reverse orientation (see Figs. 4 and 5A). −173(−173/−358)F2 and −173(−173/−358)F3 were generated by inserting two or three copies of the −173/−358 SacII fragment into −173-AQP5-Luc. TK(−173/−358) was generated by cloning the −173/−358 SacII fragment into TK-Luc, which is pGL2-Basic containing the thymidine kinase promoter from pRL-TK. pcDNA3/GATA-6 was generated by cloning the 2.6-kb mouse full-length GATA-6 cDNA into Xhol/XbaI sites of pcDNA3 (Invitrogen). pPacO and pPacSp1 were kindly provided by Dr. John D. Noti (Sayre, Pennsylvania). pPacO/GATA-6 was generated by cloning GATA-6 cDNA from pcDNA3/GATA-6 in the BamHI site of pPacO. pCMV/Sp1 is as previously described (22). D238 Renilla was generated from pRL-SV40 (Promega) by deleting a 238-bp fragment containing GATA-6-binding sites by digestion with ScaI and HindIII.
Mutagenesis of GATA-6 binding site in AQP5 promoter and GATA-6 DNA binding domain in GATA-6 expression plasmid.
The Quik Change II Site Directed mutagenesis kit (Stratagene, La Jolla, CA) was used with the primers 5′-GCGGCTTGGACTGTCCTCCCGCAACTGTCACACCACAAC-3′ and 5′-GTTGTGGTGTGACAGTTGCGGGAGGACAGTCCAAGCCGC-3′ to mutate amino acids cysteine and alanine at positions 293 and 294 (which are responsible for binding of GATA-6 to DNA) to serine and arginine to generate the mouse GATA-6 DNA-binding mutant (pcDNA3/GATA-6 mBD). To generate mutations in the AQP5 promoter, putative GATA-6-binding site nos. 2, 8, and 9 were mutated individually or in combination to generate Mut2-AQP5, Mut8-AQP5, Mut9-AQP5, and Mut2+8+9-AQP5 (see Fig. 4) from the parental −1716-AQP5-Luc plasmid. Primer pairs are as follows: no. 2 forward 5′-CGACGTAGGAATGGGAGGCCGGGCAGAAATAAAG-3′ and reverse 5′-CTTTATTTCTGCCCGGCCTCCCATTCCTACGTCG-3′; no. 8 forward 5′-CGGATTGGGAGCGAGGCGCTGTCCTC-3′ and reverse 5′-GAGGACAGCGCCTCGCTCCCAATCCG-3′; no. 9 forward 5′-GCTGTCCTCAGAGCGCATCACTCTGG-3′ and reverse 5′-CCAGAGTGATGCGCTCTGAGGACAGC-3′ to delete the ATA, TAA, and AAT nucleotides in the respective putative GATA-6-binding sites. Mutant clones were sequenced to verify mutations.
Transient transfection assays.
NIH 3T3 or MLE-15 cells were plated at 6 × 104 cells/well in 24-well plates 24 h before transfection. Cells were cotransfected with 0.75 μg of the series of deleted AQP5-luciferase constructs and 0.25 μg of either the GATA-6 expression plasmid pcDNA3/GATA-6 or control vector pcDNA3 using Superfect reagent (Qiagen, Valencia, CA). MLE-15 cells express both AQP5 and GATA-6, whereas NIH 3T3 cells express only low levels of GATA-6. Both cell types were therefore evaluated, since it was expected that activation would be greater in cells with low endogenous levels of GATA-6. To test the effects of mutation of GATA-binding sites in the AQP5 promoter or internal deletion of −173/−358 on the ability of GATA-6 to trans-activate AQP5, equimolar amounts of −1716-, Mut2-, Mut8-, Mut9-, Mut2+8+9-, and −1716(Δ−173/−358)-AQP5-Luc were transfected using Superfect reagent into NIH 3T3 cells. To test enhancer function of the −173/−358 AQP5 fragment, −173-, −173(−173/−358)F1-, −173(−173/−358)R-, −173(−173/−358)F2-, −173(−173/−358)F3-AQP5-Luc, or (−173/−358)-TK-Luc and TK-Luc were transfected in equimolar amounts using Superfect reagent into MLE-15 and NIH 3T3 cells. To analyze whether GATA-6 activation of AQP5 requires binding of GATA-6 to DNA, 0.75 μg of −358-AQP5-Luc, 0.25 μg of pcDNA3/GATA-6, or GATA-6 DNA binding mutant pcDNA3/GATA-6 mBD were cotransfected into NIH 3T3 cells. Results were normalized to transfection with the minimal AQP5 promoter luciferase construct (−173-AQP5-Luc). To evaluate synergistic interactions between GATA-6 and Sp1, −1716-AQP5-Luc (0.75 μg), pcDNA3/GATA-6 (37.5 ng), and pCMV/Sp1 (100 ng) singly or in combination, or corresponding empty expression vectors, were transfected into NIH 3T3 cells. Synergy between GATA-6 and Sp1 is identified by calculating the interaction response (IR), which compares the effect of two expression vectors cotransfected together with the additive effect of each of the expression vectors cotransfected separately as defined by the equation IR = log[(GATA-6 + Sp1)/(GATA-6) + (Sp1)]. Values of −0.1 to 0.1 are defined as additive, >0.1 synergistic and less than −0.1 antagonistic (15). In all transfections, Renilla luciferase or D238 Renilla were included to normalize for transfection efficiency. Firefly and Renilla luciferase activity were determined with the Dual-Luciferase reporter assay system (Promega). For transfections of Sp1 in S2 cells, 105 cells were plated in 24-well plates 24 h before transfection. −358-AQP5-Luc (0.75 μg) and increasing amounts (0–100 ng) of pPacSp1 were transfected using calcium phosphate precipitation. Empty vector (pPacO) was used to equalize amounts of DNA in each transfection. β-Galactosidase (β-gal) was used to normalize for transfection efficiency. Cells were harvested after 24 h for measurement of luciferase activity and β-gal activity using the Dual Light System Assay (TROPIX, Bedford, MA).
Sp1 small-interfering RNA transfection.
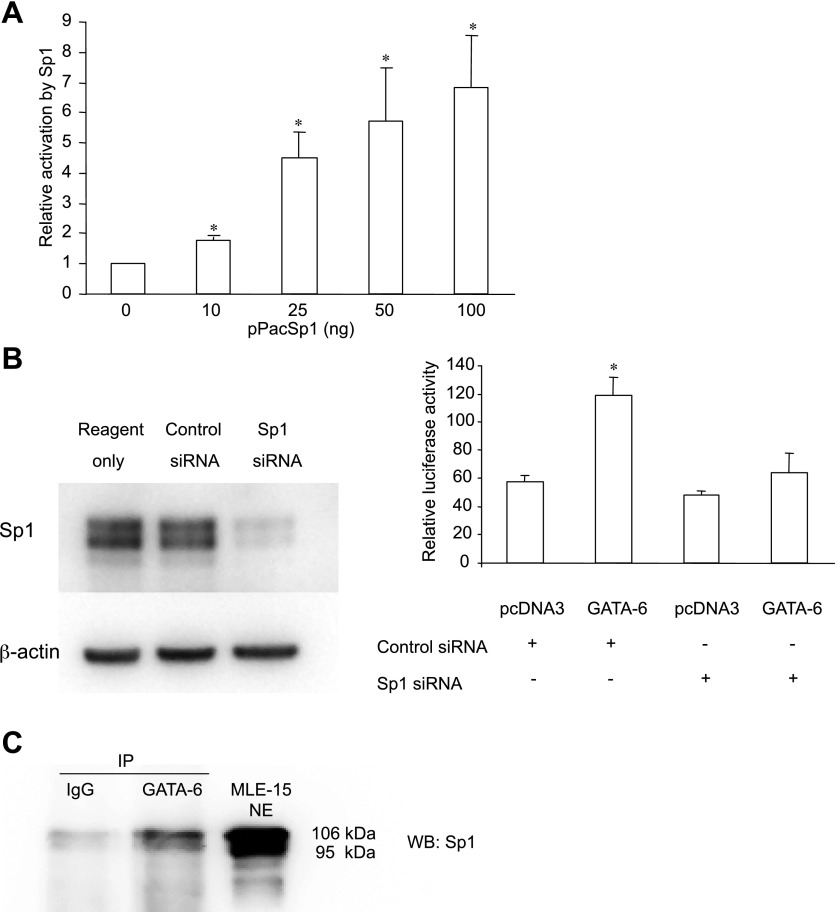
NIH 3T3 cells were plated at 4.5 × 104 cells/well in 24-well plates. After 18 h, medium was removed and replaced with 0.5 ml DMEM. For DNA transfection, 1.5 μg of −358-AQP5-Luc and 0.15 μg of either pcDNA-3 or pcDNA3/GATA-6 were complexed with 2 μl of Lipofectamine 2000 (Invitrogen) in 100 μl of Opti-MEM medium (Invitrogen) and added to each well. Small-interfering RNA (siRNA) targeting the mouse Sp1 gene (sequence: 5′-CCAAUUACAGAACCAGCAAGUUCU-3′) or control nontargeting siRNA complexed with 2 μl Lipofectamine 2000 in 100 μl of Opti-MEM was added concurrently. After 6 h, 50 μl of FBS were added to each well. Cells were harvested after 48 h in passive lysis buffer (Promega) for luciferase reporter assay and in 2% SDS lysis buffer for Sp1 Western analysis. Luciferase values were normalized to total protein concentration measured by the Bio-Rad protein assay kit.
Coimmunoprecipitation.
MLE-15 nuclear extract (1,000 μg) in Low IP Buffer (Active Motif) was incubated with 8 μg of goat antibody against GATA-6 (sc-7244; Santa Cruz Biotechnology) at 4°C overnight. Prewashed protein A/G beads (100 μl at 50%; Santa Cruz Biotechnology) were then added to the mixture and incubated for 4 h. Beads were washed five times with PBS. Isolated protein complexes were denatured for 5 min at 95°C, analyzed by 7.5% SDS-PAGE, and transferred to PVDF membranes, followed by immunoblotting with anti-Sp1 antibody (Santa Cruz Biotechnology) with detection by enhanced chemiluminescence (Pierce).
Statistical analysis.
Data are shown as means ± SE for n no. of observations. We used z-tests to determine whether ratiometric data were different from control. When comparing multiple groups of data, one-way ANOVA (with post hoc Tukey's test) was used to determine significant differences. P < 0.05 was considered significant.
RESULTS
Expression of GATA-6 during transdifferentiation of rat AEC in vitro.
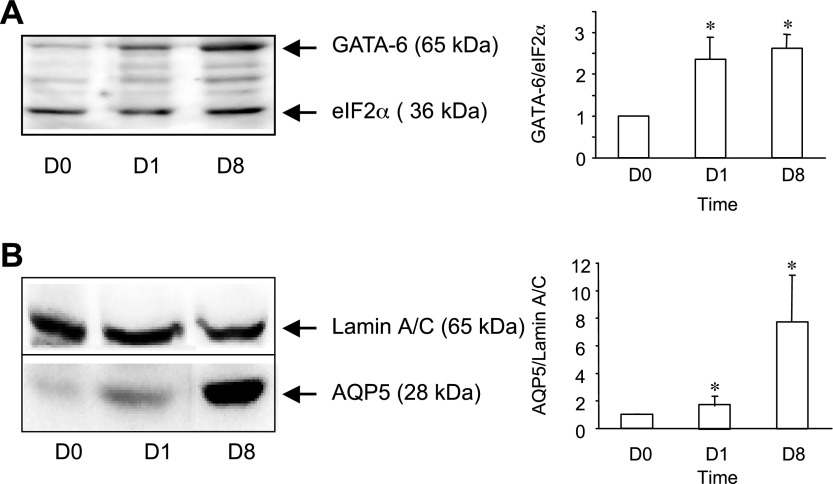
To investigate the role of GATA-6 in regulating AEC cell type-specific gene expression, we first evaluated expression of GATA-6 as a function of time in a well-characterized in vitro model in which AT2 cells in primary culture undergo transdifferentiation to an AT1 cell-like phenotype. Freshly isolated rat AT2 cells (day 0) were plated in serum-free medium on polycarbonate filters, and GATA-6 expression was analyzed by Western blotting (Fig. 1A) on days 0, 1, and 8. GATA-6 protein increased 263 ± 33% between day 0 and day 8. Concurrently, levels of the AT1 cell marker AQP5 increased approximately eightfold (Fig. 1B), consistent with transition to an AT1 cell-like phenotype.
Fig. 1.
Expression of GATA-6 and aquaporin-5 (AQP5) during alveolar epithelial cell (AEC) transdifferentiation in vitro. Protein was harvested from freshly isolated AT2 cells (D0) or from AEC on day 1 (D1) and day 8 (D8) in primary culture. Representative Western blot and densitometric analyses for GATA-6 (A) and AQP5 (B) are shown. Data represent means ± SE (n = 3 experiments). *P < 0.05 compared with day 0 (D0).
Transcriptional activation of AQP5 promoter by GATA-6.
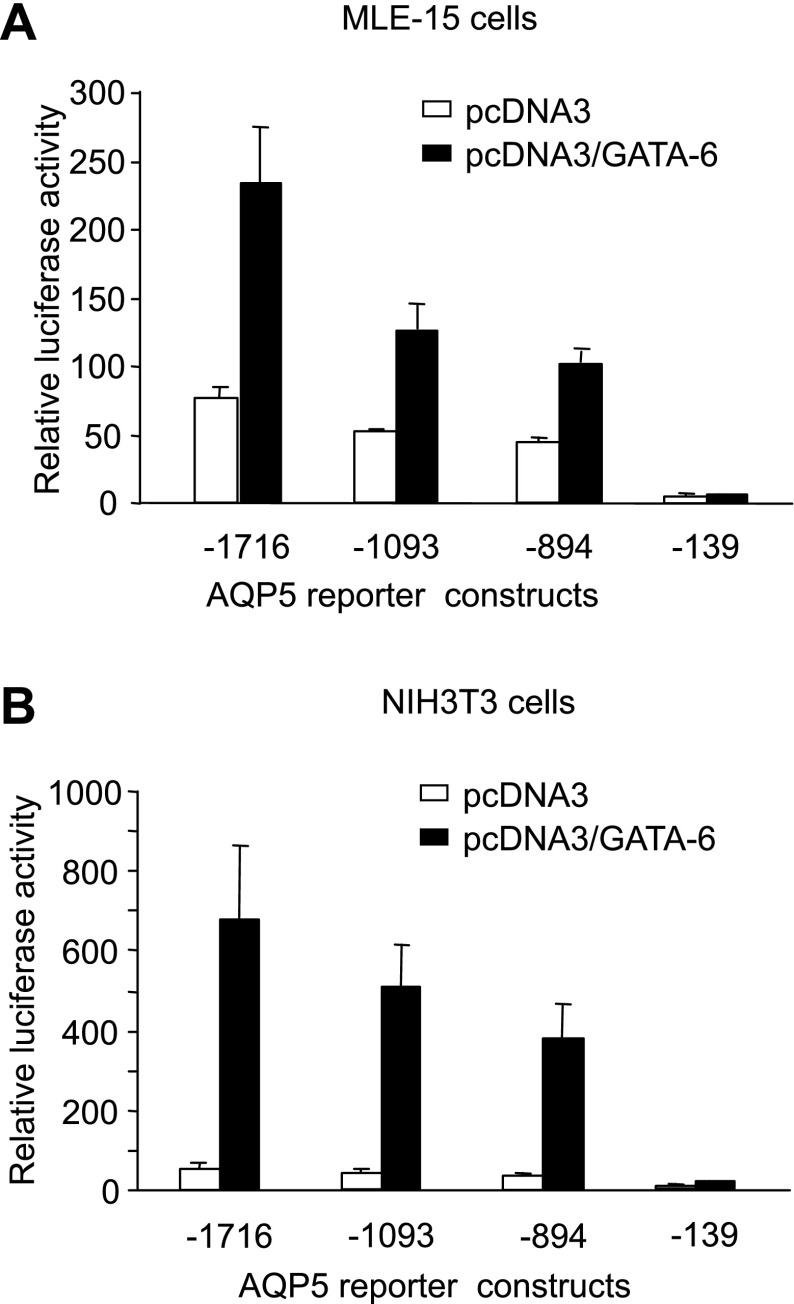
We previously generated a series of luciferase reporter constructs with progressive 5′-deletions (−1,716, −1,097, −894, and −139) of the 5′-proximal sequences regulating the AT1 cell-enriched rat AQP5 gene (3). Cotransfections in MLE-15 and NIH 3T3 cells of these AQP5-luciferase constructs and the GATA-6 expression plasmid pcDNA3/GATA-6 demonstrated trans-activation of the rat AQP5 promoter by GATA-6 compared with the control vector pcDNA3 (Fig. 2). Greatest activation of the AQP5 promoter by GATA-6 was seen with −1716-AQP5-Luc (∼13-fold in NIH 3T3 cells and 3-fold in MLE-15 cells). Trans-activation of AQP5 by GATA-6 declines progressively with serial deletion of the AQP5 promoter in both cell types, suggesting the presence of functional GATA-6 binding sites within the deleted regions. Activation of AQP5 by GATA-6 is greater in NIH 3T3 cells than in MLE-15 cells, likely because of the higher endogenous expression levels of GATA-6 in MLE-15 cells (36).
Fig. 2.
Transcriptional activation of AQP5 promoter by GATA-6. Cotransfections in MLE-15 (A) and NIH 3T3 (B) cells with a series of 5′-deletion constructs of AQP5-luciferase and the GATA-6 expression plasmid pcDNA3/GATA-6, demonstrate trans-activation of the AQP5 promoter by GATA-6 compared with the empty vector pcDNA3. Firefly luciferase activity was measured 24 h after transfection and normalized to Renilla luciferase activity. Data represent means ± SE (n = 4).
GATA-6 interacts with cognate binding sites in the AQP5 promoter.
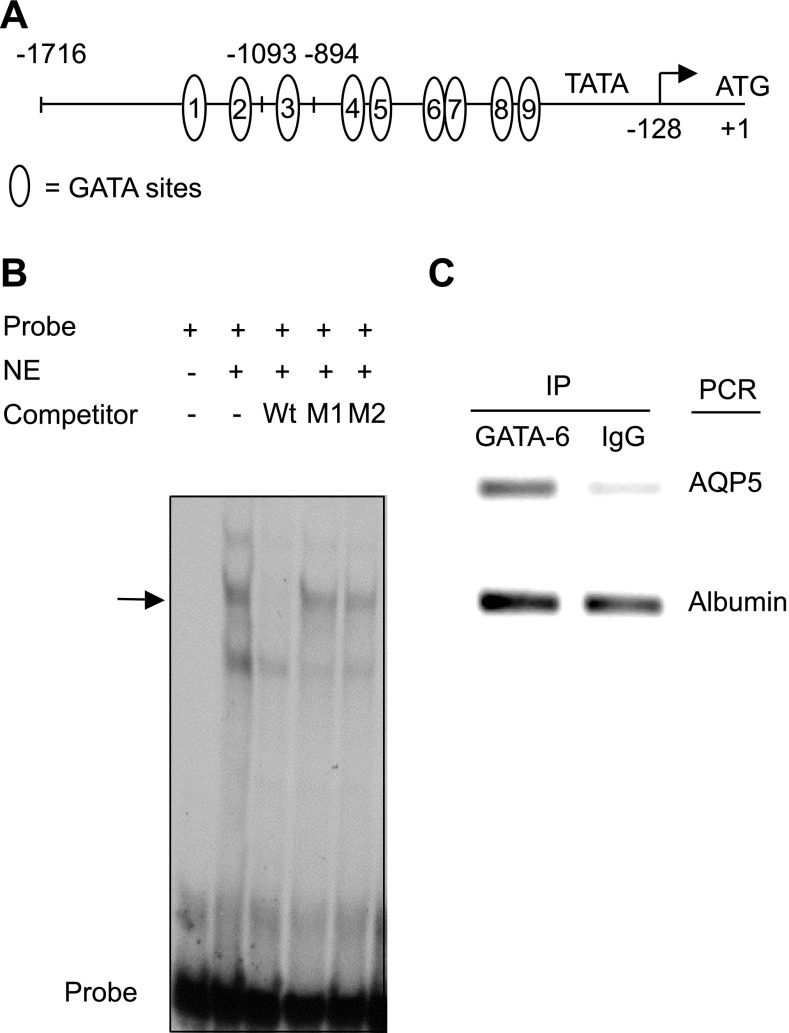
GATA-6 interacts with a DNA sequence containing a core GATA motif, with the order of site preference being GATA > GATT > GATC (35). Analysis of the 1,716-bp AQP5 promoter sequence reveals nine putative GATA-6-binding sites hereby designated nos. 1 through 9 (Fig. 3A). EMSA using oligonucleotide probes spanning these putative GATA consensus sites with nuclear extracts from NIH 3T3 cells transfected with pcDNA3/GATA-6 demonstrated shifted complexes using probes spanning sites 2 (−1,197/−1,174) (Fig. 3B), 8 (−624/−641), and 9 (−601/−620) (data not shown). Binding to site 2 could be competed by wild-type but not mutant cold competitor oligonucleotides. Because GATA-6 binding site 2 is conserved between rat and mouse, we selected this site for further in vivo characterization of GATA-6 binding by chromatin immunoprecipitation (ChIP). As shown in Fig. 3C, the PCR product spanning GATA-6 binding site 2 was amplified from the DNA-protein complex immunoprecipitated by anti-GATA-6 antibody, indicating occupancy of this binding site by GATA-6 and suggesting a role for direct binding of GATA-6 to DNA in activation of AQP5 transcription.
Fig. 3.
GATA-6 interacts with conserved binding site in AQP5 promoter. A: proximal −1,716-bp fragment of the 5′-flanking region of the AT1 cell-specific rat AQP5 gene contains 9 putative GATA-6-binding sites upstream of the transcription start site. The transcriptional start site is designated as −128 relative to the ATG (+1) (3). B: electrophoretic mobility shift assay (EMSA) was performed using nuclear extract (NE) from NIH 3T3 cells transfected with pcDNA3/GATA-6 and a 33P-labeled oligonucleotide probe. A DNA-protein complex (arrow) is formed with probe 2 (spanning −1,197/−1,174 in the AQP5 promoter). Complexes are competed by 100× wild-type (Wt) but not mutated cold competitor (M1 and M2). C: soluble chromatin from MLE-15 cells was immunoprecipitated with GATA-6 antibody or IgG. Coprecipitated DNA was analyzed by PCR using a primer pair specific for the no. 2 conserved GATA-binding site in the AQP5 promoter/enhancer. Albumin was used as nonspecific binding control.
GATA-6-mediated transcriptional activation of AQP5 promoter with mutated or deleted GATA-6-binding sites.
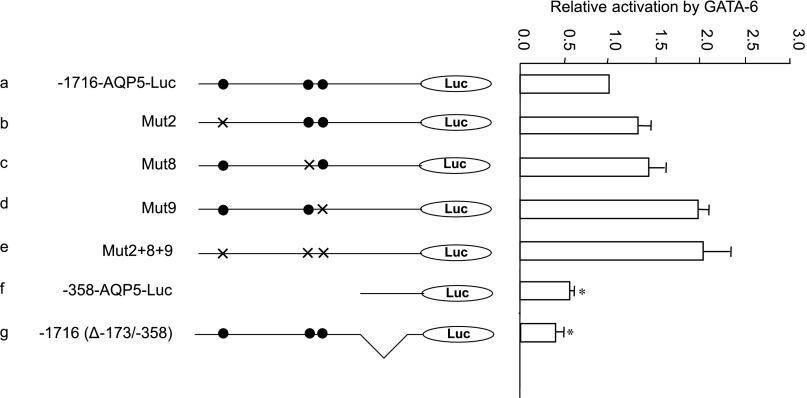
To further assess whether activation of AQP5 transcription by GATA-6 is mediated by direct binding to DNA, GATA-binding sites 2, 8, and 9 of the parental −1716-AQP5-Luc were mutated singly or in combination and cotransfected with pcDNA3/GATA-6 or pcDNA3 in NIH 3T3 cells. In a series of transient transfection assays, the GATA binding site mutants were as active as the parent construct, even when all these sites were mutated simultaneously (Fig. 4, b–e). Furthermore, deletion of the entire −1,716/−358 promoter sequence that encompasses all nine GATA sites did not abolish GATA-6-mediated activation (Fig. 4f). In fact, −358-AQP5-Luc still retains >50% of the GATA-6-mediated activation observed with the parent construct. These data suggest that other elements located within the −358/−128 fragment may play a role in GATA-6-mediated trans-activation of AQP5 via protein-protein interactions. To address this possibility, we deleted the −358/−173 sequence from the parental plasmid to yield −1716(Δ−173/−358)-Luc (Fig. 4g). Transient transfection assays in NIH 3T3 cells showed ∼60% reduction of GATA-6-mediated trans-activation of the AQP5 promoter with internal deletion of this fragment. In fact, GATA-6-mediated trans-activation of AQP5 was reduced from 23-fold in −1716-AQP5-Luc to 9-fold in −1716(Δ−173/−358), further suggesting that the −358/−173 region is important for transcriptional regulation of the AQP5 promoter by GATA-6 despite the absence of putative GATA consensus sites.
Fig. 4.
GATA-6-mediated transcriptional activation of AQP5 promoter-reporter constructs containing mutated or deleted GATA-6-binding sites. NIH 3T3 cells were transiently transfected with reporter constructs (shown on left) together with pcDNA3/GATA-6 or empty vector pcDNA3. GATA-6-mediated activation of the parental plasmid (a) is defined as 1.0. Activation of each of the mutant constructs (b–g) is shown relative to activation of the parental plasmid 1,716-bp-AQP5-luciferase (−1716-AQP5-Luc). Firefly luciferase is normalized to Renilla luciferase activity. Data represent means ± SE (n ≥ 3). *P < 0.05 compared with −1716-AQP5-Luc. •, Wild-type GATA site; ×, mutated GATA site.
−358/−173 AQP5 fragment functions as a transcriptional enhancer.
Internal deletion of −358/−173 also reduced basal transcriptional activity relative to the parental −1716-AQP5-Luc construct, suggesting that, consistent with previous observations, this region functions as a transcriptional enhancer of AQP5 (3). To further examine putative enhancer function of the −358/−173 fragment, this sequence was cloned upstream of the minimal promoter −173-AQP5-Luc construct in both forward and reverse orientations. In addition, two and three copies of the −358/−173 fragment were cloned upstream of −173-AQP5-Luc or TK-Luc (Fig. 5A). In transient transfection assays in both MLE-15 (Fig. 5B) and NIH 3T3 (Fig. 5C) cells, one copy of the −358/−173 AQP5 fragment enhanced transcriptional activity of −173-AQP5-Luc when placed in either forward or reverse orientations. In contrast, this fragment did not enhance activity of the TK viral promoter, indicating promoter specificity of this enhancer element. Transcriptional activity of −173-AQP5-Luc was not further enhanced by the insertion of additional copies of the −358/−173-AQP5 fragment in MLE-15 cells. In NIH 3T3 cells, insertion of two and three copies of the −358/−173 AQP5 fragment upstream of the AQP5 minimal promoter led to an increase in promoter activity. These data confirm that the −358/−173 fragment functions as an enhancer of AQP5 transcription. Lack of increased transcriptional activity in MLE-15 cells with multiple copies of the enhancer fragment may be due to depletion of other cell-specific TF required for AQP5 transcription in MLE-15 cells and not in NIH 3T3 cells (which do not express endogenous AQP5).
Fig. 5.
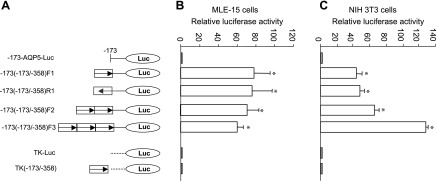
Transcriptional enhancement by −358/−173 AQP5 fragment in MLE-15 and NIH 3T3 cells. A: schematic diagram of DNA constructs showing insertion of single or multiple copies of −358/−173 AQP5 fragment in either forward or reverse orientations upstream of the AQP5 (−173-AQP5-Luc) or TK (TK-Luc) minimal promoters. B: transient transfections were performed in MLE-15 cells using the constructs shown in A. C: transient transfections were performed in NIH 3T3 cells using the constructs shown in A. Data represent means ± SE (n = 3). *P < 0.05 compared with −173-AQP5-Luc. Firefly luciferase was normalized to Renilla luciferase activity.
Activation of −358-AQP5-Luc by GATA-6.
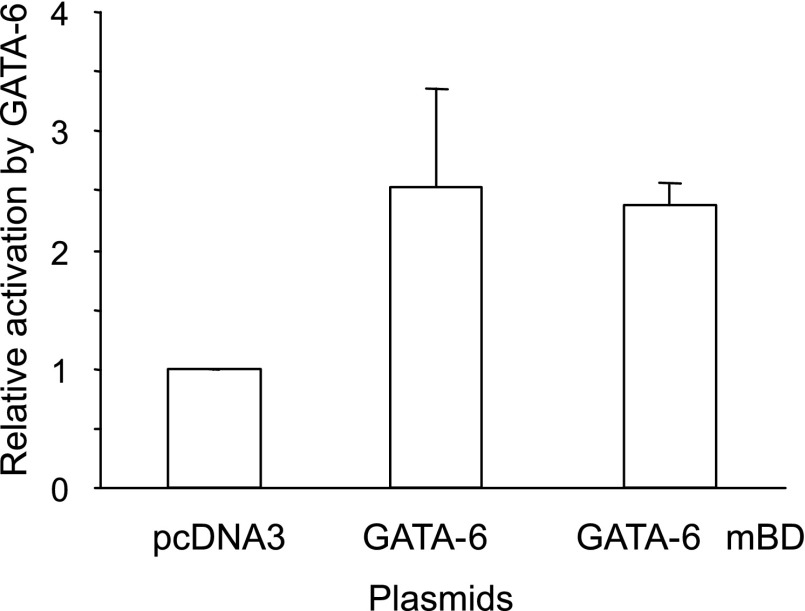
Analysis of the AQP5 promoter sequence between nucleotides −358 and −128 relative to the ATG (which includes the minimal promoter and the −173/−358 enhancer fragment) revealed multiple putative Sp1, but no putative GATA, consensus sites (3). This suggests that GATA-6 activation of this promoter/enhancer region may be mediated not through direct binding of GATA-6 to DNA but rather indirectly through interactions between GATA-6 and Sp1. To further investigate this possibility, we generated pGATA-6 mBD, a GATA-6 expression plasmid, in which Cys293 and Ala294 are mutated to disrupt the interaction of GATA-6 with DNA (45). In transient transfection assays in NIH 3T3 cells, both GATA-6 and GATA-6 mBD similarly activate −358-AQP5-Luc ∼2.5-fold (Fig. 6), further suggesting that GATA-6-mediated activation of this enhancer region is independent of its binding to DNA.
Fig. 6.
GATA-6 activation of −358/−173 AQP5 enhancer fragment. GATA-6 and a GATA-6 DNA-binding mutant (GATA-6 mDB) were cotransfected with the −358/−173 transcriptional enhancer in NIH 3T3 cells. Firefly luciferase was normalized to D234 Renilla luciferase activity. Data represent means ± SE (n = 6).
Trancriptional activation of −358-AQP5-Luc by Sp1.
To further explore the possibility that GATA-6 activation of AQP5 is mediated through interactions with Sp1, we evaluated the ability of Sp1 alone to activate −358-AQP5-Luc. In transient transfection assays in S2 cells (which lack Sp1), cotransfected Sp1 activates transcription of −358-AQP5-Luc in a dose-dependent manner up to approximately sevenfold with 100 ng of Sp1 (Fig. 7A), confirming that Sp1 can directly activate AQP5.
Fig. 7.
GATA-6 activates −358-AQP5 promoter by interaction with Sp1. A: in transient transfection assays in S2 cells that lack Sp1, Sp1 activates −358-AQP5-Luc in a dose-dependent manner. Firefly luciferase was measured 24 h after transfection and normalized to β-galactosidase activity. Data represent means ± SE. *P < 0.05 compared with 0 ng of Sp1 (n = 3). B: NIH 3T3 cells were transfected with either Sp1-specific or control nontargeting small-interfering RNA (siRNA). Simultaneously, cells were transfected with −358-AQP5-Luc and pcDNA3/GATA-6 or empty vector pcDNA3. After 48 h, cells were harvested for Western analysis and luciferase assay. Firefly luciferase was normalized to total protein. Data represent means ± SE (n ≥ 3). *P < 0.05 compared with all other constructs. C: nuclear extract from MLE-15 cells was immunoprecipitated (IP) using anti-GATA-6 antibody or IgG. Sp1 associated with GATA-6 was detected by anti-Sp1 antibody as shown in this representative Western blot (WB). NE from MLE-15 cells is used as positive control for Sp1 Western blot.
Effect of Sp1 siRNA on trans-activation of −358-AQP5-Luc by GATA-6.
To further investigate the role of Sp1 in GATA-6 activation of −358-AQP5-Luc, cotransfection of −358-AQP5-Luc with pcDNA3/GATA-6 was performed in NIH 3T3 cells in which Sp1 activity was inhibited by Sp1 siRNA. As shown in Fig. 7B, siRNA for Sp1 reduced expression of endogenous Sp1 in NIH 3T3 cells by ∼65% compared with control nontarget siRNA. Furthermore, GATA-6-mediated activation of AQP5 transcription was almost completely abolished following knockdown of Sp1. These data further suggest that GATA-6 activation of AQP5 is mediated through interaction with Sp1.
Coimmunoprecipitation of GATA-6 and Sp1.
Interaction between GATA-6 and Sp1 was investigated by coimmunoprecipitation in MLE-15 cells. As shown in Fig. 7C, Sp1 was identified in the protein complex immunoprecipitated by anti-GATA-6 antibody, but not by IgG control.
Synergistic activation of −358-AQP5-Luc by GATA-6 and Sp1.
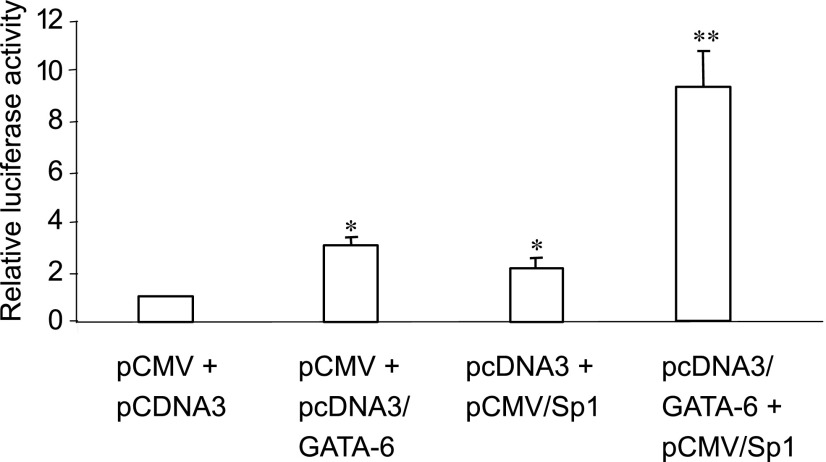
To further investigate interactions between GATA-6 and Sp1, cotransfection assays were performed in NIH 3T3 cells with GATA-6 and Sp1 separately or in combination together with −358-AQP5-Luc. As shown in Fig. 8, GATA-6 alone activates −358-AQP5-Luc approximately threefold, whereas Sp1 activates −358-AQP5-Luc approximately twofold. Cotransfection of GATA-6 and Sp1 increases reporter activity approximately ninefold. The calculated interaction response is 0.23, demonstrating synergistic activation of −358-AQP5-Luc by GATA-6 and Sp1, consistent with interaction between these TF as suggested by siRNA and coimmunoprecipitation experiments.
Fig. 8.
Synergistic activation of −358-AQP5 promoter by GATA-6 and Sp1. GATA-6 and Sp1 singly and in combination were cotransfected with −358-AQP5-Luc in NIH 3T3 cells. Synergistic activation of the AQP5 reporter construct is observed with cotransfection of both GATA-6 and Sp1. Firefly luciferase was measured 24 h after transfection and normalized to D234 Renilla luciferase activity. Data represent means ± SE (n = 9). *P < 0.05 compared with pCMV + pcDNA3. **P < 0.05 compared with all other constructs.
DISCUSSION
Mice expressing a GATA-6-Engrailed dominant-negative fusion protein in the distal lung epithelium during development lack detectable AT1 cells and demonstrate attenuated expression of the AT1 cell-enriched gene AQP5, suggesting an important role for GATA-6 in regulation of AT1 cell differentiation (45). GATA-6 was also shown to activate the AQP5 promoter in vitro, suggesting a role for GATA-6 in regulating gene expression in AT1 cells. To gain further insight into transcriptional regulation of gene expression in AT1 cells, we investigated mechanisms underlying GATA-6-mediated activation of AQP5. We demonstrated upregulation of GATA-6 in AEC in primary culture concurrent with an increase in AQP5 and transition to an AT1 cell-like phenotype, suggesting that temporal and spatial regulation of GATA-6 contributes to activation of AQP5 transcription in AT1 cells, whereas GATA-6 activates transcription of a −1,716-bp AQP5-luciferase construct in both MLE-15 and NIH 3T3 cells.
Previous studies demonstrated that GATA-6 can regulate gene expression by binding directly to the promoter/enhancer of target genes via its DNA-binding domain, as well as indirectly through interactions with other cell-specific and ubiquitous TF that in turn bind to DNA (8, 11, 26, 33). Because the proximal −1,716-bp AQP5 promoter region encompasses a number of putative GATA-binding sites, we initially evaluated the ability of GATA-6 to activate AQP5 through direct binding to DNA. Although we demonstrated binding of GATA-6 by both EMSA and ChIP to putative GATA-binding sites, mutation of these sites, individually or in combination, did not prevent GATA-6-mediated activation of the AQP5 promoter. Furthermore, a deletion construct in which all putative GATA-6-binding sites were removed still retained >50% of GATA-6-mediated activation compared with the parent construct, suggesting that interactions between GATA-6 and other TF (e.g., Sp1) that themselves interact with DNA may play a critical role in regulating AQP5 transcription.
Further characterization of the proximal AQP5 promoter identified a GC-rich enhancer element located between −358 and −173 bp that encompasses multiple putative Sp1 consensus sites (3). Because functional Sp1 sites in the proximal AQP5 promoter have been previously suggested to be important in activation of AQP5 by all-trans-retinoic acid (31) and because GATA-6 is known to interact combinatorially with other TF, including Sp1, we explored the role of interactions between GATA-6 and Sp1 in mediating activation of AQP5. In transient transfection assays, both basal and GATA-6-mediated activation of an AQP5 luciferase reporter were significantly reduced after internal deletion of this proximal enhancer element from the parental plasmid −1716-AQP5-Luc. Transient transfections in S2 cells that lack endogenous Sp1 confirmed the ability of Sp1 to directly activate the AQP5 promoter. In addition, coimmunoprecipitation showed an association between GATA-6 and Sp1 in MLE-15 cells, whereas knockdown of Sp1 using siRNA reduced GATA-6-mediated activation of −358-AQP5-Luc. Cotransfections further demonstrated synergistic activation of −358-AQP5-Luc by GATA-6 and Sp1. In the absence of putative GATA-6 consensus motifs in the proximal enhancer, these data suggest that interaction between GATA-6 and Sp1 plays an important role in GATA-6-mediated activation of AQP5 transcription.
Combinatorial interactions between GATA factors and other TF contribute to their level of activity and regulation of cell-specific gene expression (30, 39, 40, 42, 43). In particular, functional interactions between GATA factors and Sp1, a member of the Sp-multigene family and Krüppel-like zinc finger superfamily, have been shown to be important for regulation of tissue-specific expression (13, 20, 23). In this regard, GATA-1 has been shown to interact with Sp1 to regulate the erythropoietin receptor promoter (9), whereas GATA-6 modulates tissue-specific transcription of the human gene for P450c17 by direct interaction with Sp1 (11). In the case of the P450c17 promoter, there was no requirement for direct binding of GATA-6 to the promoter, whereas transcriptional synergy for the erythropoietin receptor promoter was dependent on the integrity of both GATA and Sp1 binding sites at low concentration of GATA-1 and Sp1 but independent of the GATA-binding site at high concentrations (26). GATA-2 and Sp1 have also been shown to cooperate functionally in regulation of endothelial nitric oxide synthase expression (46). Although EMSA provided evidence for binding of GATA-2 and Sp1 to their own cognate sites, the GATA cis-element was only found to be operational when the Sp1 site was intact. Several other examples of cooperative interactions between GATA factors and Sp1 have been described (15), supporting a role for cooperative GATA-Sp1 interactions in regulating tissue- and cell-specific gene expression. Interactions between GATA factors and Sp1 can be GATA-motif dependent or independent, and in the latter case Sp1 serves to recruit the GATA factor to the promoter.
In the current study, mutation of putative GATA-binding sites in −1716-AQP5-Luc individually or in combination did not reduce transcriptional activity and was in fact associated with a slight increase in luciferase activity. Similar findings were observed in studies of the P450c17 promoter in which mutating the GATA binding site was suggested to increase the availability of GATA factors to interact with Sp1, resulting in increased activation (11). Together with residual activation of the internally deleted construct −1716(Δ−173/−358)-Luc by GATA-6, these results suggest that there may be additional functional Sp1 sites upstream of −358 that are involved in regulation of AQP5 transcriptional activity by GATA-6.
AQP5 is expressed in salivary and lacrimal gland, but within distal lung is specifically expressed in AT1 cells (18, 29). During development, AQP5 is first expressed at embryonic day 17.5, concurrent with the appearance of AT1 cells. In addition, AQP5 is upregulated during transition to the AT1 cell phenotype, concurrent with acquisition of a type I cell-like morphology (4). This close temporal and spatial relationship to the type I cell phenotype both in vitro and in vivo suggests its utility as a marker of AT1 cells with which to investigate mechanisms of transcriptional regulation in AT1 cells. A role for Sp1 in regulation of AQP5 expression by all-trans-retinoic acid has been previously suggested (31) through increased binding of Sp1 to an Sp1/Sp3-binding element in the basal rat AQP5 promoter proximal to the enhancer region characterized in the current study (31).
Several studies have demonstrated that Sp1 transcriptional activity can be modulated by epigenetic modifications that involve either chromatin remodeling or promoter methylation (12, 17). Histone deacetylase (HDAC) inhibitors have been shown to enhance expression of several genes by acting through the Sp1 site to inhibit HDAC activity recruited by Sp1 (10, 12, 16). In a recent report, Nomura et al. (31) demonstrated that hypomethylation of the proximal AQP5 promoter was associated with increased binding of Sp1 and increased expression of AQP5 under basal conditions, suggesting that differential methylation of CpG islands in the proximal AQP5 promoter may play a role in regulating its expression by modulating access to and binding of Sp1 in a cell-specific fashion. The role of epigenetic modifications of the AQP5 promoter in recruitment of GATA-6 leading to synergistic interactions and increased binding at Sp1 sites remains to be determined.
In summary, we have demonstrated that GATA-6-mediated activation of AQP5 is largely mediated through interactions with Sp1. GATA-6 and Sp1 synergistically activate a truncated −358-AQP5-Luc fragment that contains Sp1 consensus binding sites but no GATA-6 motifs. Synergistic activation was almost completely abolished by depleting Sp1 using siRNA, further suggesting that GATA-6-mediated activation of AQP5 occurs independently of GATA-6 binding to DNA. This is further supported by the finding that a GATA-6 protein in which the DNA-binding domain has been mutated retains the ability to activate AQP5 to a similar extent as the native protein. Although we cannot exclude the possibility that the proximal −358 fragment encompasses cryptic GATA-binding sites, almost complete abrogation of GATA-6-mediated activation following depletion of Sp1 strongly suggests that the GATA effects on the proximal promoter are mediated predominantly through interactions with Sp1. This is further supported by demonstration of an association between endogenous GATA-6 and Sp1 in MLE-15 cells and suggests that DNA-bound Sp1 functions to recruit GATA-6 to cooperatively activate AQP5. Consistent with these findings, in vitro interactions have been previously demonstrated between GATA-6 and the zinc finger domains of Sp1 (26). Furthermore, the increase in GATA-6 levels during transition from the AT2 to AT1 cell phenotype likely facilitates synergistic interactions between GATA-6 and Sp1, leading to increased AQP5 expression in AT1 cells. Sp1 has also been shown to play a role in regulation of expression of T1α, another AT1 cell-enriched gene (7, 34), suggesting that recruitment of TF to the proximal promoter through interactions with Sp1 may be an important mechanism mediating transcriptional regulation and cell-specific gene expression in AT1 cells.
GRANTS
This work was supported by the Hastings Foundation and National Institutes of Health Research Grants DE-10742, DE-14183, HL-38578, HL-38621, HL-38658, HL-56590, HL-60231, HL-62659, HL-64365, and HL-73471.
Acknowledgments
We note with appreciation the expert technical assistance of Juan Ramon Alvarez and Monica Flores. E. D. Crandall is Hastings Professor and Kenneth T. Norris Jr. Chair of Medicine, B. Frenkel holds the LaBriola Chair in Genetic Orthopaedic Research, and Z. Borok holds the Edgington Chair in Medicine.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Borgnia M, Nielsen S, Engel A, Agre P. Cellular and molecular biology of the aquaporin water channels. Annu Rev Biochem 68: 425–458, 1999. [DOI] [PubMed] [Google Scholar]
- 2.Borok Z, Danto SI, Zabski SM, Crandall ED. Defined medium for primary culture de novo of adult rat alveolar epithelial cells. In Vitro Cell Dev Biol Anim 30A: 99–104, 1994. [DOI] [PubMed] [Google Scholar]
- 3.Borok Z, Li X, Fernandes VF, Zhou B, Ann DK, Crandall ED. Differential regulation of rat aquaporin-5 promoter/enhancer activities in lung and salivary epithelial cells. J Biol Chem 275: 26507–26514, 2000. [DOI] [PubMed] [Google Scholar]
- 4.Borok Z, Lubman RL, Danto SI, Zhang XL, Zabski SM, King LS, Lee DM, Agre P, Crandall ED. Keratinocyte growth factor modulates alveolar epithelial cell phenotype in vitro: expression of aquaporin 5. Am J Respir Cell Mol Biol 18: 554–561, 1998. [DOI] [PubMed] [Google Scholar]
- 5.Bruno MD, Korfhagen TR, Liu C, Morrisey EE, Whitsett JA. GATA-6 activates transcription of surfactant protein A. J Biol Chem 275: 1043–1049, 2000. [DOI] [PubMed] [Google Scholar]
- 6.Burch JB Regulation of GATA gene expression during vertebrate development. Semin Cell Dev Biol 16: 71–81, 2005. [DOI] [PubMed] [Google Scholar]
- 7.Cao YX, Ramirez MI, Williams MC. Enhanced binding of Sp1/Sp3 transcription factors mediates the hyperoxia-induced increased expression of the lung type I cell gene T1alpha. J Cell Biochem 89: 887–901, 2003. [DOI] [PubMed] [Google Scholar]
- 8.Charron F, Paradis P, Bronchain O, Nemer G, Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol Cell Biol 19: 4355–4365, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin K, Oda N, Shen K, Noguchi CT. Regulation of transcription of the human erythropoietin receptor gene by proteins binding to GATA-1 and Sp1 motifs. Nucleic Acids Res 23: 3041–3049, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davie JR Inhibition of histone deacetylase activity by butyrate. J Nutr 133: 2485S–2493S, 2003. [DOI] [PubMed] [Google Scholar]
- 11.Fluck CE, Miller WL. GATA-4 and GATA-6 modulate tissue-specific transcription of the human gene for P450c17 by direct interaction with Sp1. Mol Endocrinol 18: 1144–1157, 2004. [DOI] [PubMed] [Google Scholar]
- 12.Huang W, Tan D, Wang X, Han S, Tan J, Zhao Y, Lu J, Huang B. Histone deacetylase 3 represses p15(INK4b) and p21(WAF1/cip1) transcription by interacting with Sp1. Biochem Biophys Res Commun 339: 165–171, 2006. [DOI] [PubMed] [Google Scholar]
- 13.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol 4: 206, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keijzer R, van Tuyl M, Meijers C, Post M, Tibboel D, Grosveld F, Koutsourakis M. The transcription factor GATA6 is essential for branching morphogenesis and epithelial cell differentiation during fetal pulmonary development. Development 128: 503–511, 2001. [DOI] [PubMed] [Google Scholar]
- 15.Kiela PR, LeSueur J, Collins JF, Ghishan FK. Transcriptional regulation of the rat NHE3 gene. Functional interactions between GATA-5 and Sp family transcription factors. J Biol Chem 278: 5659–5668, 2003. [DOI] [PubMed] [Google Scholar]
- 16.Kim S, Kang JK, Kim YK, Seo DW, Ahn SH, Lee JC, Lee CH, You JS, Cho EJ, Lee HW, Han JW. Histone deacetylase inhibitor apicidin induces cyclin E expression through Sp1 sites. Biochem Biophys Res Commun 342: 1168–1173, 2006. [DOI] [PubMed] [Google Scholar]
- 17.Kim YH, Lim JH, Lee TJ, Park JW, Kwon TK. Expression of cyclin D3 through Sp1 sites by histone deacetylase inhibitors is mediated with protein kinase C-delta (PKC-delta) signal pathway. J Cell Biochem 101: 987–995, 2007. [DOI] [PubMed] [Google Scholar]
- 18.King LS, Nielsen S, Agre P. Aquaporins in complex tissues. I. Developmental patterns in respiratory and glandular tissues of rat. Am J Physiol Cell Physiol 273: C1541–C1548, 1997. [DOI] [PubMed] [Google Scholar]
- 19.Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol 13: 4011–4022, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolell KJ, Crawford DL. Evolution of Sp transcription factors. Mol Biol Evol 19: 216–222, 2002. [DOI] [PubMed] [Google Scholar]
- 21.Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development 126: 723–732, 1999. [PubMed] [Google Scholar]
- 22.Li C, Ling X, Yuan B, Minoo P. A novel DNA element mediates transcription of Nkx2.1 by Sp1 and Sp3 in pulmonary epithelial cells. Biochim Biophys Acta 1490: 213–224, 2000. [DOI] [PubMed] [Google Scholar]
- 23.Li L, He S, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochem Cell Biol 82: 460–471, 2004. [DOI] [PubMed] [Google Scholar]
- 24.Liu C, Glasser SW, Wan H, Whitsett JA. GATA-6 and thyroid transcription factor-1 directly interact and regulate surfactant protein-C gene expression. J Biol Chem 277: 4519–4525, 2002. [DOI] [PubMed] [Google Scholar]
- 25.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol 13: 3999–4010, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol Cell Biol 15: 2437–2447, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molkentin JD The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem 275: 38949–38952, 2000. [DOI] [PubMed] [Google Scholar]
- 28.Morrisey EE, Ip HS, Lu MM, Parmacek MS. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol 177: 309–322, 1996. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen S, King LS, Christensen BM, Agre P. Aquaporins in complex tissues. II. Subcellular distribution in respiratory and glandular tissues of rat. Am J Physiol Cell Physiol 273: C1549–C1561, 1997. [DOI] [PubMed] [Google Scholar]
- 30.Nishida W, Nakamura M, Mori S, Takahashi M, Ohkawa Y, Tadokoro S, Yoshida K, Hiwada K, Hayashi K, Sobue K. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J Biol Chem 277: 7308–7317, 2002. [DOI] [PubMed] [Google Scholar]
- 31.Nomura J, Horie I, Seto M, Nagai K, Hisatsune A, Miyata T, Isohama Y. All-trans retinoic acid increases expression of aquaporin-5 and plasma membrane water permeability via transactivation of Sp1 in mouse lung epithelial cells. Biochem Biophys Res Commun 351: 1048–1053, 2006. [DOI] [PubMed] [Google Scholar]
- 32.Peterkin T, Gibson A, Loose M, Patient R. The roles of GATA-4, -5 and -6 in vertebrate heart development. Semin Cell Dev Biol 16: 83–94, 2005. [DOI] [PubMed] [Google Scholar]
- 33.Pikkarainen S, Tokola H, Kerkela R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res 63: 196–207, 2004. [DOI] [PubMed] [Google Scholar]
- 34.Ramirez MI, Rishi AK, Cao YX, Williams MC. TGT3, thyroid transcription factor I, and Sp1 elements regulate transcriptional activity of the 1.3-kilobase pair promoter of T1alpha, a lung alveolar type I cell gene. J Biol Chem 272: 26285–26294, 1997. [DOI] [PubMed] [Google Scholar]
- 35.Sakai Y, Nakagawa R, Sato R, Maeda M. Selection of DNA binding sites for human transcriptional regulator GATA-6. Biochem Biophys Res Commun 250: 682–688, 1998. [DOI] [PubMed] [Google Scholar]
- 36.Shaw-White JR, Bruno MD, Whitsett JA. GATA-6 activates transcription of thyroid transcription factor-1. J Biol Chem 274: 2658–2664, 1999. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu R, Yamamoto M. Gene expression regulation and domain function of hematopoietic GATA factors. Semin Cell Dev Biol 16: 129–136, 2005. [DOI] [PubMed] [Google Scholar]
- 38.Umenishi F, Carter EP, Yang B, Oliver B, Matthay MA, Verkman AS. Sharp increase in rat lung water channel expression in the perinatal period. Am J Respir Cell Mol Biol 15: 673–679, 1996. [DOI] [PubMed] [Google Scholar]
- 39.van Wering HM, Huibregtse IL, van der Zwan SM, de Bie MS, Dowling LN, Boudreau F, Rings EH, Grand RJ, Krasinski SD. Physical interaction between GATA-5 and hepatocyte nuclear factor-1alpha results in synergistic activation of the human lactase-phlorizin hydrolase promoter. J Biol Chem 277: 27659–27667, 2002. [DOI] [PubMed] [Google Scholar]
- 40.Vanpoucke G, Goossens S, De Craene B, Gilbert B, van Roy F, Berx G. GATA-4 and MEF2C transcription factors control the tissue-specific expression of the alphaT-catenin gene CTNNA3. Nucleic Acids Res 32: 4155–4165, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viger RS, Taniguchi H, Robert NM, Tremblay JJ. Role of the GATA family of transcription factors in andrology. J Androl 25: 441–452, 2004. [DOI] [PubMed] [Google Scholar]
- 42.Wada H, Hasegawa K, Morimoto T, Kakita T, Yanazume T, Abe M, Sasayama S. Calcineurin-GATA-6 pathway is involved in smooth muscle-specific transcription. J Cell Biol 156: 983–991, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wada H, Hasegawa K, Morimoto T, Kakita T, Yanazume T, Sasayama S. A p300 protein as a coactivator of GATA-6 in the transcription of the smooth muscle-myosin heavy chain gene. J Biol Chem 275: 25330–25335, 2000. [DOI] [PubMed] [Google Scholar]
- 44.Wikenheiser KA, Vorbroker DK, Rice WR, Clark JC, Bachurski CJ, Oie HK, Whitsett JA. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci USA 90: 11029–11033, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang H, Lu MM, Zhang L, Whitsett JA, Morrisey EE. GATA6 regulates differentiation of distal lung epithelium. Development 129: 2233–2246, 2002. [DOI] [PubMed] [Google Scholar]
- 46.Zhang R, Min W, Sessa WC. Functional analysis of the human endothelial nitric oxide synthase promoter. Sp1 and GATA factors are necessary for basal transcription in endothelial cells. J Biol Chem 270: 15320–15326, 1995. [DOI] [PubMed] [Google Scholar]
- 47.Zheng J, Kitajima K, Sakai E, Kimura T, Minegishi N, Yamamoto M, Nakano T. Differential effects of GATA-1 on proliferation and differentiation of erythroid lineage cells. Blood 107: 520–527, 2006. [DOI] [PubMed] [Google Scholar]