Abstract
Certain guanine-rich sequences are capable of forming higher order structures known as G-quadruplexes. Moreover, particular genomic regions in a number of highly divergent organisms are enriched for such sequences, raising the possibility that G-quadruplexes form in vivo and affect cellular processes. While G-quadruplexes have been rigorously studied in vitro, whether these structures actually form in vivo and what their roles might be in the context of the cell have remained largely unanswered questions. Recent studies suggest that G-quadruplexes participate in the regulation of such varied processes as telomere maintenance, transcriptional regulation and ribosome biogenesis. Here we review studies aimed at elucidating the in vivo functions of quadruplex structures, with a particular focus on findings in yeast. In addition, we discuss the utility of yeast model systems in the study of the cellular roles of G-quadruplexes.
Keywords: G-quadruplex, yeast, telomere, transcription, chromatin
Introduction
The genomes and transcriptomes of many organisms, including those as diverse as E. coli and humans, contain a number of G-rich sequences that, at least in vitro and perhaps in vivo, are capable of forming structures known as G-quadruplexes (G4-DNA and G4-RNA, respectively). These structures are composed of stacked associations of G-quartets, which are planar assemblies of four Hoogsteen-bonded guanines (Figs. 1A and B) [1, 2]. G4 structures can arise through the interactions of guanines present on a single nucleic acid strand (intra-molecular) or multiple strands (inter-molecular). Beyond hydrogen bonding among guanines, the stability of quadruplexes derives from π-orbital interactions among stacked quartets as well as coordination by quartets of centrally located cations (e.g. Na+ or K+). Thus a minimum of two adjacent quartets, but ideally three or more, is required for stable quadruplex formation. G4 structures are stable under physiologic salt and pH conditions in vitro, and some have higher melting temperatures than the duplex DNA that would be formed by providing the complementary strand. There is a high degree of polymorphism among different G4 structures. In principal, 16 different quartet structures can form, which are distinguished by the patterns of glycosidic bond angle of the guanines [3]. Further, the number of stacked quartets, the number and polarity of the phosphodiester backbone strands from which the guanines derive, the type of coordinated cations, and the length, sequence and connectivity of intervening loops may vary [1].
Figure 1.
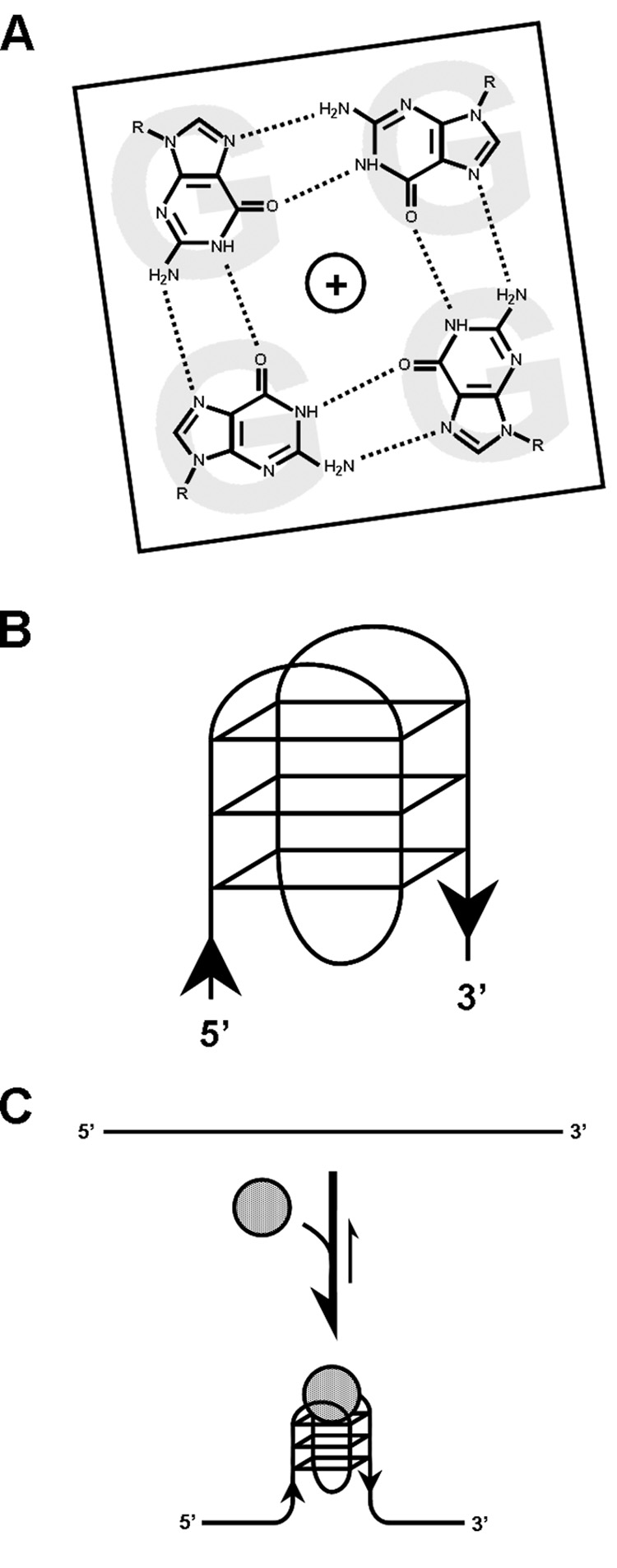
G-quadruplex structure and ligand binding. (A) Structure of a G-quartet, with hydrogen bonds between guanines indicated by dotted lines. (B) Schematic of one type of unimolecular G-quadruplex that is composed of three planar G-quartet structures, as shown. The orientation (5’->3’) of the phosphodiester backbone is indicated by black arrows. Many other types of G-quadruplex folds are possible. (C) Binding of certain G-quadruplex interacting factors (grey circles), be they proteins or small molecular weight ligands, can promote and/or stabilize the formation of G-quadruplexes from nucleic acid strands with quadruplex-forming potential (QFP). Further, they might inhibit access of other factors to G-quadruplexes.
Although the structures of G-quadruplexes have been well studied in vitro, if, when and where they form in vivo and how they might affect cell biology have remained key questions. The structural heterogeneity of quadruplexes makes it difficult to obtain universal rules to predict their formation or probes to test for their presence. Nonetheless, a good deal of information demonstrating or strongly suggesting their functions in vivo has emerged in recent years. For example, telomeric G4-DNA has been proven to exist in Stylonichia lemnae [4, 5], and sequences with intramolecular quadruplex-forming potential (QFP) have been shown to be highly overrepresented in the promoter regions of diverse organisms and to be connected with control of gene expression [6–12]. In addition, a number of small molecule ligands have been identified that bind to and stabilize quadruplexes (Fig. 1C) [13–21], and some of these have been found to affect expression from QFP-containing loci, indicating that QFP sequences can adopt G4 conformations.
Here we review findings pertaining to the in vivo functions of G-quadruplexes, with an emphasis on findings in yeast. We begin by highlighting the ways in which yeast model systems can help identify and dissect the cellular roles of quadruplex structures. For readers particularly interested in findings outside of yeast, we recommend these outstanding reviews [22, 23]. Yeast genetic tools have significant potential for revealing the full extent to which G-quadruplexes regulate biological processes, as well as for revealing underlying mechanisms.
Genetic systems for the analysis of G-quadruplexes in yeast
S. cerevisiae offers several genetic systems that could facilitate exploration of the in vivo functions of G-quadruplexes. Although none is unique to yeast, the ease with which they can be carried out in this single celled eukaryote make it an ideal choice for these studies. We first describe these systems, and in the second half of the review describe findings obtained from their use.
Synthetic enhancement and suppressor screens
A powerful method for exploring the function of a gene is to ask how phenotypes caused by mutations in the gene are influenced by mutations at other loci. Synthetic enhancement refers to two mutations that together yield a phenotype more severe than that of the sum of the phenotypes caused by the individual mutations (Fig. 2A). In the case of null alleles, the corresponding genes typically function in distinct biological pathways that operate in parallel to achieve similar outcomes. In the case of partial loss of function alleles, synthetic enhancement may instead identify factors that work together in a protein complex or facilitate different steps in a pathway. In contrast to enhancer mutations, suppressor mutations lessen the severity of a phenotype caused by the first mutation (Fig 2B), and can reflect activation of a step in a biological pathway downstream of impairment by the first mutation, activation of a process that directly counteracts the effects of the first mutation, or activation of a parallel pathway that compensates for the effects of first mutation. While the interpretation of synthetic enhancement and suppressor screens is thus sometimes not straightforward, such screens can provide important clues to the biological functions of particular genes and illuminate the network of pathways in which they function. Instead of asking how two mutations interact with one another, it is also possible to ask how the effects of a drug interact with different mutations. For example, mutations that enhance the effect of the drug might identify factors that metabolize the drug, destabilize the drug target, or participate in a biochemical pathway that compensates for a toxic effect of the drug. The development in recent years of advanced strategies for combining mutations and for drug screening, together with microarray-based readouts, have dramatically improved the ease, accuracy and precision with which these screens can be accomplished in yeast [24–27]. For example, the epistatic mini array profiling (E-MAP) technique identifies pairs of mutations that have quantitatively similar enhancing or suppressing genetic interactions and can thus provide information on which proteins cooperate in particular biochemical pathways [28]. An E-MAP with all pairwise combinations of 743 mutant alleles of genes involved in chromosome biology yielded numerous insights into the mechanisms of chromosome maintenance [24]. In another example, a wealth of chemical-genetic interactions were obtained by exposing a pool of all viable yeast deletion mutants, each marked with a unique sequence tag, to 82 separate compounds; the relative growth of each mutant in presence of each compound was assessed by quantitative PCR amplification of the tags and microarray hybridization [29].
Figure 2.
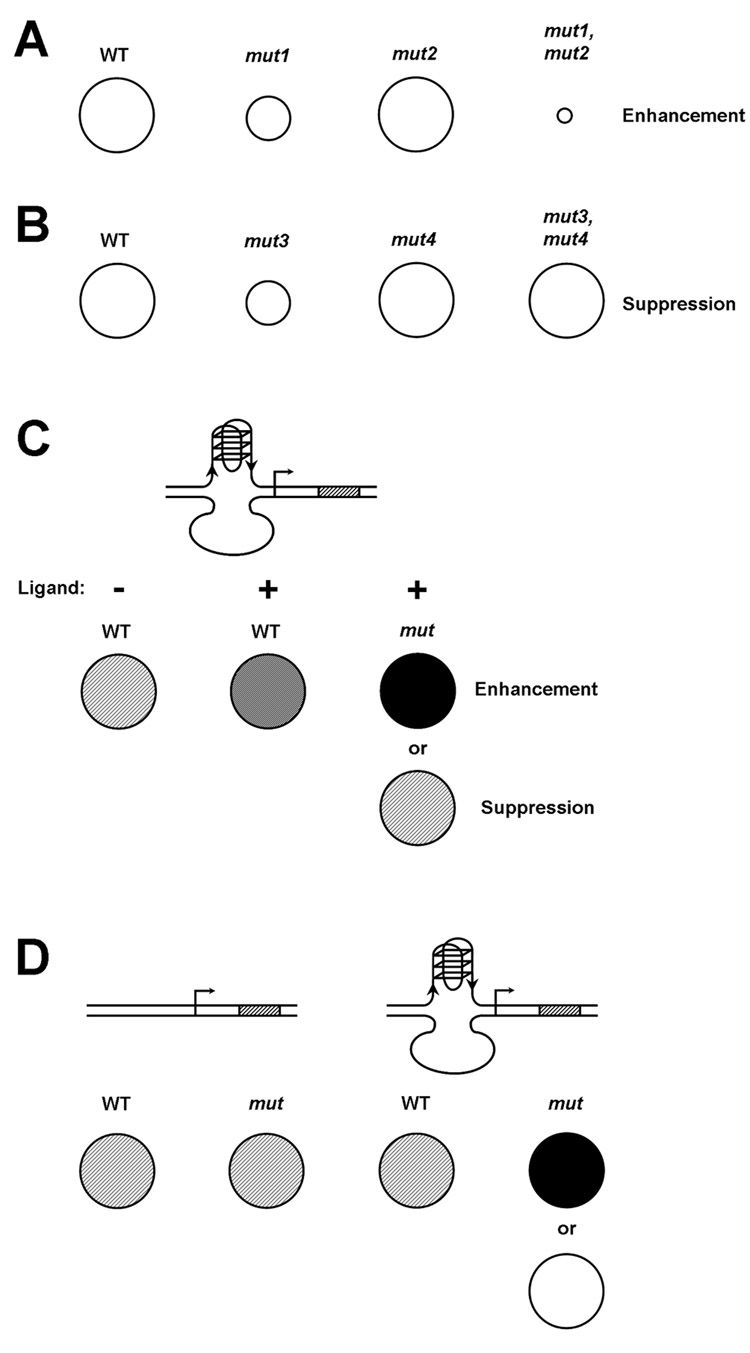
Genetic tools for analyzing G-quadruplexes in yeast. (A) Circles represent yeast colonies that might be obtained from a typical synthetic enhancement screen. The relative sizes of colonies reflect the ability of yeast cells to grow given varying genetic conditions. For example, cells in which the fictional MUT1 gene has been disrupted (mut1) grow poorly as compared with wild-type (WT) cells (indicated by the small circle, representing colony size). Combining the mut1 mutation with the mut2 mutation, which has only a very subtle growth defect, results in synthetic enhancement of the sickness displayed by the mut1 single mutant. (B) In this example of genetic suppression, another mutation (mut3) also confers a growth disadvantage to cells. When combining the mut3 mutation with a mutant allele of another gene, mut4, the latter mutation suppresses the toxicity observed in mut3 cells, allowing improved growth of the double mutant. Similarly, synthetic enhancement or suppression, caused by particular mutations, of the impaired growth caused by a G-quadruplex binding ligand could identify factors that function with G-quadruplexes. (C) Synthetic screens could also utilize transcriptional reporter constructs driven by promoters containing QFP regulatory sequences. In this example, the reporter gene encodes β-galactosidase (β-gal), which produces a blue pigment when cells are grown on the appropriate substrate (level of expression indicated by degree of shading of the circles). Mutations can be obtained that alter the effect of a quadruplex-selective ligand on expression of the reporter gene. In this case, the ligand enhances transcription of the β-gal locus downstream of the promoter quadruplex, and mutations that further upregulate β-gal expression are considered to be “enhancing”. Conversely, those that counteract the activity of the ligand, thereby repressing β-gal, are considered to be “suppressive”. (D) A screen for mutations that affect transcription from a reporter construct with promoter QFP but not from an otherwise identical construct lacking promoter QFP could be used to identify intrinsic factors that influence quadruplexes.
One can envision several ways in which enhancer and suppressor screens might be used to investigate the biology of G-quadruplexes. First, mutations that enhance or suppress the toxic effects of small molecule G4 binding ligands could point to processes that are strongly influenced by G-quadruplexes. If the toxicity is mediated by the effect of the ligand on G-quadruplexes, factors related to quadruplex function could be identified. However, the possibility that "off-target" effects, unrelated to G4 structures, might cause toxicity needs to be considered. This problem could be addressed by conducting parallel screens using G-quadruplex ligands from different chemical families to discern pathways consistently perturbed by quadruplex ligands. Second, similar screens that rely not on toxicity of quadruplex ligands, but rather on their ability to perturb a particular quadruplex-related function could be designed. A straightforward example is testing for mutations that influence the activation or repression by a quadruplex ligand of the expression of a particular reporter gene that contains a regulatory element with G4-forming potential (Fig 2C). Third, by screening for mutations that influence a process in a fashion dependent on the presence of a G-quadruplex, factors that naturally (i.e. in the absence of exogenous ligand) influence quadruplex activities could be identified. For example, again using the transcriptional regulation paradigm to illustrate this idea, two versions of a reporter gene could be engineered: one under the control of a regulatory element with quadruplex-forming potential and the second lacking this element but otherwise identical (Fig 2D). Mutations that influenced expression of the first reporter but not the second would do so through the QFP element and would therefore identify factors that might affect quadruplex formation or function. In addition, overexpression of proteins that reduce quadruplex formation (e.g. helicases) could be used to further test the hypothesis that the QFP element functions as a G-quadruplex. A fourth type of screen could involve a search for mutations influencing phenotypes caused by selective loss of G4-related functions of particular proteins. No such selective mutations are yet known, but the mapping of the quadruplex binding domain of the RecQ helicases to the RQC domain [30] opens the possibility that point mutations within this domain, and by analogy similar domains in other proteins that interact with G-quadruplexes, might have the selective defects desired. In addition, as small molecules and proteins that bind with selectivity to subclasses of quadruplexes are identified, it should be possible to use such agents in combination with the four approaches just described in order to learn about the functions of these individual quadruplex subclasses.
Microarray-based gene expression analyses
Given the enrichment of sequences with QFP in promoter regions in several organisms, as well as other lines of evidence demonstrating that these sequences can modulate gene expression (see below), it is of great interest to determine the extent to which this regulation takes place on a genome-wide scale. Microarray-based expression analyses provide a relatively straightforward way to accomplish this, and such analyses can be carried out in all of the major model organisms as well as in human cells. The effects of quadruplex ligands, or mutations in the genes encoding quadruplex-interacting factors, on global gene expression could be determined. Genes with altered expression could then be compared with the set of genes possessing quadruplex-forming potential to determine if there is a statistically significant association between these sets. Such an association would imply that the ligands or mutations act via a quadruplex-based mechanism to modulate gene expression. Further, it may be possible to find associations between particular kinds of G4 forming sequences and responses to different classes of ligands; while it is not clear at this point that it will be possible to use ligands that are selective for particular G-quadruplex subclasses to tailor gene expression, such findings would provide a starting point for the investigation of this possibility. Because any particular QFP sequence might or might not form a G4 configuration in vivo, an important advantage of the genome-wide approach is that it allows the question of whether QFP sequences, in general, are associated with gene expression changes related to perturbations in factors that affect G-quadruplex formation or function. The yeast genome has a QFP density that is approximately an order of magnitude less than the human genome. For example, approximately 40% and 1.5% of human and yeast upstream promoter sequences have QFP, using the same definition of QFP in both organisms [6, 9]. The smaller number of sequences with QFP in yeast might provide advantages for certain analyses. In particular, it might be more straightforward to discern associations between QFP and responsiveness to factors that affect quadruplexes than it would be in humans, because changes in the expression of fewer genes would be expected, facilitating analysis of the resulting gene expression patterns. A first examination of global gene expression changes caused by perturbation of factors that bind or unwind G-quadruplexes in yeast was published recently, and the results are reviewed below [9].
Affinity purification and tiling array-based analyses of genomic G-quadruplex distribution
Chromatin immunoprecipitation has provided a powerful tool for understanding the distribution of transcription factors and chromatin modifications throughout the genome. Similar approaches might be taken to survey the distribution of quadruplexes on a genome-wide scale. Chromatin could be prepared, fragmented, fractionated based on affinity of the fragments for a quadruplex-binding ligand, and then fragments retained by the ligand could be identified. Selected genomic regions could be analyzed by PCR-based techniques, but a genome-wide view could be provided by using available tiling arrays that cover the yeast genome at 5 bp resolution [31]. Before such approaches can succeed, several obstacles must first be overcome. One difficulty is that the best method for isolating quadruplexes from other genomic sequences is not yet certain. Although antibodies specific for certain G-quadruplexes have been generated, the diversity of G4 folds makes antibody-based capture a non-ideal way to purify quadruplexes in a universal fashion. The interaction of small molecule ligands, while lower-affinity than typical antibody-antigen interactions, might be more universal. Many of these ligands interact with the outer quartet surfaces or intercalate between quartets of quadruplex stacks, making them perhaps relatively insensitive to particular loop structures of intra-molecular quadruplexes and thus relatively universal quadruplex ligands. Nonetheless, there is evidence that they can discriminate to some extent among different quadruplex folds [21, 32, 33]. Affinity chromatography using the selective quadruplex ligand N-methyl mesoporphyrin (NMM) coupled to acrylic beads has been used successfully to select quadruplexes from complex mixtures of nucleic acids [19, 34], and so use of this ligand and probably others should be possible. Another approach would be to use well-defined quadruplex binding domains of proteins, but again, knowledge of the specificity of these domains is at an early stage. Ultimately, use of a variety of ligands will likely be required to probe the full range of possible quadruplexes. A second obstacle is that during the isolation of chromatin and selection with ligands it is possible that quadruplexes might be artifactually formed or lost, and would thus fail to reflect in vivo quadruplexes. Crosslinking of chromatin might minimize this problem, but it will be essential to demonstrate that the level of quadruplexes that are detected depends on the functional status of factors known to affect quadruplexes in the cells from which the chromatin is derived. For example, showing that the apparent quadruplex levels increase in chromatin isolated from sgs1 mutants, which lack the Sgs1p quadruplex-unwinding helicase and would thus be expected have higher steady-state quadruplex levels (Table 1; and see below), would support the interpretation that these quadruplexes actually exist in vivo.
Table 1.
S. cerevisiae proteins that have been reported to interact with G-quadruplexes
| Yeast protein | Mammalian homolog(s) | Known function(s) | Quadruplex-related function(s) | Functional connection with quadruplex binding | Quadruplexes recognized and binding affinities | References |
|---|---|---|---|---|---|---|
| Rap1p | Rap1 | Involved in regulation of telomere length, transcriptional silencing and transcriptional activation | Binds to and stabilizes G4-DNA Promotes formation of G4-DNA |
No known connection | Parallel, anti-parallel inter-molecular; KD = 3 × 10−8 M for G4-DNA; 1000-fold stronger than for dsDNA | [109, 110] |
| Hop1p | HIM-3 family proteins (HORMAD1) | Required for generation of the synaptonemal complex that mediates association of homologous chromosomes during meiosis | Binds to and stabilizes G4-DNA Promotes formation of G4-DNA |
Facilitation of G4-DNA formation by Hop1p might aid alignment of homologous chromosomes during meiosis, but unproven. | Parallel, anti-parallel inter-molecular; KD = 2 × 10−10 M for G4-DNA; 1000-fold stronger than for dsDNA | [97, 111, 112] |
| Mre11p | MRE11 | DNA exo- and endonuclease that plays an essential role in DNA double strand break repair, meiotic recombination and telomere maintenance | Binds to and cleaves G4-DNA | Implicated in the maintenance of telomeres by providing the proper substrate for telomerase; relationship to G4-cleavage is unknown. | Parallel tetramer; KD ~ 7 × 10−8 M; ~12-fold stronger than for ssDNA | [107] |
| Kem1p/Sep1p | mXRN1, Dhm2 | 5’->3’ exonuclease involved in pairing of homologous chromosomes during meiosis and maintenance of telomeres | Binds to and cleaves G4-DNA | In vitro studies suggest a role in cleaving G4-DNA between paired chromosomes to allow migration to opposite poles; might also cleave G4-DNA within telomeres to allow access to telomerase. | Parallel tetramer; KD ~ 10−8 M; at least 100-fold stronger than for ssDNA | [98, 113–115] |
| Stm1p (G4p2, Mpt4p, Sto1p) | None identified | Potential role in telomere capping, also facilitates translation under nutritional stress, and promotes apoptosis-like cell death | Binds to and stabilizes G4-DNA and G4-RNA | Might play a role in telomere capping via G4-DNA binding; no direct evidence for this. | Parallel, anti-parallel inter-molecular; KD = 2 × 10−8 M for G4-DNA; 1000-fold stronger than for dsDNA or ssDNA | [57–59, 116, 117] |
| Sgs1p | RecQ helicases (including WRN and BLM) | RecQ-family helicase involved in maintenance of genomic stability including recombinational repair and telomere maintenance | Binds to and unwinds G4-DNA (15-fold greater activity than Holliday-junction unwinding); possesses conserved RQC domain that specifically binds G4-DNA | Plays a role in maintaining stability of G-rich regions of the genome including telomeres and rDNA, but unclear if G4-DNA unwinding is involved in these activities. | Parallel tetramer, anti-parallel dimer; KD = 5 × 10−9 M for G4-DNA; 8 × 10−9 M for Holliday junction substrates | [30, 40, 89, 118, 119] |
| Arc1p (G4p1) | p43 | Binds tRNA and functions as a co-factor for the methionyl- and glutamyl-tRNA synthetases | Binds to and stabilizes G4-DNA and G4-RNA | No apparent connection between quadruplex binding and function | Parallel, anti-parallel; KD = 5 × 10−8 M for G4-DNA; 1000-fold stronger than for dsDNA or ssDNA | [58, 120, 121] |
| Cdc13p | Pot1 | Essential telomere capping protein; inhibits exonucleolytic degradation at telomeres and regulates telomerase access | Destabilizes G4-DNA. | Might be important for destabilizing G4-DNA during telomere extension by telomerase | Parallel inter-molecular (anti-parallel not tested); KD = 1.5 × 10−8 for S. cerevisiae telomere repeat in G4-DNA form and 9 × 10−9 in ssDNA form | [63] |
Mutational analyses
The approaches described above may reveal associations between particular nucleic acid sequences with quadruplex-forming potential, or proteins with quadruplex-binding activity, and biological events hypothesized to be impacted by G-quadruplexes (e.g. transcriptional stimulation or repression of a QFP-rich promoter by a small-molecule quadruplex ligand). To determine whether any such association reflects a bona fide G-quadruplex function would require additional testing. A logical next step would be to then disrupt quadruplex formation or binding via site-directed mutagenesis, followed by a test for loss of the hypothesized quadruplex-dependent events. This approach could be combined with biophysical studies of the native and mutated QFP sequences, or the native and mutated quadruplex-binding proteins, to compare their intrinsic ability to form or bind quadruplexes, respectively. Ideally, a good correspondence between the in vitro and in vivo analyses would be obtained. We note, however, that the quadruplex-forming ability of nucleic acids in vitro might differ from in vivo settings, where other factors (e.g. chromatin proteins) might facilitate or impede quadruplex formation. Given such a scenario, the in vivo tests might prove more definitive. Because G-quadruplexes might interact with chromatin factors (see below), it might be particularly important that point mutations that perturb quadruplex formation be introduced within a native genomic context, and the relative ease with which this can be accomplished in yeast are a benefit of this system.
Evidence for G-quadruplex functions in vivo
Many observations suggest, and in some cases demonstrate, roles for G-quadruplexes in different aspects of cell biology. Each of the sections below describes one such aspect, beginning with general examples from several organisms and then focusing on findings from yeast. Table 1 provides a summary of various yeast proteins implicated in G-quadruplex metabolism.
G-quadruplexes and telomere metabolism
The 3' ends of most eukaryotic telomeres terminate in a guanine-rich single stranded overhang. Consequently, telomere ends can have high G-quadruplex forming potential. Indeed, telomere sequences from several organisms ranging from yeast to humans readily adopt G4-DNA conformations in vitro. Recently, it was demonstrated that G4-DNA can be detected at telomeres in Stylonichia lemnae cells using highly specific G4-DNA antibodies and immunofluorescence microscopy [5]. Importantly, the possibility that the observed staining was an artifact of G4-DNA formation catalyzed by the antibodies was ruled-out by the subsequent demonstration that the staining depends on expression of TEBPβ[4], a telomere-binding protein with a high-degree of similarity to the beta subunit of the telomere-binding protein in Oxytricha, which itself has been shown to promote G4-DNA in vitro [35]. Other observations suggest that G4-DNA might form at mammalian telomeres. These include demonstrations that loss of the Werner, Bloom or RTEL helicases results in defects in telomere maintenance in vivo [36–38]. The Werner and Bloom helicases are particularly adept at unwinding G-quadruplex substrates in vitro [39–42], while RTEL is homologous to the C. elegans DOG-1 protein that prevents deletions in G-rich sequences [37, 43]. Further, the human telomere binding protein POT1, which binds the single stranded telomere overhang, inhibits G4-DNA formation in vitro [44], while treatment of cells with the quadruplex-selective ligand telomestatin displaces POT1 and uncaps telomeres (i.e. causes them to be recognized as DNA breaks) [45, 46]. Therefore, POT1 binding and G4-DNA formation at the telomere are likely mutually exclusive states. However, the distribution of telomeres between these states in untreated cells is currently unknown.
In yeast, several observations suggest roles for G-quadruplexes in telomere metabolism. In S. cerevisiae, the telomere repeat DNA is 300–350 bp in length and follows the consensus 5′-[(TG)0–6TGGGTGTG(G)]n-3′ [47]. An average of 8 nt exists between each GGG run, and the intervening sequence may contain GG dinucleotides [47]; GG runs may contribute to quadruplex formation, albeit more weakly than GGG runs. Yeast telomere repeats form G4-DNA in vitro [48, 49], although the types of G4-DNA structures formed by natural yeast telomere repeats have not been well characterized. The telomere 3' single strand overhang is short (~10–15 nt in G1, and up to 22 nt in S-phase) [50] and thus might have limited intra-molecular quadruplex forming potential under most conditions. But G4-DNA might form, perhaps transiently, when duplex telomere repeats become single-stranded, for example during telomere replication or recombination. Yeast telomere repeats at the ends of a linear plasmid were found to mediate physical interaction of the ends [51]. This report mentioned unpublished work indicating that the methylation of guanine N7 does not block the association, suggesting that the associations are mediated by a non-G4-DNA structure. These results do not, however, address telomere G-DNA formation in other settings. We note also that the irregular nature of the telomere repeat does not preclude quadruplex formation; this is particularly true of intra-molecular quadruplexes where guanines not involved in quadruplexes can exist in the loops.
A potential in vivo link between telomere G4-DNA and telomere maintenance in yeast was uncovered in studies using a temperature sensitive point mutant, cdc13-1, of the telomere capping protein Cdc13p. Cdc13p inhibits exonucleolytic degradation of the C-rich telomere strand and also regulates telomerase access to telomeres [52–55]. At non-permissive temperatures, this mutant accumulates long stretches of G-rich ssDNA at telomeres, an environment conducive to forming G4-DNA [56]. Overexpression of the quadruplex-binding protein Stm1p rescues cdc13-1 temperature sensitivity [57]. Stm1p has weak homology with the ciliate TEBPβ telomere binding proteins which, as discussed above, bind to and promote formation of G4-DNA in vivo [4, 5, 57]. While Stm1p itself has not been shown to bind G4-DNA in vivo, it shows such activity in vitro, and additionally, is known to bind guanine-rich telomeric and subtelomeric DNA in vitro [58, 59]. Furthermore, overexpression of the RecQ helicase Sgs1p, which efficiently unwinds G4-DNA substrates [40, 60], abolishes rescue by Stm1p overexpression [57]. And G4-forming sequences at the ends of DNA molecules can inhibit recognitions of the ends by checkpoint proteins in vitro [61]. Together, these findings suggest a role for G4-DNA in telomere capping. This capping might occur only in the absence of functional Cdc13p, or alternatively might play a role in normal yeast cells under conditions where Cdc13p is not bound. It would be interesting determine if telomeric quadruplexes can be detected physically in the cdc13-1 mutants rescued by Stm1p overexpression. Antibodies specific for yeast telomeric G4-DNA structures have been generated recently [62], which might provide a valuable reagent for future studies. We also point out that Cdc13p itself (in addition to its mammalian homolog Pot1) is known to destabilize G4-DNA in vitro, which might be a biologically relevant function given that G4-DNA structures are known to inhibit telomerase action, and would need to be disrupted in order for yeast to maintain their telomeres through a telomerase-based mechanism [44, 63, 64].
A screen was recently performed to identify yeast single-locus deletion mutants that are either resistant or sensitive to growth inhibition by the highly-selective quadruplex-interacting compound NMM [9, 19]. Remarkably, stm1 mutants were among the NMM-resistant class, consistent with the notion that Stm1p stabilizes G4-DNA and thus provides a target for NMM action. A related mutation, cgi121, yielded NMM-sensitivity. Cgi121p is a member of the KEOPS telomere protein complex, and cgi121 mutation also rescues cdc13-1 temperature sensitivity [65]. A possible explanation for these observations is that loss of Cgi121p somehow stabilizes G4-DNA, similar to Stm1p overexpression, and that the observed NMM-sensitivity is a result of increased G4-DNA targets for the drug [9]. In addition to the stm1 and cgi121 mutations being connected with telomere metabolism, gene ontology (GO) categorization of the mutants from the screen revealed that mutations in genes encoding telomere maintenance proteins were significantly overrepresented in both the NMM-sensitive and NMM-resistant categories. Although further work is required to test this idea, these findings suggest that NMM toxicity might be caused, in part, by stabilization of telomeric G4-DNA or interference with its function.
Genomic distribution of sequences with quadruplex forming potential
In addition to telomeres, numerous regions in bacterial and eukaryotic genomes have the potential to form intramolecular G-quadruplexes. These regions include the promoter regions of single copy genes, the ribosomal DNA (rDNA), certain minisatellites and the immunoglobulin (Ig) heavy chain switch regions [4, 66–69]. Multiple search algorithmshave been employed to identify these regions, which generally require four runs of three or more Gs separated by loops of any sequence, with the entire pattern falling within a window of defined size [7, 9–12]. Depending on the algorithm employed, different names have been given to the identified sequences, e.g. QFS (potential quadruplex forming sequence), G4P (sequence with G4-DNA forming potential) and QFP (sequence with quadruplex-forming potential); here, we have used only QFP for simplicity. Although, in general, QFP sequences with longer G-runs and shorter loops between the G-runs are more likely to form G-quadruplexes spontaneously in vitro, it is not yet possible to predict with certainly the true potential of an arbitrary sequence to form G-quadruplexes. Moreover, rules for quadruplex formation are likely different in vivo, where their formation might be modulated by both inhibitory and stimulatory factors. Therefore, it is largely unknown which QFP sequences actually form G-quadruplexes in vivo. Recent studies have been aimed at determining whether sequences that have QFP form G4-DNA in vitro and in vivo, or if these sequences instead serve another unknown function (e.g. binding by transcription factors that recognize duplex sequences with QFP).
G-quadruplexes and transcription
QFP sequences have been identified in human, chicken, yeast, and bacterial genomes at greater than random frequency near transcriptional promoters, raising the possibility that they play a role in transcriptional regulation (Fig.3A) [6–9, 11, 12]. There is a particular enrichment of these sequences in the promoters of mammalian oncogenes [6, 10], and for some of these, there is good evidence that the QFP sequences can affect transcription. A prominent example is c-MYC, which has an upstream QFP sequence that represses transcription. This repression is augmented by addition of the porphyrin G4-DNA ligand TMPyP4 to cells [70–72]. Although TmPyP4 has little selectivity for binding G4-DNA in comparison to duplex DNA, repression of c-MYC by the ligand is abolished by point mutations that ought to abrogate quadruplex formation by the QFP sequence. Given this result, one of two scenarios is possible; that these mutations affect binding of a sequence specific transcription factor to duplex DNA or that TmPyP4 exerts its effects on c-MYC expression through a bona fide quadruplex target. Likely, experiments are underway that should discriminate between these possibilities. Similar inhibition of KRAS expression by TMPyP4 and of c-KIT by several selective trisubstituted alloisoxazines has been reported [21, 73], although these effects have not been shown to be dependent on the QFP sequences.
Figure 3.
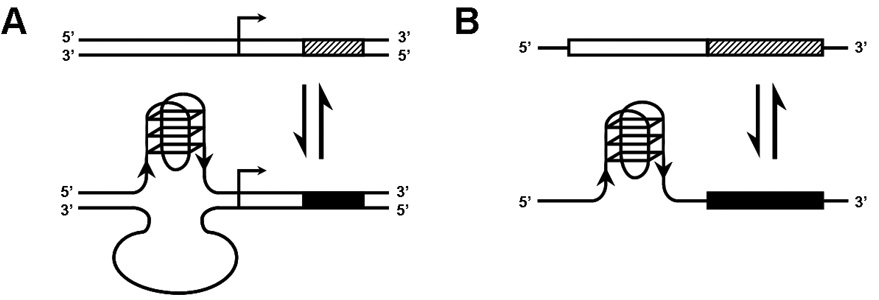
Modulation of gene expression by G-quadruplexes. (A) A model of how quadruplex formation at DNA promoter regions with QFP might modulate transcription from downstream ORFs. G-quadruplex formation might repress transcription by occluding transcriptional activators and/or creating binding sites for transcriptional repressors. Conversely, quadruplex formation might also serve to activate transcription in some cases by blocking repressors and/or recruiting activators. In either case, the state of transcription of the relevant ORF is changed from one state (cross-hatched rectangle) to another (solid black rectangle) by quadruplex formation. (B) A model of how RNA quadruplex formation in the 5’UTR (white rectangle) of mRNA transcripts might affect its translation or stability. As in (A), G-quadruplex formation could positively or negatively modulate gene expression.
Genome-wide approaches examining the relationship between QFP and transcription in S. cerevisiae have yielded findings that support those obtained in other organisms, and have also yielded novel insights. QFP sequences are present at above random frequencies in open reading frames (ORFs) and, to a greater extent, in promoter regions. Further, there are strong correlations between promoter QFP and reduced occupancy of histones H2A, H3 and H2A.Z [9]. H2A and H3 are histone components of canonical nuclesomes, while H2A.Z is a H2A variant that is present at promoters that are poised for transcriptional activation. These observations suggest that G4-DNA formation might exclude nucleosomes, thereby facilitating transcriptional initiation at such sites. This is consistent with another recent report that QFP is enriched more than 200-fold at nuclease-hypersensitive sites near human promoters, which typically have reduced histone occupancy [6]. The correlation between QFP and reduced nucleosome occupancy was not observed in yeast ORFs, implying a possible connection to the initiation of transcription. While binding of the Rap1p transcription factor is correlated with low histone occupancy in yeast, and Rap1p binding sites resemble QFP sequences, the possibility that Rap1p explains the nucleosome exclusion was ruled-out by removing known Rap1p target genes from the analysis [9, 74]. Still, it remains possible that the binding of factors other than Rap1p to duplex DNA at these regions causes the low histone occupancy, and it also remains to be determined whether QFP sequences, themselves, cause nucleosome exclusion or if they are only proximal to such regions.
A possible connection between G-quadruplexes and chromatin was also revealed by the screen for enhancers of NMM toxicity described above. Among null mutations conferring enhanced sensitivity to NMM, there was highly significant enrichment for loci encoding factors involved in nucleosome remodeling or modification. These included several members of the RSC and SWI/SNF nucleosome remodeling complexes, the ADA histone acetyltransferase complex, and histone H2A.Z (encoded by HTZ1). Perhaps G4-DNA helps remove nucleosomes, consistent with the reduced histone occupancy of promoters with QFP, but binding of G4-DNA by NMM interferes with this activity. Interestingly, htz1 mutants, like NMM treated cells, have a heightened requirement for SWI/SNF complex activity [75, 76], raising the possibility that H2A.Z and G4-DNA might have overlapping roles in chromatin modification. It will be important to test this idea further, using well-defined quadruplex-forming sequences in in vitro and in vivo settings.
Gene expression analyses have demonstrated that manipulations in yeast that are predicted to affect G-quadruplex homeostasis preferentially affect loci with QFP, thus supporting the model that QFP sequences can form bona fide quadruplexes that, in turn, modify gene expression. The first of these manipulations, treatment of cells with the quadruplex-selective ligand NMM, causes upregulation of loci with QFP upstream or downstream of promoters [9]. Binding of G-quadruplexes by NMM might upregulate gene expression in a number of ways. Stabilization of G4-DNA by NMM might upregulate gene expression by occluding repressive factors, such as nucleosomes, or by enabling the binding of transcriptional activators. Alternatively, NMM binding might displace repressors that bind to G4-DNA that forms even in the absence of ligand. In the case of loci with QFP located downstream of the promoter and on the sense strand, the binding of NMM to G4-RNA structures in mRNA transcripts might allow for their stabilization. A second manipulation, deletion of the SGS1 gene, which encodes a G4-DNA-unwinding helicase, caused preferential downregulation of loci with QFP in their ORFs [77, 78]. Remarkably, this association was particularly true for the template strand, raising the possibility that quadruplexes unresolved by Sgs1p provide an impediment progression of RNA polymerase. Future studies should help to determine the precise molecular mechanisms underlying the observed NMM-induced and Sgs1p-mediated upregulation of gene expression in loci with QFP.
An argument against the possibility that G4-DNA forms and affects transcription in vivo is that genes exist within duplex DNA, and the equilibrium between the duplex form and the G4-DNA form (which requires melting of the duplex before it can form) might lie too far in the direction of the duplex. While the energetics that govern this equilibrium will differ among various QFP sequences, and will doubtless differ for naked DNA compared with chromatin, several arguments can be made supporting the notion that such sequences do, indeed, form G-quadruplexes. First, the c-KIT oncogene contains a 20 nucleotide QFP sequence within a transcriptional activation element, and this sequence has been shown to form G4-DNA in vitro. Remarkably, G-quadruplexes were determined to account for roughly 30% of the species formed under physiologic conditions by annealing the strand containing this QFP sequence, and flanked by a total of 76 nt of additional DNA, to its complementary strand [79]. Although a conclusive demonstration that the system was at equilibrium was not provided, these findings argue that the G-quadruplex conformation has stability similar to the duplex form. Second, DNA is typically negatively supercoiled in vivo, which favors duplex unwinding and thus should facilitate G4-DNA formation. Third, QFP sequences located downstream from promoters might give rise to G4-DNA after passage of the first RNA polymerase, affecting the subsequent activity of the promoter. Fourth, G4-DNA might form only transiently, e.g. during replication, but affect the binding of factors (e.g. histones) that could have a persistent effect on transcription. Finally, simple thermodynamics of DNA alone might not dictate the equilibrium between duplex and quadruplex forms in vivo. There are numerous examples in biology of processes that require the input of energy to accomplish beneficial tasks (e.g. proofreading by DNA polymerases); perhaps the regulatory benefits of G4-DNA outweigh any costs of its formation and therefore proteins have evolved to favor its formation under specific conditions and in particular genomic regions.
G-quadruplexes and translational regulation
Another mechanism by which quadruplexes might affect gene expression is through G4-RNA structures in mRNA (Fig 3B). Compared with DNA, a substantial portion of total cellular RNA is single stranded and therefore, energetically more likely to form non-canonical secondary and tertiary structures, including quadruplexes [80]. A pivotal study by Darnell et al. demonstrated that the FMRP protein, which is absent in the fragile X mental retardation syndrome, binds to a class of G-rich mRNAs that form intramolecular G4-RNA [81]. A significant proportion of these mRNAs were also found to have reduced or increased polysome occupancy in fragile X patient cells, which strongly suggests that binding of FMRP to these targets regulates their translation. Further, Khateb et al. demonstrated that the 5’ untranslated region (UTR) of the FMRP transcript contains QFP-rich CGG repeats that, when amplified to the extent observed in fragile X premutation carriers, inhibit translation of FMRP in both cell-free extracts and cultured human cells [82]. Moreover, expression of the G-quadruplex destabilizing proteins hnRNP A2 or CBF-A relieves this inhibition. While there is debate as to whether CGG repeats actually form G-quadruplex or only hairpin structures, the effects of hnRNP A2 and CBF-A on FMRP translation are consistent with a quadruplex-based regulatory mechanism. In addition, Kumari et al. identified a G4-RNA-forming element in the 5’ UTR of the human NRAS proto-oncogene that is capable of inhibiting translation in a cell-free in vitro translation system [83]. To extend their findings, the authors also used sequence analyses to predict the existence of 2,922 other sequences with QFP in the 5’ UTRs of human gene transcripts, raising the possibility regulation by G4-RNA has broad effects on gene expression. An important caveat of all of the above studies is that it has not yet been proven rigorously that G-quadruplex formation per se, and not some other function of the quadruplex-forming sequences (e.g. binding to protein factors), is responsible for modulation of translation.
In yeast, there is a peak of QFP in ORFs just downstream from the start of translation; these QFP sequences are found preferentially in the template (i.e. anti-sense) strand and so would be underrepresented in mRNAs. However, this strand asymmetry is limited to short QFP sequences and actually is reversed for QFP sequences with longer loops. Because G4-DNA with shorter loop regions are generally more stable than those with long loops [84, 85], short-loop G4-RNA might be avoided because it could prove strongly inhibitory to translation. Conversely, G4-RNA with long loop regions might be in dynamic equilibrium with other RNA structures that are permissive for translation, and thus represent a mechanism by which translation could be regulated.
G-quadruplexes and nucleolar function
As mentioned above, QFP sequences have been identified within the repeated rDNA loci present in the nucleoli of both yeast and humans. Interestingly, this QFP is highly concentrated in the sense strand, and thus G-quadruplexes might form both in the rDNA genes and their rRNA transcripts. Consistent with a role for quadruplexes in nucleolar function, the protein nucleolin, which is involved in both the synthesis and maturation of ribosomes, binds to quadruplexes with very high affinity [67]. Interestingly, treatment of cultured human cells with a synthetic quadruplex aptamer (AS1411), which binds to nucleolin, results in the mislocalization of nucleolin to the cytoplasm, as well as the altered activity of nucleolin-containing protein complexes [86]. Further evidence for a role of quadruplexes in ribosome metabolism has been provided by studies in S. cerevisiae, using microarray-based techniques to assay the effect of NMM treatment on gene expression [9]. In addition to regulation of QFP loci by NMM (see above), the quadruplex ligand also caused a highly significant downregulation of genes connected with nucleolar function, including those involved in rRNA processing and ribosome biogenesis. This finding is intriguing given the high density of QFP sequences within the rDNA and the 25S and 18S rRNAs, and the observation that these rRNAs were downregulated in response to NMM treatment. Thus, NMM might inhibit rRNA transcription, stability, or processing by binding rDNA or rRNA quadruplexes, and the downregulation of other loci having nucleolar function would be a secondary consequence of these quadruplex-related effects of the ligand. While the NMM studies do not address whether G-quadruplexes form naturally in the nucleolus or if they are formed only in the presence of ligand, the binding specificity of nucleolin and the evolutionary conservation of QFP in the rDNA loci suggests that G-quadruplexes play some role in ribosome biogenesis.
G-quadruplexes and DNA replication
Another consequence of the strand bias of QFP in the rDNA repeats is that the strand likely to give rise to G4-DNA will most often be replicated by lagging strand DNA synthesis. Interestingly, the same is true of the QFP present at telomeres. It is possible that the increased levels of single stranded DNA constituting the lagging strand might facilitate G4-DNA formation, thereby generating potential blocks to the completion of replication. Consistent with this idea, studies in human cells have shown that the absence of the WRN DNA helicase, which like its yeast homologue Sgs1p, is particularly adept at unwinding G4-DNA [39, 40, 42, 87], results in loss of the telomere strand that is replicated by lagging strand synthesis [36]. Telomere defects related to replication in sgs1 mutants might have a similar basis [88, 89]. Further, the unwinding of telomere quadruplexes during Stylonichia telomere replication [4, 5], and the CGG repeat-induced pausing of DNA polymerases in vitro [90, 91], are consistent with G4-DNA providing an impediment to replication. However, telomere repeat DNA from the fission yeast S. pombe causes replication pausing regardless of its orientation to the replication fork [92], suggesting that the G4-DNA at telomeres might represent a difficult substrate for both leading and lagging strand synthesis. Therefore, it is possible that the natural orientation of telomere and rDNA QFP sequences helps ensure their full replication, because G4-DNA formed after the passage of the replication fork on the lagging strand template might be less deleterious than G4-DNA formed ahead of the fork on the leading strand template. In the latter case, the replication fork might collapse, while in the former case, the ability to prime DNA synthesis from a downstream Okazaki fragment could allow for continued replication fork progression, and subsequent removal of quadruplexes and completion of lagging strand synthesis by gap repair. It would be interesting to test if QFP sequences at other genomic loci are preferentially replicated by lagging strand synthesis, or if deleterious consequences (e.g. DNA breaks) occur more frequently when QFP sequences are copied by leading strand synthesis.
G-quadruplexes and DNA recombination
G-quadruplexes have long been hypothesized to play roles in DNA recombination, particularly given the transient single-stranded intermediates that occur in many recombination transactions. G4-DNA might play a role in class switch recombination, a highly-specialized form of recombination that takes place during antibody isotype switching in mammalian cells, and involves recombination between switch (S) regions, which contain G-rich sequences confined largely to one strand [93, 94]. Transcription of these regions is required for this process, and produces a stable R-loop that facilitates G4-DNA formation of the G-rich non-template strand, yielding a structure called a G-loop. Studies of these switch regions on plasmids transcribed in E. coli have shown that G-loop formation is elevated in recQ mutants, which lack the quadruplex unwinding helicase RecQ, therefore providing strong evidence that the observed G-loops are formed in vivo [66]. Furthermore, the Mutsα protein, which facilitates recombination in S regions, binds with high affinity to G4-DNA, supporting the biological relevance of quadruplexes in switch recombination [95]. In addition, there is some evidence that certain chromosomal translocations leading to oncogene activation might occur preferentially in QFP-containing regions that give rise to G-loops [96]. While the authors suggest that these translocations are not targeted to G4-DNA itself, but rather to flanking single stranded DNA, it is possible that quadruplex formation facilitates these translocation events by stabilizing the adjacent single stranded target sites.
Studies in yeast suggest possible roles for G4-DNA in homologous recombination during meiosis. Two proteins involved in meiotic recombination, Hop1p and Kem1p, have been shown to have G4-DNA specific activities. Hop1p is a member of the synaptonemal complex that mediates association of homologous chromosomes during meiosis. In vitro, Hop1p binds to and catalyzes the formation of G4-DNA, and promotes the pairing of double stranded DNA molecules via quadruplex structures [97]. Kem1p is a nuclease that can cleave single stranded DNA 5' from G4-DNA structures in vitro, and kem1 mutants are unable to complete meiosis [98, 99]. However, it is not yet clear how the in vitro activities of these proteins on G4-DNA relate to their in vivo functions.
A key factor in the initiation of meiotic recombination, as well as recombination in vegetative cells, is the MRX complex, which is composed of Mre11p, Rad50p, and Xrs2p [100, 101]. The complex recognizes DNA double strand breaks and then Mre11p-dependent nuclease activity processes DNA ends to generate 3’ single stranded overhangs [102]. Rad50p and Xrs2p function, respectively, to tether the DNA ends and to enhance the nucleolytic activity of Mre11p [103–105]. The 3’ single stranded protrusions generated by Mre11p then invade homologous sequences [106]. Interestingly, Mre11p has been shown to bind G4-DNA with greater affinity than single stranded or double stranded DNA substrates, and to cleave G4-DNA upstream of the guanine runs [107]. Caveats that complicate interpretation of these results include the weak affinity of Mre11p for G4-DNA (~100 nM) and the inefficient cleavage activity observed in these studies. Nevertheless, consistent with these findings, loss of Mre11p also resulted in increased sensitivity to the quadruplex ligand, NMM, indicating that elevated levels of G4-DNA in mre11 mutants might enhance the toxicity of NMM [9]. rad50 mutants were found in the same NMM screen to have the opposite phenotype, suppressing the toxicity of NMM, which was surprising, given that the MRX proteins generally work together as a complex. However, the Rad50p component of the MRX complex was shown recently to attenuate the endonuclease activity of Mre11p on G-quadruplex and other substrates in vitro [108]. Therefore, it is conceivable that rad50 mutants have lower G4-DNA levels due to increased endonucleolytic cleavage of quadruplexes by Mre11p, and are therefore resistant to NMM. More studies will be required, however, to confirm the findings of the NMM screen and to test whether Mre11p and Rad50p indeed have opposing effects on G4-DNA substrates in vivo.
Perspective
A combination of biophysical, bioinformatics, genetic and cell biological approaches have yielded a remarkable series of findings that argue for the relevance of G-quadruplexes to natural biology. However, more work is required to firmly establish the roles of G4-DNA and G4-RNA in nucleic acid functions and to decipher the mechanisms by which they operate. This knowledge might provide new approaches for selectively targeting processes ranging from transcription and translation to DNA replication and recombination to telomere maintenance. Studies in multiple systems will likely be required to unlock the secrets of G-quadruplexes, and we anticipate a key role for yeast in these exciting explorations.
Acknowledgements
We thank Li-San Wang, Steve Hershman and Qijun Chen for discussions, and Alex Chavez for insightful discussions and comments on the manuscript. This work was supported by the National Institute on Aging (5R01AG021521 to F.B.J)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 3.Webba da Silva M. Geometric Formalism for DNA Quadruplex Folding. Chemistry. 2007;13:9738–9745. doi: 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- 4.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 5.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci U S A. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang LS, Brad Johnson F. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du Z, Kong P, Gao Y, Li N. Enrichment of G4 DNA motif in transcriptional regulatory region of chicken genome. Biochem Biophys Res Commun. 2007;354:1067–1070. doi: 10.1016/j.bbrc.2007.01.093. [DOI] [PubMed] [Google Scholar]
- 13.De Cian A, Lacroix L, Douarre C, Temime-Smaali N, Trentesaux C, Riou JF, Mergny JL. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Hurley LH, Wheelhouse RT, Sun D, Kerwin SM, Salazar M, Fedoroff OY, Han FX, Han H, Izbicka E, Von Hoff DD. G-quadruplexes as targets for drug design. Pharmacol Ther. 2000;85:141–158. doi: 10.1016/s0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- 15.Mergny JL, Riou JF, Mailliet P, Teulade-Fichou MP, Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Cian A, Delemos E, Mergny JL, Teulade-Fichou MP, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J Am Chem Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 17.Dixon IM, Lopez F, Tejera AM, Esteve JP, Blasco MA, Pratviel G, Meunier B. A G-quadruplex ligand with 10000-fold selectivity over duplex DNA. J Am Chem Soc. 2007;129:1502–1503. doi: 10.1021/ja065591t. [DOI] [PubMed] [Google Scholar]
- 18.Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J Am Chem Soc. 2001;123:1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 19.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 20.Moorhouse AD, Santos AM, Gunaratnam M, Moore M, Neidle S, Moses JE. Stabilization of G-quadruplex DNA by highly selective ligands via click chemistry. J Am Chem Soc. 2006;128:15972–15973. doi: 10.1021/ja0661919. [DOI] [PubMed] [Google Scholar]
- 21.Bejugam M, Sewitz S, Shirude PS, Rodriguez R, Shahid R, Balasubramanian S. Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: small molecule regulation of c-kit oncogene expression. J Am Chem Soc. 2007;129:12926–12927. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fry M. Tetraplex DNA and its interacting proteins. Front Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 23.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 24.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z, Brown GW, Ingles CJ, Emili A, Allis CD, Toczyski DP, Weissman JS, Greenblatt JF, Krogan NJ. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 25.Pan X, Yuan DS, Ooi SL, Wang X, Sookhai-Mahadeo S, Meluh P, Boeke JD. dSLAM analysis of genome-wide genetic interactions in Saccharomyces cerevisiae. Methods. 2007;41:206–221. doi: 10.1016/j.ymeth.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 27.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 28.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 29.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, Lorenz TC, Payne GS, Ishihara S, Ohya Y, Andrews B, Hughes TR, Frey BJ, Graham TR, Andersen RJ, Boone C. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 30.Huber MD, Duquette ML, Shiels JC, Maizels N. A conserved G4 DNA binding domain in RecQ family helicases. J Mol Biol. 2006;358:1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Hesselberth JR, Fields S. Genome-wide identification of spliced introns using a tiling microarray. Genome Res. 2007;17:503–509. doi: 10.1101/gr.6049107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ragazzon P, Chaires JB. Use of competition dialysis in the discovery of G-quadruplex selective ligands. Methods. 2007;43:313–323. doi: 10.1016/j.ymeth.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez R, Pantos GD, Goncalves DP, Sanders JK, Balasubramanian S. Ligand-Driven G-Quadruplex Conformational Switching By Using an Unusual Mode of Interaction. Angew Chem Int Ed Engl. 2007;46:5405–5407. doi: 10.1002/anie.200605075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Geyer CR, Sen D. Recognition of anionic porphyrins by DNA aptamers. Biochemistry. 1996;35:6911–6922. doi: 10.1021/bi960038h. [DOI] [PubMed] [Google Scholar]
- 35.Fang G, Cech TR. The beta subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell. 1993;74:875–885. doi: 10.1016/0092-8674(93)90467-5. [DOI] [PubMed] [Google Scholar]
- 36.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 37.Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, Tam PP, Nagy A, Lansdorp PM. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 38.Du X, Shen J, Kugan N, Furth EE, Lombard DB, Cheung C, Pak S, Luo G, Pignolo RJ, DePinho RA, Guarente L, Johnson FB. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol Cell Biol. 2004;24:8437–8446. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 40.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 42.Mohaghegh P, Karow JK, Brosh Jr RM, Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- 44.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci U S A. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomez D, Wenner T, Brassart B, Douarre C, O'Donohue MF, El Khoury V, Shin-Ya K, Morjani H, Trentesaux C, Riou JF. Telomestatin-induced telomere uncapping is modulated by POT1 through G-overhang extension in HT1080 human tumor cells. J Biol Chem. 2006;281:38721–38729. doi: 10.1074/jbc.M605828200. [DOI] [PubMed] [Google Scholar]
- 46.Gomez D, O'Donohue MF, Wenner T, Douarre C, Macadre J, Koebel P, Giraud-Panis MJ, Kaplan H, Kolkes A, Shin-ya K, Riou JF. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 47.Forstemann K, Lingner J. Molecular basis for telomere repeat divergence in budding yeast. Molecular & Cellular Biology. 2001;21:7277–7286. doi: 10.1128/MCB.21.21.7277-7286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giraldo R, Suzuki M, Chapman L, Rhodes D. Promotion of parallel DNA quadruplexes by a yeast telomere binding protein: a circular dichroism study. Proc Natl Acad Sci U S A. 1994;91:7658–7662. doi: 10.1073/pnas.91.16.7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel PK, Hosur RV. NMR observation of T-tetrads in a parallel stranded DNA quadruplex formed by Saccharomyces cerevisiae telomere repeats. Nucleic Acids Res. 1999;27:2457–2464. doi: 10.1093/nar/27.12.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larrivee M, LeBel C, Wellinger RJ. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18:1391–1396. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wellinger RJ, Ethier K, Labrecque P, Zakian VA. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 52.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 53.Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Current Biology. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- 54.Meier B, Driller L, Jaklin S, Feldmann HM. New function of CDC13 in positive telomere length regulation. Mol Cell Biol. 2001;21:4233–4245. doi: 10.1128/MCB.21.13.4233-4245.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 56.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayashi N, Murakami S. STM1, a gene which encodes a guanine quadruplex binding protein, interacts with CDC13 in Saccharomyces cerevisiae. Mol Genet Genomics. 2002;267:806–813. doi: 10.1007/s00438-002-0712-3. [DOI] [PubMed] [Google Scholar]
- 58.Frantz JD, Gilbert W. A yeast gene product, G4p2, with a specific affinity for quadruplex nucleic acids. J Biol Chem. 1995;270:9413–9419. doi: 10.1074/jbc.270.16.9413. [DOI] [PubMed] [Google Scholar]
- 59.Van Dyke MW, Nelson LD, Weilbaecher RG, Mehta DV. Stm1p, a G4 quadruplex and purine motif triplex nucleic acid-binding protein, interacts with ribosomes and subtelomeric Y' DNA in Saccharomyces cerevisiae. J Biol Chem. 2004;279:24323–24333. doi: 10.1074/jbc.M401981200. [DOI] [PubMed] [Google Scholar]
- 60.Han H, Bennett RJ, Hurley LH. Inhibition of unwinding of G-quadruplex structures by Sgs1 helicase in the presence of N,N'-bis[2-(1-piperidino)ethyl]-3,4,9,10- perylenetetracarboxylic diimide, a G-quadruplex-interactive ligand. Biochemistry. 2000;39:9311–9316. doi: 10.1021/bi000482r. [DOI] [PubMed] [Google Scholar]
- 61.Tsai YC, Qi H, Liu LF. Protection of DNA ends by telomeric 3' G-tail sequences. J Biol Chem. 2007;282:18786–18792. doi: 10.1074/jbc.M702171200. [DOI] [PubMed] [Google Scholar]
- 62.Dong Q. [Generation and characterization of the monoclonal antibody and scFv against yeast telomeric guanine-quadruplex DNA] Fen Zi Xi Bao Sheng Wu Xue Bao. 2006;39:482–488. [PubMed] [Google Scholar]
- 63.Lin YC, Shih JW, Hsu CL, Lin JJ. Binding and partial denaturing of G-quartet DNA by Cdc13p of Saccharomyces cerevisiae. J Biol Chem. 2001;276:47671–47674. doi: 10.1074/jbc.M104989200. [DOI] [PubMed] [Google Scholar]
- 64.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 65.Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, Emili A, Greenblatt JF, Harrington L, Lydall D, Durocher D. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell. 2006;124:1155–1168. doi: 10.1016/j.cell.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 66.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanakahi LA, Sun H, Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J Biol Chem. 1999;274:15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 68.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 69.Weitzmann MN, Woodford KJ, Usdin K. DNA secondary structures and the evolution of hypervariable tandem arrays. J Biol Chem. 1997;272:9517–9523. doi: 10.1074/jbc.272.14.9517. [DOI] [PubMed] [Google Scholar]
- 70.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat Chem Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simonsson T, Henriksson M. c-myc Suppression in Burkitt's lymphoma cells. Biochem Biophys Res Commun. 2002;290:11–15. doi: 10.1006/bbrc.2001.6096. [DOI] [PubMed] [Google Scholar]
- 73.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 76.Flaus A, Rencurel C, Ferreira H, Wiechens N, Owen-Hughes T. Sin mutations alter inherent nucleosome mobility. Embo J. 2004;23:343–353. doi: 10.1038/sj.emboj.7600047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Etzioni S, Yafe A, Khateb S, Weisman-Shomer P, Bengal E, Fry M. Homodimeric MyoD preferentially binds tetraplex structures of regulatory sequences of muscle-specific genes. J Biol Chem. 2005;280:26805–26812. doi: 10.1074/jbc.M500820200. [DOI] [PubMed] [Google Scholar]
- 78.Fry RC, Sambandan TG, Rha C. DNA damage and stress transcripts in Saccharomyces cerevisiae mutant sgs1. Mech Ageing Dev. 2003;124:839–846. doi: 10.1016/s0047-6374(03)00144-1. [DOI] [PubMed] [Google Scholar]
- 79.Shirude PS, Okumus B, Ying L, Ha T, Balasubramanian S. Single-Molecule Conformational Analysis of G-Quadruplex Formation in the Promoter DNA Duplex of the Proto-Oncogene C-Kit. J Am Chem Soc. 2007 doi: 10.1021/ja070497d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sacca B, Lacroix L, Mergny JL. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 82.Khateb S, Weisman-Shomer P, Hershco-Shani I, Ludwig AL, Fry M. The tetraplex (CGG)n destabilizing proteins hnRNP A2 and CBF-A enhance the in vivo translation of fragile X premutation mRNA. Nucleic Acids Res. 2007;35:5775–5788. doi: 10.1093/nar/gkm636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5' UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J Am Chem Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 85.Risitano A, Fox KR. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Teng Y, Girvan AC, Casson LK, Pierce WM, Jr, Qian M, Thomas SD, Bates PJ. AS1411 alters the localization of a complex containing protein arginine methyltransferase 5 and nucleolin. Cancer Res. 2007;67:10491–10500. doi: 10.1158/0008-5472.CAN-06-4206. [DOI] [PubMed] [Google Scholar]
- 87.Li B, Comai L. Requirements for the nucleolytic processing of DNA ends by the Werner syndrome protein-Ku70/80 complex. J Biol Chem. 2001;276:9896–9902. doi: 10.1074/jbc.M008575200. [DOI] [PubMed] [Google Scholar]
- 88.Azam M, Lee JY, Abraham V, Chanoux R, Schoenly KA, Johnson FB. Evidence that the S.cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence. Nucleic Acids Res. 2006;34:506–516. doi: 10.1093/nar/gkj452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee J, Kozak M, Martin J, Pennock E, Johnson FB. Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLOS Biology. 2007 doi: 10.1371/journal.pbio.0050160. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kamath-Loeb AS, Loeb LA, Johansson E, Burgers PM, Fry M. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J Biol Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- 91.Usdin K, Woodford KJ. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 93.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 94.Maizels N. Immunoglobulin gene diversification. Annu Rev Genet. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- 95.Larson ED, Duquette ML, Cummings WJ, Streiff RJ, Maizels N. MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr Biol. 2005;15:470–474. doi: 10.1016/j.cub.2004.12.077. [DOI] [PubMed] [Google Scholar]
- 96.Duquette ML, Huber MD, Maizels N. G-rich proto-oncogenes are targeted for genomic instability in B-cell lymphomas. Cancer Res. 2007;67:2586–2594. doi: 10.1158/0008-5472.CAN-06-2419. [DOI] [PubMed] [Google Scholar]
- 97.Muniyappa K, Anuradha S, Byers B. Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol Cell Biol. 2000;20:1361–1369. doi: 10.1128/mcb.20.4.1361-1369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu Z, Gilbert W. The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: implication of in vivo functions for this novel DNA structure. Cell. 1994;77:1083–1092. doi: 10.1016/0092-8674(94)90447-2. [DOI] [PubMed] [Google Scholar]
- 99.Bahler J, Hagens G, Holzinger G, Scherthan H, Heyer WD. Saccharomyces cerevisiae cells lacking the homologous pairing protein p175SEP1 arrest at pachytene during meiotic prophase. Chromosoma. 1994;103:129–141. doi: 10.1007/BF00352322. [DOI] [PubMed] [Google Scholar]
- 100.Assenmacher N, Hopfner KP. MRE11/RAD50/NBS1: complex activities. Chromosoma. 2004;113:157–166. doi: 10.1007/s00412-004-0306-4. [DOI] [PubMed] [Google Scholar]
- 101.D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 102.Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 103.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc Natl Acad Sci U S A. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen L, Trujillo KM, Van Komen S, Roh DH, Krejci L, Lewis LK, Resnick MA, Sung P, Tomkinson AE. Effect of amino acid substitutions in the rad50 ATP binding domain on DNA double strand break repair in yeast. J Biol Chem. 2005;280:2620–2627. doi: 10.1074/jbc.M410192200. [DOI] [PubMed] [Google Scholar]
- 106.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ghosal G, Muniyappa K. Saccharomyces cerevisiae Mre11 is a high-affinity G4 DNA-binding protein and a G-rich DNA-specific endonuclease: implications for replication of telomeric DNA. Nucleic Acids Res. 2005;33:4692–4703. doi: 10.1093/nar/gki777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ghosal G, Muniyappa K. The characterization of Saccharomyces cerevisiae Mre11/Rad50/Xrs2 complex reveals that Rad50 negatively regulates Mre11 endonucleolytic but not the exonucleolytic activity. J Mol Biol. 2007;372:864–882. doi: 10.1016/j.jmb.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 109.Giraldo R, Rhodes D. The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. Embo J. 1994;13:2411–2420. doi: 10.1002/j.1460-2075.1994.tb06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 111.Anuradha S, Muniyappa K. Meiosis-specific yeast Hop1 protein promotes synapsis of double-stranded DNA helices via the formation of guanine quartets. Nucleic Acids Res. 2004;32:2378–2385. doi: 10.1093/nar/gkh559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen YT, Venditti CA, Theiler G, Stevenson BJ, Iseli C, Gure AO, Jongeneel CV, Old LJ, Simpson AJ. Identification of CT46/HORMAD1, an immunogenic cancer/testis antigen encoding a putative meiosis-related protein. Cancer Immun. 2005;5:9. [PubMed] [Google Scholar]
- 113.Liu Z, Lee A, Gilbert W. Gene disruption of a G4-DNA-dependent nuclease in yeast leads to cellular senescence and telomere shortening. Proc Natl Acad Sci U S A. 1995;92:6002–6006. doi: 10.1073/pnas.92.13.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bashkirov VI, Solinger JA, Heyer WD. Identification of functional domains in the Sep1 protein (= Kem1, Xrn1), which is required for transition through meiotic prophase in Saccharomyces cerevisiae. Chromosoma. 1995;104:215–222. doi: 10.1007/BF00352186. [DOI] [PubMed] [Google Scholar]
- 115.Shobuike T, Sugano S, Yamashita T, Ikeda H. Cloning and characterization of mouse Dhm2 cDNA, a functional homolog of budding yeast SEP1. Gene. 1997;191:161–166. doi: 10.1016/s0378-1119(97)00053-x. [DOI] [PubMed] [Google Scholar]
- 116.Ligr M, Velten I, Frohlich E, Madeo F, Ledig M, Frohlich KU, Wolf DH, Hilt W. The proteasomal substrate Stm1 participates in apoptosis-like cell death in yeast. Mol Biol Cell. 2001;12:2422–2432. doi: 10.1091/mbc.12.8.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nelson LD, Musso M, Van Dyke MW. The yeast STM1 gene encodes a purine motif triple helical DNA-binding protein. J Biol Chem. 2000;275:5573–5581. doi: 10.1074/jbc.275.8.5573. [DOI] [PubMed] [Google Scholar]
- 118.Sun H, Bennett RJ, Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Weitao T, Budd M, Campbell JL. Evidence that yeast SGS1, DNA2, SRS2, and FOB1 interact to maintain rDNA stability. Mutat Res. 2003;532:157–172. doi: 10.1016/j.mrfmmm.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 120.Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt EC. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. Embo J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 121.Frantz JD, Gilbert W. A novel yeast gene product, G4p1, with a specific affinity for quadruplex nucleic acids. J Biol Chem. 1995;270:20692–20697. doi: 10.1074/jbc.270.35.20692. [DOI] [PubMed] [Google Scholar]