Summary
Toll-like receptor (TLR) responses are regulated to avoid toxicity and achieve coordinated responses appropriate for the cell environment. We found that Notch and TLR pathways cooperated to activate canonical Notch target genes, including transcriptional repressors Hes1 and Hey1, and to increase production of canonical TLR-induced cytokines TNF, IL-6 and IL-12. Cooperation by these pathways to increase target gene expression was mediated the Notch pathway component and transcription factor RBP-J, which also contributed to lethality after endotoxin injection. TLR- and Notch-induced Hes1 and Hey1 attenuated IL-6 and IL-12 production. This Hes1- and Hey1-mediated feedback inhibitory loop was abrogated by interferon-γ (IFN-γ), which blocked TLR-induced activation of canonical Notch target genes by inhibiting Notch2 signaling and downstream transcription. These findings identify new immune functions for RBP-J, Hes and Hey proteins and provide insights into mechanisms by which Notch, TLR and IFN-γ signals are integrated to modulate specific effector functions in macrophages.
Introduction
Toll-like receptors (TLRs) recognize conserved microbial structures and are important in activating innate immunity and regulating the transition from innate to acquired immune responses. Activation of macrophages with TLR ligands leads to production of inflammatory cytokines such as TNF and interleukin-1 (IL-1) and also cytokines of the IL-6 and IL-12 family, namely IL-6, IL-12, IL-23 and IL-27, that regulate T cell differentiation (Medzhitov, 2007). TLR2 and TLR4 recognize bacterial lipopeptides and lipopolysaccharides (LPS) and induce cytokine production via the downstream signaling molecules IκB kinases (IKKs) and mitogen activated protein kinases (MAPKs), which in turn activate transcription factors nuclear factor kappa B (NF-κB) and activator protein-1 (AP-1) (Kawai and Akira, 2007). In addition, emerging evidence suggests that TLRs also regulate transcription by inducing chromatin modifications in a gene-specific manner (Anest et al., 2003; Saccani et al., 2002; Yamamoto et al., 2003).
Unrestrained activation of TLR responses can lead to excessive inflammation and tissue damage, and contribute to pathogenesis of inflammatory disorders such as septic shock. Therefore, TLR signaling is subject to negative regulation and feedback inhibition (Liew et al., 2005). TLRs induce expression of counter-regulatory cytokines such as IL-10 and numerous signaling inhibitors (Lang and Mansell, 2007). TLRs also induce expression of transcriptional repressors, such as ATF3, that feed back and suppress expression of specific subsets of TLR-inducible genes (Gilchrist et al., 2006). In addition, TLRs induce remodeling and post-translational modification of chromatin (histone ‘marks’) that can either activate or silence gene expression (Foster et al., 2007; Ramirez-Carrozzi et al., 2006; Saccani et al., 2002). Silencing by such epigenetic modifications during endotoxin tolerance, which is likely mediated by transcriptional repressors (Foster et al., 2007), plays a key role in specifically restraining potentially toxic inflammatory cytokine expression, while allowing beneficial expression of host defense genes. Thus, selective regulation of subsets of TLR-inducible genes allows fine-tuning of distinct biological functions induced by TLRs.
The Notch signaling pathway regulates cell differentiation, proliferation, survival and development. In mammalian cells, there are four Notch receptors (Notch 1-4) and five Notch ligands (Jagged1, Jagged2, Delta-like 1 (DLL1), DLL3, and DLL4). Ligation of Notch receptors by their ligands leads to a two-step proteolytic cleavage of Notch by a disintegrin and metalloprotease (ADAM) family proteases and the intracellular γ-secretase complex that releases the Notch intracellular domain (NICD). NICD translocates to the nucleus and binds to the DNA-binding protein recombinant recognition sequence binding protein at the Jκ site (RBP-J, also named CSL or CBF1). This interaction results in displacement of RBP-J-associated transcriptional corepressors and assembly of a transcriptional activation complex that drives expression of Notch target genes (Bray, 2006). Among the best characterized direct Notch target genes are hairy and enhancer of split (Hes) and hairy and enhancer of split with YRPW motif (Hey) families of basic helix-loop-helix transcriptional repressors. Hes and Hey proteins function as feedback inhibitors of Notch-induced gene expression (Fischer and Gessler, 2007). Although RBP-J plays a key role in canonical Notch signal transduction, Notch can signal independently of RBP-J and RBP-J can be activated by alternative signaling pathways (Martinez Arias et al., 2002).
In the immune system, Notch signaling regulates multiple steps of T and B cell development (Tanigaki and Honjo, 2007), T cell activation (Eagar et al., 2004), regulatory T cell function (Ostroukhova et al., 2006) and T helper cell differentiation (Amsen et al., 2007; Amsen et al., 2004; Fang et al., 2007; Maillard et al., 2005; Osborne and Minter, 2007; Skokos and Nussenzweig, 2007). Notch ligands and receptors are induced on dendritic cells and macrophages by TLRs and various stimuli, and previous work has demonstrated a role for antigen presenting cell (APC)-expressed DLL in promoting T helper 1 (Th1) cell and Jagged in promoting Th2 cell differentiation. Investigation of Notch responses has focused predominantly on lymphocytes, and knowledge about the effects of Notch signaling in myeloid lineage cells is more limited. The Notch pathway has been implicated in dendritic cell (DC) differentiation and survival (Caton et al., 2007; Ohishi et al., 2001; Weijzen et al., 2002; Yamada et al., 2003), and TLRs have been suggested to indirectly activate noncanonical Notch pathways leading to NF-κB activation and TNF production by inducing Notch receptor and ligand expression on myeloid cells (Fung et al., 2007; Monsalve et al., 2006; Palaga et al., 2008).
The potent macrophage activating cytokine IFN-γ synergizes with TLRs to induce augmented production of inflammatory cytokines (Schroder et al., 2004). Mechanisms underlying this synergy and crosstalk between IFN-γ and TLR responses have not been resolved and are under active investigation. We previously demonstrated that IFN-γ suppresses TLR-induced expression of AP-1 proteins and IL-10 (Hu et al., 2007; Hu et al., 2006). In this study, we investigated whether IFN-γ suppresses expression of subsets of TLR-inducible genes that can be distinguished by their functions or regulatory requirements. We discovered that in primary macrophages TLRs rapidly induced expression of canonical Notch target genes, including the transcriptional repressors Hes1 and Hey1, by a direct mechanism that did not require upregulation of Notch receptors or ligands. Robust gene expression required synergy between TLR and canonical Notch pathways and thus the Notch pathway regulates expression of a subset of TLR-inducible genes. IFN-γ inhibited expression of these Notch-dependent TLR-inducible genes by suppressing intracellular NICD2 and subsequent Notch-dependent transcription, thus revealing a new mechanism by which IFN-γ inhibits gene expression. Furthermore, by using a genetic approach we demonstrated that RBP-J contributed to TLR-mediated production of IL-6 and IL-12. Production of IL-6 and IL-12 family cytokines was suppressed by Hes1 and Hey1, thereby revealing a new TLR-induced feedback inhibitory loop and identifying the first function of Hes1 and Hey1 in immunity. Overall, our results identify direct cooperation between TLR and Notch pathways that is modulated by IFN-γ to regulate specific components of TLR responses with distinct biological functions.
Results
TLRs Directly Induce Expression of Canonical Notch Target Genes
We extended our previous analysis of IFN-γ-mediated inhibition of TLR-induced gene expression (Hu et al., 2007; Hu et al., 2006) by using microarray analysis to define the subset of TLR-inducible genes that are inhibited by IFN-γ in primary human macrophages. Strikingly, the group of TLR-induced and IFN-γ-suppressed genes included the well established Notch target genes HES1, HEY1, IL2RA, IL7R, BATF and MYC (Table S1). These genes contain RBP-J sites in their promoters and are direct targets of the Notch pathway (Adler et al., 2003; Garcia-Peydro et al., 2006; Johansen et al., 2003; Maier and Gessler, 2000; Maillard et al., 2005; Nakagawa et al., 2000; Weng et al., 2006). Although IL2RA, IL7R, BATF and MYC expression is known to be induced by many pathways, expression of HES1 and HEY1 is regulated predominantly by Notch in most systems (Louvi and Artavanis-Tsakonas, 2006), and the functions of these proteins in immune responses remain to be determined.
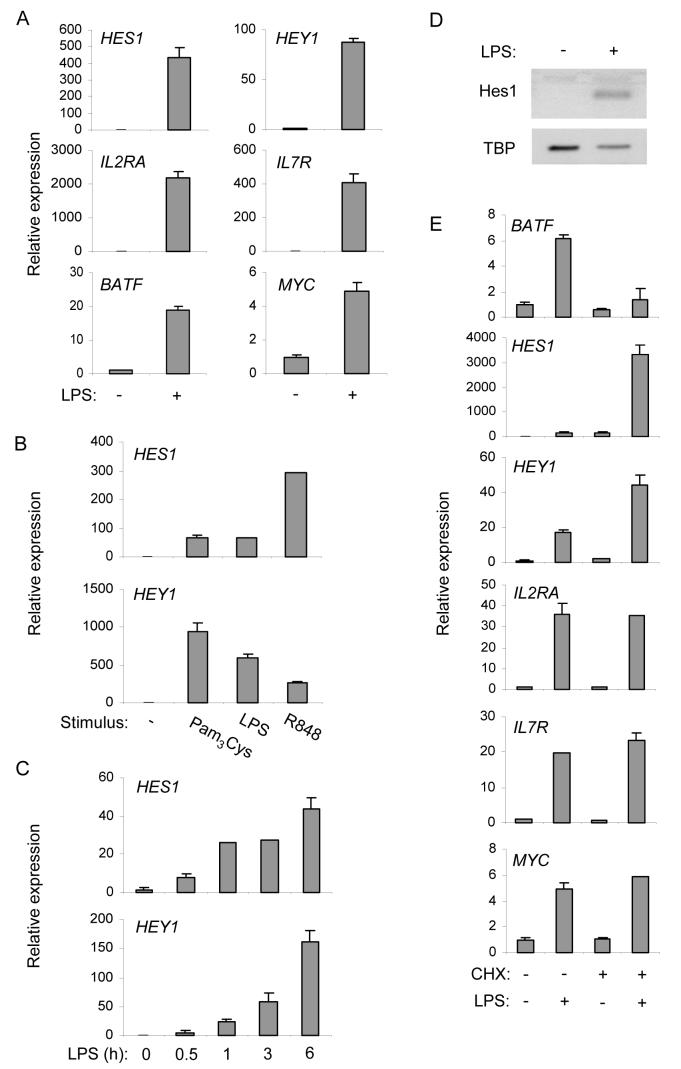
Activation of Notch target gene expression by TLR ligands was analyzed using quantitative real time PCR. Robust induction of BATF, HES1, HEY1, IL2RA, and IL7R mRNA was consistently observed in primary human macrophages derived from more than 20 blood donors, although the amount of induction relative to baseline expression was variable among different blood donors (a representative experiment using LPS is shown in Figure 1A). Expression of the canonical Notch target genes HES1 and HEY1 was effectively induced by Pam3Cys (a TLR2 ligand), LPS (TLR4 ligand), and R848 (TLR7-TLR8 ligand) (Figure 1B), indicating that activation of Notch target genes is a common feature of macrophage TLR responses. Induction of HES1 and HEY1 mRNA was apparent 0.5 h after LPS stimulation, and amounts of mRNA increased in a time-dependant manner with somewhat faster kinetics for HES1 (Figure 1C); in some donors an oscillatory but progressively increasing pattern of HES1 expression was observed, consistent with previous reports (Hirata et al., 2002; Yoshiura et al., 2007) (Figure S1). Induction of nuclear Hes1 protein was observed in macrophages treated for 6 h with LPS (Figure 1D) and Pam3Cys (data not shown). These results show that TLRs induce expression of Notch target genes, including the canonical Notch targets HES1 and HEY1 that are widely used as reporters of Notch pathway activity.
Figure 1.
TLRs induce Notch target gene expression
(A) Human primary macrophages were stimulated with 10 ng/ml of LPS for 3 h. mRNA expression of the indicated genes was measured by quantitative real time PCR and normalized relative to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA. Relative expression was calculated by setting expression of untreated samples as 1. Data are shown as means + SD of triplicate determinants and are representative of more than twenty experiments using cells derived from different human donors.
(B) Human macrophages were treated with the indicated TLR agonists for 3 h. Hes1 and Hey1 mRNA was measured by real time PCR. Data are shown as means + SD of triplicate determinants and are representative of two independent experiments.
(C) Human macrophages were stimulated with 10 ng/ml of LPS for the indicated periods. Data are shown as means + SD of triplicate determinants and one representative experiment out of three is shown.
(D) Human macrophages were treated with 10 ng/ml of LPS for 6 h. Nuclear extracts were analyzed using immunoblotting with antibodies against Hes1, followed by probing the same filter with antibodies against TBP. One representative experiment out of five performed is shown.
(E) Human macrophages were treated with 15 μg/ml CHX prior to stimulation with 10 ng/ml of LPS for 3 h, and mRNA of the indicated genes was measured using real time PCR. Data are shown as means + SD of triplicate determinants and are representative of four independent experiments.
TLRs induce expression of Notch ligands and receptors (Amsen et al., 2004; Fung et al., 2007; Monsalve et al., 2006; Palaga et al., 2008), which could potentially mediate indirect TLR-induced expression of Notch target genes. However, induction of HES1, HEY1, IL2RA, IL7R, and MYC by LPS occurred prior to induction of cell surface expression of Notch ligands (data not shown) and was intact in the presence of the protein synthesis inhibitor cycloheximide (CHX) (Figure 1E), and thus did not require de novo synthesis of Notch ligands or other proteins. These results show direct activation of Notch target genes by TLR pathway.
TLR-induced Gene Expression is Regulated by the Notch Pathway
We next tested the regulation of Notch signaling in human macrophages and whether TLR-induced activation of Notch target genes is dependent on the Notch pathway. Notch activation involves proteolytic release of NICD by the γ—secretase complex, and thus expression of NICD is used to monitor Notch pathway activity. Human macrophages constitutively expressed Notch1, Notch2, Jagged1 and DLL1, with minimal or no detectable expression of other Notch receptors or ligands (data not shown). Consistent with basal expression of ligands and receptors and previous observations of basal Notch activity in macrophages and DCs in vivo (Caton et al., 2007), we detected basal Notch activity, as evidenced by expression of NICD1 and NICD2 (Figure 2A, lane 1 and Figure S2B, lane 1). The basal expression of NICD1 was suppressed by two different γ—secretase inhibitors, compound E and L685458 (Figure 2A, lane 3 and 4), consistent with basal activation of Notch receptors in human macrophages. Basal Notch signaling was also manifested by basal expression of low amounts of HES1 and HEY1 mRNA (data not shown). LPS stimulation did not increase expression of NICD1 or NICD2 above baseline amounts within the time frame of these experiments, 1-6 h of stimulation where robust induction of Notch target genes was observed (Figure 2A, lane 2 and Figure S2). Similar results were obtained when cells were treated with Pam3Cys (data not shown). These data show that TLR signals did not directly activate canonical Notch signaling and suggest that TLRs may engage a parallel pathway that works together with Notch-mediated signals to induce the early phase of Notch target gene expression, which occurs independently of new protein synthesis.
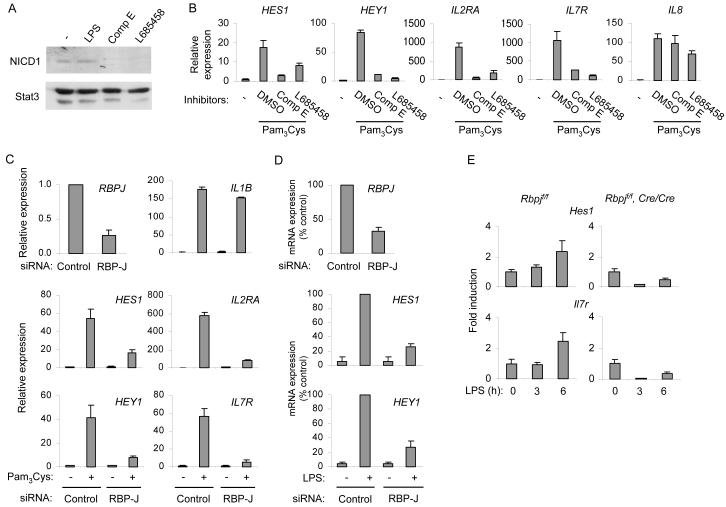
Figure 2.
Notch signaling is necessary for TLR induction of Notch target genes
(A) Human macrophages were activated with 10 ng/ml of LPS for 1 h (lane 2), or incubated with γ-secretase inhibitors Compound E (Comp E; 10 μM) or L685458 (5 μM) (lanes 3 and 4). Whole cell extracts were assayed for NICD1 by immunoblotting. Stat3 served as a loading control. One representative experiment out of two performed is shown.
(B) Human macrophages were pretreated with DMSO vehicle control or γ-secretase inhibitors for 2 d and subsequently stimulated with 10 ng/ml of Pam3Cys for 3 h. mRNA was measured by real time PCR. Data are shown as means + SD of triplicate determinants and are representative of five independent experiments.
(C, D) Primary human macrophages were transfected with control non-targeting or RBPJ-specific short interfering RNAs. 4 d post transfection, cells were stimulated with TLR ligands (10 ng/ml) for 3 h, and mRNA was measured using real time PCR. (C) Macrophages were activated with Pam3Cys. One representative experiment out of three performed is shown. Results are presented as mean + SD of triplicate determinants. (D) Macrophages were stimulated with LPS. Percentage of maximal expression was calculated relative to mRNA amounts in LPS-stimulated control cells and data are expressed as means + SD of three independent experiments.
(E) Bone marrow derived macrophages from Rbpjflox/flox (f/f) mouse and Rbpjf/f, Cre/Cre littermate controls were stimulated with 1 ng/ml of LPS for the indicated periods. RNA was extracted and mRNA was measured using real time PCR. Unstimulated controls within each genotype were set as 1. Data are shown as means + SD of triplicate determinants and are representative of three independent experiments. The difference in Hes1 expression between control and RBP-J-deficient macrophages at 6 h was statistically significant (P = 0.004 by paired Student’s t test).
We investigated the requirement for the Notch pathway in TLR-induced gene activation by inhibiting γ—secretase. Treatment of macrophages with γ—secretase inhibitors did not result in increased cell death or altered morphology (data not shown) and had minimal effects on TLR induction of IL8, a prototypical NF-κB target gene (Figure 2B, right panel). Notably, two different γ—secretase inhibitors suppressed TLR-induced expression of HES1, HEY1, IL2RA, and IL7R, suggesting that Notch signaling was necessary for gene induction. To obtain additional evidence supporting a role for the Notch pathway in gene activation, we used RNA interference to knock down expression of RBP-J, a master regulator of Notch target genes, in primary human macrophages. Although expression of Notch target genes was robustly induced in cells transfected with control, non-targeting siRNA, knockdown of RBPJ expression (Figure 2C, top left panel) resulted in diminished activation of HES1, HEY1, IL2RA, and IL7R by Pam3Cys and LPS (Figure 2C and 2D). These results were corroborated using an additional non-targeting control siRNA and two additional RBPJ-specific siRNAs (Figure S3). Induction of IL1B by TLR2 was not affected by knockdown of RBPJ expression (Figure 2C, upper panels), showing that decreased RBPJ expression did not result in global suppression of TLR responses.
Next, we wished to corroborate these results in mouse macrophages deficient in RBP-J. Because RBP-J deficient mice are not viable (Oka et al., 1995), we used macrophages from mice with myeloid-specific deletion of the floxed Rbpj gene (Tanigaki et al., 2002) that exhibited an approximately 80% reduction in RBP-J mRNA. Stimulation of control murine macrophages (obtained from littermate controls) resulted in a modest but reproducible increase in Hes1 expression (Figure 2E). This lower amplitude induction of Hes1 is most likely explained by low basal Notch receptor expression by murine macrophages in vitro (Fung et al., 2007; Monsalve et al., 2006; Palaga et al., 2008) and is consistent with amounts of Hes1 induction observed in other murine systems (Hirata et al., 2002; Yoshiura et al., 2007). Induction of Hes1 and also Il7r expression by LPS was abrogated in macrophages expressing low amounts of RBP-J (Figure 2E). Overall, results obtained from chemical inhibitor studies, RNA interference, and gene-deficient mice show that a subset of TLR-inducible genes is dependent on components of the Notch signaling pathway.
Notch Target Gene Induction is Dependent on Canonical TLR Signaling Pathways
We next investigated the interplay between Notch and canonical TLR signaling in the induction of Notch target genes in macrophages. Treatment of human macrophages with exogenous Jagged1 or DLL1 to maximally engage Notch receptors resulted in modest upregulation of HES1, HEY1, and IL7R expression relative to strong induction by LPS (Figure 3A). IL6 was not induced by Notch ligands, indicating that the induction of HES1, HEY1, and IL7R by these factors was not secondary to contamination with LPS (Figure 3A, lower panel). These results suggest that input from TLRs and synergy between Notch and TLR pathways is necessary to achieve full scale activation of Notch target genes in macrophages. This notion was further corroborated by transfection experiments with a Hes1 promoter-driven reporter gene (Ong et al., 2006). Cotransfection of RAW264.7 murine macrophage-like cells that do not exhibit basal Notch activity with the Hes1 reporter construct and NICD1 increased reporter gene activity, as expected (Figure 3B). Hes1 promoter activity was augmented when cells were stimulated with LPS only when NICD was expressed (Figure 3B), further supporting the notion that both Notch and TLR signals are required for maximal gene induction, and that these signals interact independently of Notch ligand induction.
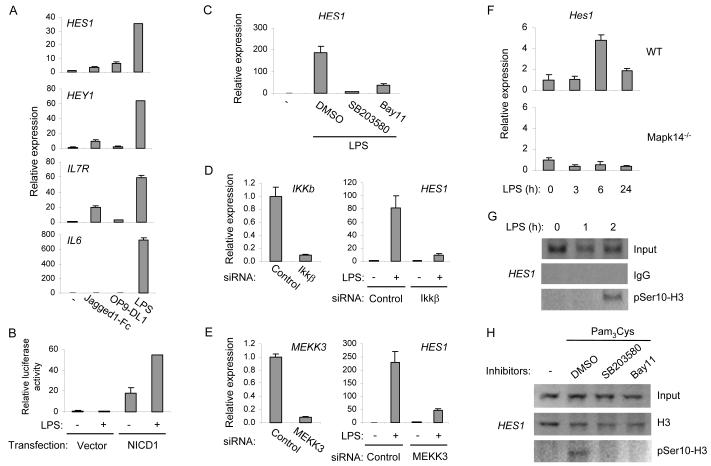
Figure 3.
TLRs contribute to Notch target gene activation by IKK- and p38-dependent pathways
(A) Human macrophages were treated with 10 μg/ml of Jagged1-Fc, cocultured with OP9-DL1 cells, or stimulated with 10 ng/ml of LPS for 3 h. mRNA was measured by real time PCR. Similar results were obtained when cells were treated with plate-bound Jagged1-Fc. Results are shown as means + SD of triplicate determinants and are representative of three independent experiments.
(B) RAW264.7 cells were cotransfected with a Hes1 reporter gene construct and an expression plasmid encoding NICD1 or a control empty vector. One day post transfection, cells were stimulated with 1 μg/ml of LPS for 6 h. Results were shown as normalized firefly luciferase activity relative to internal control and expressed as mean + SD from duplicate transfections. One representative experiment out of four performed is shown.
(C) Human macrophages were pretreated with DMSO vehicle control, 10 μM of Bay11-7082 (Bay11), or 10 μM of SB203580 for 30 min and stimulated with 10 ng/ml of LPS for 3 h. mRNA was measured using real time PCR. Results are shown as means + SD of triplicate determinants and are representative of seven independent experiments.
(D and E) Primary human macrophages were transfected with control or IKKβ-specific siRNAs (D) or MEKK3-specific siRNAs (E) using an Amaxa nucleofector. 4 d after transfection, cells were stimulated with LPS (10 ng/ml) for 3 h, and mRNA was measured using real time PCR. Data are shown as means + SD of triplicate determinants and are representative of three independent experiments.
(F) Bone marrow derived macrophages from control or Mapk14-deficient mice were stimulated with 10 ng/ml of LPS for the indicated time periods. mRNA was measured using real time PCR. Unstimulated controls within each genotype were set as 1. Data shown as means + SD of triplicate determinants and one experiment representative of two is shown.
(G) ChIP was perform with LPS-treated primary human macrophages using anti-phospho-serine 10 histone H3 (pSer10-H3) or rabbit IgG as a control. Immunoprecipitated DNA was analyzed by semiquantitative PCR with Hes1 promoter-specific primers.
(H) ChIP was done as in (G) with 30 min preincubation of inhibitors and 2 h of Pam3Cys stimulation. Data in (G) and (H) are representative of three independent experiments.
To delineate the pathway by which TLRs induce Notch target genes, chemical inhibitors of TLR signaling components were applied to human primary macrophages prior to stimulation. Within the time frame of these experiments (3 h), no apparent cell death or toxicity was observed with inhibitor treatments, and the efficacy of inhibitors was verified by their ability to suppress known TLR-inducible NF-κB and MAPK target genes such as IL6 and MKP1 (data not shown). Pharmacological inhibition of IKKs or the p38 MAPK consistently attenuated TLR-mediated induction of Notch target genes such as HES1, IL2RA and IL7R (Figure 3C, Figure S4, and Table S2). To further establish a role for IKKs and p38 in gene induction, we used RNAi to knock down components of these pathways in human macrophages and also used macrophages from gene-deficient mice. Knockdown of IKKα did not alter HES1 induction (data not shown) whereas knockdown of IKKβ (Figure 3D, left panel) strongly attenuated TLR-stimulated HES1 expression (Figure 3D). MEKK3 is a MAPKKK required for p38 activation downstream of TLRs (Huang et al., 2004). RNA interference-mediated MEKK3 knockdown resulted in reduced LPS-induced HES1 expression (Figure 3E). Furthermore, HES1 induction was abolished in macrophages with targeted deletion of the Mapk14 gene that encodes p38α (Figure 3F), an isoform of p38 that is critical for TLR responses in myeloid cells (Kang et al., 2008). Thus, both pharmacological and genetic evidence demonstrated that expression of Notch target genes was induced by TLR signaling via IKKβ- and p38-dependent pathways that worked in collaboration with basal Notch signals. Induction of IL2RA and IL7R genes by IKK- and MAPK-dependent pathways is well established, but a role for these pathways in inducing ‘dedicated’ Notch target genes such as Hes1 has not been previously appreciated.
Next, we sought to investigate the pathways downstream of IKKs and p38 that lead to Hes1 activation. We chose to study Hes1 because it is a classic Notch target gene and was very sensitive to MAPK and IKK inhibition. Hes1 is not known to be regulated by NF-κB or p38-activated transcription factors (Bray, 2006) and thus we examined phosphorylation of histone H3 at serine 10, a histone modification that is induced by both IKKs and p38 and is linked with recruitment of RNA polymerase II and transcriptional activation (Anest et al., 2003; Saccani et al., 2002; Yamamoto et al., 2003). Chromatin immunoprecipitation (ChIP) assays with primary human macrophages showed that TLR stimulation induced phosphorylation of histone H3 on serine 10 at the HES1 promoter (Figure 3G). This inducible phosphorylation was substantially reduced in macrophages treated with inhibitors of IKKs or p38, while these inhibitors did not alter total histone H3 occupancy on the HES1 promoter (Figure 3H). Taken together, the results support a link between TLR signaling and the HES1 locus and imply that TLRs contributed to Notch target gene activation in part by inducing histone H3 phosphorylation (Berger, 2007).
IFN-γ Inhibits Notch Responses in Macrophages
One striking characteristic of TLR-inducible Notch target genes is that they are suppressed by IFN-γ (Table S1 and Figure 4A), and we wished to further characterize the effects of IFN-γ. IFN-γ-mediated inhibition of Hes1 protein expression was confirmed by immunoblotting (Figure 4B). To determine whether IFN-γ affects transcription or post-transcriptional regulation, we measured expression of primary transcripts, a well-accepted measure of transcription rate (Murray, 2005). IFN-γ suppressed basal expression of HES1 and HEY1 primary transcripts (Figure 4C), thereby showing that IFN-γ inhibited Notch-dependent transcription. The addition of LPS resulted in additional RNA polymerase II recruitment to the HES1 promoter, and polymerase recruitment was suppressed by IFN-γ (Figure 4D). These results suggest that IFN-γ targets the Notch pathway and suppresses Notch-dependent transcription.
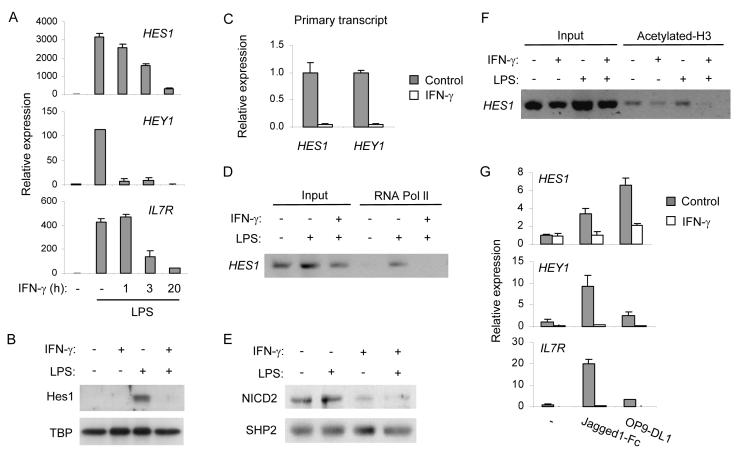
Figure 4.
IFN-γ inhibits Notch responses in macrophages
(A) Human macrophages were primed with 100 U/ml of IFN-γ for the indicated periods and stimulated with 10 ng/ml of LPS for 3 h. mRNA was measured using real time PCR.
(B) Human macrophages were treated with IFN-γ (100 U/ml) overnight, stimulated with 10 ng/ml of LPS, and nuclear Hes1 protein was measured by immunoblotting.
(C) Human macrophages were left untreated or treated with 100 U/ml of IFN-γ overnight. Primary transcripts were measured using real time PCR.
(D) Control or IFN-γ primed human macrophages were stimulated with 10 ng/ml of LPS for 2 h. RNA polymerase II (RNA Pol II) recruitment to Hes1 promoter was assessed by ChIP.
(E) Control or IFN-γ primed human primary macrophages were stimulated with 10 ng/ml of LPS for 6 h. Whole cell extracts were subjected to western blotting using an antibody that recognizes NICD2 (upper panel). The same filter was blotted with anti-SHP2 antibody (lower panel).
(F) Control or IFN-γ primed human macrophages were stimulated with 10 ng/ml of LPS for 2 h. Histone H3 K9K14 acetylation at the HES1 locus was assessed by ChIP.
(G) Human control or IFN-γ-treated macrophages were stimulated with Notch ligands for 3 h. mRNA was measured by real time PCR.
Data in A-G are representative of at least three independent experiments; in A, C and G data are shown as means + SD of triplicate determinants.
Next, we investigated the mechanisms by which IFN-γ inhibits TLR-inducible and Notch-dependent responses. Because IFN-γ does not attenuate and could augment canonical TLR4-induced signaling (Hu et al., 2005), we reasoned that IFN-γ may inhibit the Notch pathway. This notion was tested by examining the effects of IFN-γ on basal Notch signaling. IFN-γ did not inhibit Notch1 or Notch2 mRNA expression (data not shown) and we then assessed the effects of IFN-γ on amounts of NICD1 and NICD2, proteolytic fragments of Notch that mediate transcriptional activation. IFN-γ had minimal effects on the amounts of NICD1 (Figure S5A), but strikingly decreased cellular NICD2 amounts (Figure 4E) without suppressing full length Notch2 protein expression (Figure S5B). Decreased NICD2 would be predicted to result in diminished histone acetylation at Notch target promoters (Bray, 2006). Indeed, we found that IFN-γ suppressed H3 K9K14 acetylation at the HES1 locus (Fig. 4F, lane 8), whereas methylation of H3 K27 or phosphorylation of H3 serine 10 were not affected (data not shown). These results indicate that IFN-γ blocked Notch signaling at least in part by suppressing NICD2, the active fragment of the most highly expressed Notch receptor in human macrophages. The notion that IFN-γ suppressed the Notch pathway was supported by evidence that IFN-γ inhibited the induction of HES1 and HEY1 expression by the Notch ligands Jagged1 and DLL1 (Figure 4G). Collectively, these results indicate that IFN-γ targets the Notch pathway by inhibiting Notch signaling and downstream Notch-dependent transcription.
Role of Notch component RBP-J in TLR-induced cytokine responses
Having identified a role of TLR signaling in regulation of canonical Notch target genes, we sought to investigate the reciprocal regulation, namely the role of Notch components in canonical TLR responses such as inflammatory cytokine production. Notch activity has been suggested to augment NF-κB activation and TNF production in TLR-activated macrophages based upon experiments using γ-secretase inhibitors and RNAi-mediated suppression of Notch expression in RAW264.7 cells (Fung et al., 2007; Palaga et al., 2008). We confirmed and extended these results using a genetic approach showing that deletion of RBP-J in the myeloid compartment protected mice from endotoxin lethality in vivo and attenuated TLR-induced TNF production in vitro (Figure 5A). A comparable suppression of TLR-induced TNF production was observed in human macrophages when RBPJ expression was knocked down using RNAi (Figure S6B). These results support a physiological role for the Notch pathway in TLR responses. Next, we wished to determine whether RBP-J regulates expression of cytokines other than TNF. IL-6 and IL-12 protein production was decreased in Rbpj-deficient macrophages in response to LPS stimulation (Figure 5B), and a similar decrease in IL-6 production was observed in human macrophages when RBPJ expression was knocked down (Figure S6B). In contrast, Rbpj deficiency did not affect TLR induction of IL-1β mRNA (Figure 5C), consistent with the unaltered IL-1β expression in RBP-J-low human macrophages (Figure 3A). Constitutive binding of RBP-J to a conserved binding sequence in the Il6 promoter has been reported (Plaisance et al., 1997), suggesting a direct regulation of Il6 expression by RBP-J. Consistent with this notion, we found that a mutation that selectively blocks binding of RBP-J to the Il6 promoter (Plaisance et al., 1997) diminished LPS-induced Il6 promoter-driven reporter gene activity (Figure 5D). Thus, a role for RBP-J in mediating TLR induction of Il6 expression is supported by gene deletion, RNAi, and promoter mutagenesis data. Overall, these results show that RBP-J contributes to TLR-induced cytokine gene expression and further support direct cooperation between Notch and TLR pathways in gene regulation.
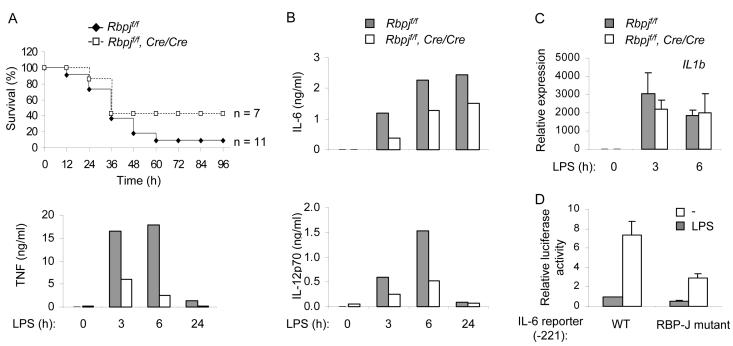
Figure 5.
RBP-J contributes to TLR-induced cytokine production
(A) Decreased TNF production and attenuated endotoxin lethality in RBP-J-deficient macrophages and mice with myeloid-specific deletion of Rbpj. Upper panel: Six to eight week old Rbpjflox/flox (f/f) and Rbpjf/f, Cre/Cre littermates were intraperitoneally injected with 1 mg of LPS and survival of animals was monitored over time. Lower panel: Bone marrow derived macrophages from Rbpjf/f mouse and Rbpjf/f, Cre/Cre littermate controls were stimulated with 1 ng/ml of LPS for the indicated periods. TNF protein amounts in culture supernatants were measured using ELISA.
(B) Control and RBP-J-deficient bone marrow derived macrophages were stimulated with 1 ng/ml of LPS for the indicated periods. IL-6 and IL-12p70 protein in culture supernatants were measured using ELISA. Data are representative of three independent experiments.
(C) Bone marrow derived macrophages were stimulated with 1 ng/ml of LPS for the indicated periods, and IL-1β mRNA was assessed using real time PCR. Data are shown as means + SD of triplicate determinants and are representative of at least three independent experiments.
(D) RAW264.7 cells were cotransfected with an IL-6 reporter construct containing 221 bases of the proximal wild-type IL-6 promoter or a construct with a T→G point mutation within the RBPJ binding site, at position −70 relative to the transcription start site. 24 h post transfection, cells were stimulated with 1 μg/ml of LPS for 6 h, and cell lysates were analyzed for luciferase activity. Results are presented as average relative luciferase activities of three independent experiments.
Feedback inhibition of cytokine production by Hes1 and Hey1
Hes1 and Hey1 function as feedback inhibitors and repressors of Notch-induced gene activation. The net effects of the Notch pathway on TLR responses will be determined by the balance between responses directly activated via RBP-J and the effectiveness and specificity of feedback inhibitors such as Hes1 and Hey1. We next tested the hypothesis that Hes1 and Hey1 may feed back and attenuate TLR-induced cytokine production. To study the intrinsic effects of the Hey1 gene in the hematopoietic compartment, we generated chimeric mice by reconstituting lethally irradiated recipients with wild-type or Hey1-/- bone marrow cells (Fischer et al., 2004). Pam3Cys-induced peritoneal exudates were significantly increased in Hey1-null bone marrow chimeras (Figure 6A), indicating an important role for Hey1 in regulating TLR-induced inflammation in vivo. To further characterize the role of Hey1 in TLR responses, bone marrow derived macrophages from chimeric mice were stimulated with Pam3Cys and gene expression and cytokine production were measured. Hey1 deficiency minimally affected TLR2-induced expression of Tnf or Il10 (Figure 6B). In contrast, TLR2-induced expression of IL-6 and IL-12 family cytokine genes, including Il6, Il12p40, Il12p35, and Il27p28, was elevated in Hey1-null macrophages (Figure 6B). The mRNA expression patterns were corroborated when the amounts of secreted IL-10, TNF, IL-6 and IL-12p70 proteins were measured (Figure 6C). These results demonstrate that Hey1 functions as a feedback inhibitor of TLR2-induced production of IL-6 and IL-12 family cytokines.
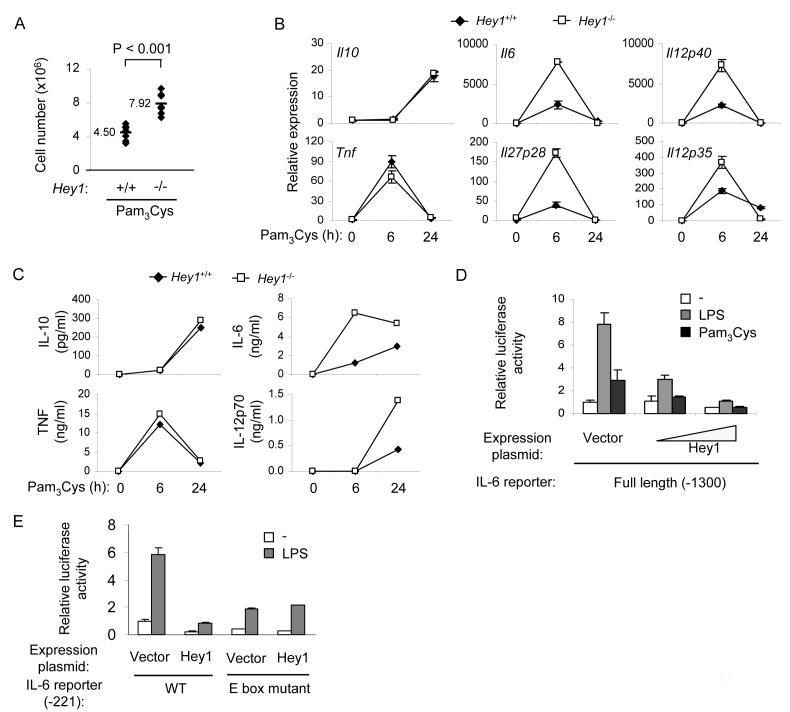
Figure 6.
Hey1 inhibits IL-6 and 12 family cytokines and regulates inflammation in vivo
(A) Peritonitis was induced in bone marrow chimeras reconstituted with Hey1+/+or Hey1-/- bone marrow cells by intraperitoneal injection of 100 μg of Pam3Cys per mouse. 1 d after Pam3Cys administration, peritoneal cells were harvested and counted. The results are cumulative from two independent experiments and n = 7 in each group. P value was calculated by unpaired Student’s t test.
(B) Bone marrow-derived murine macrophages were generated from Hey+/+ or Hey-/- bone marrow chimeras. Cells were stimulated with Pam3Cys, and mRNA was measured by real time PCR. Data are shown as means + SD of triplicate determinants and are representative of three independent experiments.
(C) Hey1+/+ or Hey1-/- bone marrow derived macrophages were stimulated with Pam3Cys and cytokine concentrations in culture supernatants were determined by ELISA. A representative experiment out of three performed is shown.
(D) RAW264.7 cells were cotransfected in duplicate with an IL-6 reporter construct containing 1300 bases of the proximal IL-6 promoter (full length) and a Hey1 expression plasmid or empty vector control. 24 h post transfection, cells were stimulated with 1 μg/ml of TLR agonists for 6 h, and cell lysates were analyzed for luciferase activity.
(E) Transfections of RAW264.7 cells were performed as in (D) with an IL-6 reporter construct containing 221 bases of the proximal wildtype IL-6 promoter or a construct with point mutations within the E box sequence (CAAATG→ACAATG). In (D) and (E) data are shown as mean + SD from duplicate transfections and are representative of three independent experiments.
Hes and Hey proteins suppress gene expression by a number of mechanisms that include binding to N boxes or suppressing E box-mediated transcription in promoters that contain tandem E boxes and RBP-J sites (Fischer and Gessler, 2007; Iso et al., 2003; Tanigaki and Honjo, 2007). We investigated the mechanism by which Hey1 inhibits IL-6 expression by analyzing the effects of Hey1 on the activity of an IL-6 promoter-driven reporter gene. We found that Hey1 suppressed TLR-induced activation of an IL-6 promoter-driven reporter gene that contains 1300 bp of promoter sequences in a dose-dependent manner in transient transfection assays (Figure 6D). These results show that Hey1 can suppress IL-6 promoter activity and suggest that transcriptional repressors such as Hey1 may directly regulate cytokine genes. The IL-6 promoter contains a potential N box sequence, but deletion of the N box did not abrogate repression of the IL-6 promoter by Hey1 (data not shown). The deletional analysis thus excluded a role for the IL-6 promoter N box-like sequence, and mapped the target of Hey1 repression to 221 bases of the proximal IL-6 promoter (Figure 6E, left). In addition to an RBP-J site (Plaisance et al., 1997), the IL-6 promoter contains a putative E box (−81 to −76 relative to transcription start site), and thus conforms to one type of promoter architecture that is inhibited by Hes and Hey proteins. Site directed mutagenesis showed that mutation of the E box sequence abrogated the ability of Hey1 to inhibit IL-6 promoter activity (Figure 6E). These results map the target sequence in the IL-6 promoter that mediates inhibition by Hey1, and suggest that Hey1 inhibits IL-6 expression by the established mechanism of inhibiting E box-RBP-J mediated transcription (Fischer and Gessler, 2007; Tanigaki and Honjo, 2007).
Next, we sought to evaluate the role of Hes1 in regulation of TLR-induced cytokine production using genetic approaches. RNAi-mediated approaches were not successful in attenuating TLR-induced Hes1 expression in human macrophages, and ablation of Hes1 in mice results in embryonic or neonatal lethality and thus function of Hes1 can not be readily studied in the adult immune system (Ishibashi et al., 1995). Instead, we generated Hes1-deficient and control fetal liver-derived macrophages from day 16.5 embryos from the same pregnancy. In Hes1-null macrophages, induction of TNF mRNA by TLR2 was comparable to that of wild-type controls, indicating that Hes1 deficiency did not globally alter TLR2 responses (Figure 7A, upper panel). In contrast to TNF, TLR2-induced IL-6 and IL-27 p28 mRNA expression was augmented in Hes1-deficient macrophages (Figure 7A). Consistent with increased amounts of IL-6 mRNA, Hes1-deficient cells produced higher amounts of IL-6 protein than did wild-type macrophages (Figure 7B). Similar to the effects of Hey1, Hes1 suppressed TLR-induced IL-6 reporter activity (Figure 7C). Overall, our results support a model of integrated TLR and Notch signaling where Hes1 and Hey1 fine tune TLR responses via feedback inhibition of cytokine production (Figure S7).
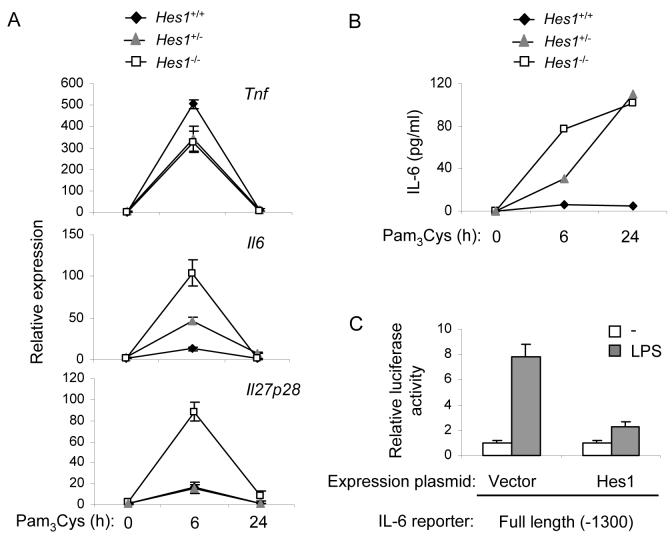
Figure 7.
Hes1 negatively regulates IL-6 expression
(A) Fetal liver-derived macrophages from wild-type, Hes1-haplodeficient or Hes1-deficient embryos were stimulated with 10 ng/ml of Pam3Cys. Cytokine mRNA expression was determined by real time PCR. Data are shown as means + SD of triplicate determinants and are representative of two independent experiments.
(B) IL-6 protein concentrations in supernatants of Pam3Cys-stimulated fetal liver-derived macrophages were analyzed by ELISA. Results are representative of two independent experiments.
(C) RAW264.7 cells were cotransfected in duplicate with the full length IL-6 reporter construct and a Hes1 expression plasmid or empty vector control. 24 h after transfection, cells were stimulated with 1 μg/ml of LPS for 6 h, and cell lysates were analyzed for luciferase activity. Data are shown as mean + SD from duplicate transfections and are representative of three independent experiments.
Discussion
The Notch pathway has been implicated in development and lymphocyte differentiation whereas TLRs play a key role in acute innate immune responses to microbial pathogens. In this study we demonstrated synergistic cooperation between the Notch pathway and acute TLR-induced signals in the activation of canonical Notch target genes such as Hes1 and Hey1, and of canonical TLR-inducible genes encoding cytokines of the IL-6 and IL-12 family. For activation of classic Notch target genes, TLRs induced an IKK- and p38-mediated signal that cooperated with canonical Notch signaling that was dependent on RBP-J. Conversely, RBP-J was required for full induction of IL-6 and IL-12 in response to TLR stimulation. Our findings support a model of cooperation between Notch and TLR pathways in the binary activation of gene expression that is mediated by RBP-J, which thus serves as an integration point for signaling by these two pathways.
Activation of canonical Notch target genes and cytokine genes was linked in a regulatory circiut, as Hes1 and Hey1 fed back to attenuate IL-6 and IL-12 expression. Ablation of Notch target gene Hey1 in hematopoietic cells resulted in augmented TLR-induced peritonitis, and diminished RBP-J expression in macrophages attenuated endotoxin lethality, thus supporting a physiological role for Notch-TLR crosstalk in inflammatory responses. In addition, IFN-γ selectively inhibited expression of TLR-inducible canonical Notch target genes by inhibiting NICD2 expression and downstream Notch-mediated transcription, thereby abrogating the Hes1-and Hey1-mediated feedback inhibitory loop and potentially augmenting cytokine production. These findings provide new insights into mechanisms by which Notch, TLR, and IFN-γ signals are integrated to modulate specific effector functions in macrophages.
The Notch pathway is best characterized as mediating cell-cell (juxtracrine) signaling between adjacent cells. In the immune system, the emphasis has been on investigation of the regulation of Notch-expressing progenitors and lymphocytes by Notch ligands Jagged and DLL that are expressed on stromal cells and APCs. Our findings highlight the importance of Notch signaling in APCs, in this case macrophages that express Notch ligands and receptors and thus have the capacity to both induce and respond to Notch signals. The Notch pathway can be activated in macrophages in vivo by Notch ligands that are constitutively or inducibly expressed by macrophages themselves, and also by Notch ligands expressed on stromal and epithelial cells in the marginal zone of the spleen, thymic epithelium and bone marrow stromal cells, or on stromal cells at inflammatory sites such as rheumatoid arthritis synovium (Caton et al., 2007; Tanigaki and Honjo, 2007).
Previous work suggests that TLR-induced Notch ligands on APCs can feed back on macrophages to augment NF-κB activation and TNF production (Amsen et al., 2004; Amsen et al., 2007; Fang et al., 2007; Fung et al., 2007; Monsalve et al., 2006; Palaga et al., 2008; Tanigaki and Honjo, 2007). In contrast, we have shown that Notch signals, when present contemporaneously with TLR stimulation, synergize with TLR signals to activate canonical Notch target genes and thereby regulate TLR responses and downstream functions, such as those regulated by Hes1 and Hey1. Thus, our work extends and modifies the current paradigm of TLR-Notch interactions to show that there is a reciprocal relationship and that Notch signaling participates in and regulates primary TLR responses. The functional outcome of TLR and Notch interaction is complex given that the key Notch transcription factor RBP-J positively regulates TLR induction of TNF, IL-6 and IL-12, whereas Notch and RBP-J target genes Hes1 and Hey1 negatively regulate TLR-induced IL-6 and IL-12 production and thereby fine tune TLR responses. The seemingly opposing actions of Notch components and Notch target genes may not be surprising given that, in the setting of developmental biology, feed back inhibition by Hes and Hey proteins is well established. The interaction of Notch and TLR pathways on cytokine gene promoters may also be direct and mediated by RBP-J, as the IL-6 promoter binds RBP-J (Plaisance et al., 1997) and we have provided evidence supporting a role for RBP-J and the IL-6 promoter RBP-J element in TLR-induced expression of IL-6.
One striking finding was that expression of Notch primary target genes was inhibited by IFN-γ. IFN-γ did not interrupt TLR4-induced proximal signaling (Hu et al., 2005) but instead suppressed NICD2 expression, Notch-dependent transcription, and TLR-induced recruitment of RNA polymerase II to Notch primary genes. IFN-γ did not inhibit expression of unprocessed Notch2 protein, suggesting that IFN-γ either prevents Notch2 cleavage or destabilizes the released intracellular NICD2 fragment. The exact mechanism of regulation of NICD2 by IFN-γ will be an interesting subject for future investigation. Because a Notch-dependent input was required for effective gene activation, suppression of Notch transcription provides a new mechanism by which IFN-γ modulates TLR responses by specifically suppressing expression of TLR-inducible canonical Notch target genes. In contrast, IFN-γ is well known for its capacity to augment TLR-induced production of cytokines including IL-6 and IL-12. There are multiple mechanisms underlying this priming effect, which appear to over-ride any negative effects on IL-6 and IL-12 expression secondary to IFN-γ-mediated decreases in Notch pathway activity. In addition, our results suggest that suppression of Hes1 and Hey1 expression by IFN-γ and interruption of this Notch-dependent feedback inhibition loop corresponds to a new mechanism by which IFN-γ augments cytokine production. Interestingly, induction of Hes4 by the Notch pathway in neural stem cells was inhibited by ciliary neurotrophic factor (CNTF), which, similar to IFN-γ, activates the Jak-STAT pathway (Androutsellis-Theotokis et al., 2006). Our findings, in conjunction with this report, suggest that cytokines that activate the Jak-STAT pathway may more broadly suppress Notch responses during cell activation and differentiation, and that the effects of IFN-γ (and potentially other cytokines) may need to be considered in investigation of the effects of the Notch pathway on T cell differentiation.
Hes and Hey inhibit transcription of primary Notch target genes in hematopoietic progenitors by targeting E box proteins, and also inhibit genes directly by binding to conserved N box sequences in gene promoters (Fischer and Gessler, 2007; Iso et al., 2003). Our results mapped inhibition of IL-6 expression to an E box-like element in the IL-6 promoter, suggesting that Hes and Hey proteins can inhibit expression of at least some cytokines by targeting E box-mediated regulation. Future work will determine whether IL-12 p40 and p35 promoters are inhibited via similar mechanisms, via binding of Hes1 and Hey1 to promoter N box elements, or by other or indirect mechanisms. The functions of Hes1 and Hey1 were not redundant in our system, possibly because these are the only 2 Hes and Hey family members expressed in macrophages (X. Hu, unpublished observations) or because Hes1 and Hey1 function more effectively as heterodimers (Iso et al., 2003). In contrast to IL-6 and IL-12, we found that Hes1 and Hey1 did not inhibit expression of the prototypic pro-inflammatory cytokines TNF and IL-1. Thus, Hes1 and Hey1 do not function to restrain TNF-mediated inflammation that is induced by TLRs and augmented by the Notch-RBP-J pathway. Our findings provide an immune function for Hes and Hey proteins and suggest that they selectively regulate cytokines that modulate the transition from innate to acquired immunity. Understanding mechanisms by which specific aspects of TLR responses are regulated is important for understanding the balance between the beneficial and potentially toxic effects of TLR stimulation.
Experimental Procedures
Cell Culture
CD14+ monocytes were purified from fresh peripheral blood mononuclear cells (PBMCs) using anti-CD14 magnetic beads (Miltenyi Biotec) and were cultured in RPMI 1640 medium with 10 % FBS (Hyclone) and 10 ng/ml of M-CSF (Peprotech). Murine bone marrow derived macrophages were obtained as described (Hu et al., 2002) and maintained in DMEM supplemented with 20 % FBS and 10 ng/ml of murine M-CSF (Peprotech). For fetal liver derived macrophages, fetal livers were harvested from E16.5 embryos of hes1+/- x hes1+/- matings (on ICR background) and homogenized liver cells were cultured in DMEM supplemented with 20 % FBS and 10 ng/ml of murine M-CSF for 2 weeks. The experiments using human and murine cells were approved by, respectively, the Hospital for Special Surgery Institutional Review Board and Institutional Animal Care and Use Committee.
Mice
Mice with myeloid-specific deletion of RBP-J were generated by crossing Rbpjflox/flox animals (Tanigaki et al., 2002) to animals with a lysozyme-driven Cre transgene on the C57/BL6 background (Jackson). F1 littermates with Rbpjflox/flox or Rbpjflox/flox, Cre/Cre genotypes were used for experiments. To generate bone marrow chimeras, donor bone marrow cells were harvested from wild type or Hey1-/- animals (on C57/BL6 background) and one fourth of total bone marrow cells was injected intravenously into each of the irradiated recipients. Recipient mice (six week old C57/BL6 mice) were obtained from Jackson Laboratory and were lethally irradiated with a single dose of 875 rads one day prior to transplantation. Chimeric mice were sacrificed 7 weeks post transplantation. Myeloid-specific p38α knockout mice were previously described (Kang et al., 2008).
Reagents
Pam3Cys was purchased from EMC microcollections and R848 was from Invivogen. Cell culture grade LPS, CHX, L685458, and SB203580 were from Sigma-Aldrich. Compound E and Bay11-7082 were obtained from Alexis Biochemicals and Calbiochem respectively. Recombinant Jagged1/Fc chimeric protein was from R&D Systems.
Analysis of mRNA and protein
Real time PCR, immunoblotting and ELISA were performed as previously described (Hu et al., 2002). Primary transcripts were measured using primers that either amplify exon-intron junctions or intronic sequences. For Hes1 and Hey1 primary transcripts, similar results were obtained using multiple primer sets targeting different regions of the same genes. For immunoblotting, anti-Hes1 (clone H-140), anti-TBP, and anti-SHP2 antibodies were from Santa Cruz Biotechnology, and anti-NICD1 and anti-Stat3 antibodies were from Cell Signaling. An anti-Notch2 antibody developed by Dr. Artavanis-Tsakonas was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242.
RNA Interference (RNAi)
Prevalidated RBPJ-, IKKB- and MEKK3-specific short interfering RNAs (siRNAs) and non-targeting control siRNAs were purchased from Dharmacon and Invitrogen. siRNAs were transfected into primary human macrophages with the Amaxa Nucleofector device set to program Y-001 using the Human Monocyte Nucleofector kit. Comparable results were obtained when macrophages were transfected with Lipofectamine RNAi Max reagent (Invitrogen).
Transient Transfection and Luciferase Assay
For Hes1 reporter gene assays, RAW264.7 cells were transfected in duplicate in 24-well plates with a Hes1 reporter construct and an expression plasmid encoding NICD1 or a control empty vector, and an internal control plasmid encoding renilla luciferase (Promega), using Lipofectamine Plus reagent from Invitrogen. On the next day, cells were stimulated with LPS for 6 h, and cell lysates were prepared and analyzed for firefly and renilla luciferase activity using the Dual-Luciferase Reporter Assay System (Promega). Results were shown as firefly normalized relative to renilla luciferase activity. For IL-6 reporter gene assays, a 1.3 kb mouse IL-6 promoter fragment was subcloned into pGL3-Basic vector (Promega). Deletion constructs were generated by cloning PCR products containing various fragments of the IL-6 promoter into pGL3-Basic vector and point mutations were generated using the QuikChange® Site-directed Mutagenesis kit (Stratagene). RAW264.7 cells were transfected with pGL3-IL6-Luc-derived reporter plasmids and an expression plasmid encoding Hes1 or Hey1 or a control vector pCMVXL4 (Origene) as described for Hes1 reporter gene assays.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed using the ChIP Assay Kit (Upstate) following the manufacturer’s instructions. 10 × 106 human primary macrophages were used per condition, and 20% of chromatin from each condition was used per IP. Antibodies against RNA polymerase II (Santa Cruz Biotechnology), anti-phospho-histone H3 Ser10 clone MC463 (Upstate), anti-acetylhistone H3 (Upstate), anti-total histone H3 (Upstate) and control rabbit IgG (Santa Cruz Biotechnology) were used. The primers used to amplify the human Hes1 promoter are: sense GGTGCCGCGTGTCTCCTCCTC; anti-sense ACCGCCCTTACCTTTCTGTGCTCA.
Microarray Data
The microarray data set has been deposited with the GEO database, accession number GSE11864.
Supplementary Material
Acknowledgements
We thank Raphael Kopan for the Hes1 reporter and NICD1 expression plasmids, Amer Beg for the IL-6 reporter construct, Tasuku Honjo for permission to use Rbpj floxed mice, and Elaine Fuchs for transfer of Rbpj floxed mice. We also thank Chao Shi for some of the RBPJ RNAi experiments, Kyung-Hyun Park-Min for critical review of the manuscript and Inez Rogatsky for helpful discussions. This work was supported by grants from the Arthritis Foundation (X.H.) and the National Institutes of Health (L.B.I.).
References
- Adler SH, Chiffoleau E, Xu L, Dalton NM, Burg JM, Wells AD, Wolfe MS, Turka LA, Pear WS. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J Immunol. 2003;171:2896–2903. doi: 10.4049/jimmunol.171.6.2896. [DOI] [PubMed] [Google Scholar]
- Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, Flavell RA. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagar TN, Tang Q, Wolfe M, He Y, Pear WS, Bluestone JA. Notch 1 signaling regulates peripheral T cell activation. Immunity. 2004;20:407–415. doi: 10.1016/s1074-7613(04)00081-0. [DOI] [PubMed] [Google Scholar]
- Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Gessler M. Delta-Notch--and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 2007;35:4583–4596. doi: 10.1093/nar/gkm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- Fung E, Tang SM, Canner JP, Morishige K, Arboleda-Velasquez JF, Cardoso AA, Carlesso N, Aster JC, Aikawa M. Delta-like 4 induces notch signaling in macrophages: implications for inflammation. Circulation. 2007;115:2948–2956. doi: 10.1161/CIRCULATIONAHA.106.675462. [DOI] [PubMed] [Google Scholar]
- Garcia-Peydro M, de Yebenes VG, Toribio ML. Notch1 and IL-7 receptor interplay maintains proliferation of human thymic progenitors while suppressing non-T cell fates. J Immunol. 2006;177:3711–3720. doi: 10.4049/jimmunol.177.6.3711. [DOI] [PubMed] [Google Scholar]
- Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Kennedy K, Hai T, Bolouri H, Aderem A. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- Hu X, Chen J, Wang L, Ivashkiv LB. Crosstalk among Jak-STAT, Toll-like receptor, and ITAM-dependent pathways in macrophage activation. J Leukoc Biol. 2007;82:237–243. doi: 10.1189/jlb.1206763. [DOI] [PubMed] [Google Scholar]
- Hu X, Herrero C, Li WP, Antoniv TT, Falck-Pedersen E, Koch AE, Woods JM, Haines GK, Ivashkiv LB. Sensitization of IFN-gamma Jak-STAT signaling during macrophage activation. Nat Immunol. 2002;3:859–866. doi: 10.1038/ni828. [DOI] [PubMed] [Google Scholar]
- Hu X, Ho HH, Lou O, Hidaka C, Ivashkiv LB. Homeostatic role of interferons conferred by inhibition of IL-1-mediated inflammation and tissue destruction. J Immunol. 2005;175:131–138. doi: 10.4049/jimmunol.175.1.131. [DOI] [PubMed] [Google Scholar]
- Hu X, Paik PK, Chen J, Yarilina A, Kockeritz L, Lu TT, Woodgett JR, Ivashkiv LB. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity. 2006;24:563–574. doi: 10.1016/j.immuni.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Huang Q, Yang J, Lin Y, Walker C, Cheng J, Liu ZG, Su B. Differential regulation of interleukin 1 receptor and Toll-like receptor signaling by MEKK3. Nat Immunol. 2004;5:98–103. doi: 10.1038/ni1014. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Johansen LM, Deppmann CD, Erickson KD, Coffin WF, 3rd, Thornton TM, Humphrey SE, Martin JM, Taparowsky EJ. EBNA2 and activated Notch induce expression of BATF. J Virol. 2003;77:6029–6040. doi: 10.1128/JVI.77.10.6029-6040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YJ, Chen J, Otsuka M, Mols J, Ren S, Wang Y, Han J. Macrophage deletion of p38alpha partially impairs lipopolysaccharide-induced cellular activation. J Immunol. 2008;180:5075–5082. doi: 10.4049/jimmunol.180.7.5075. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Lang T, Mansell A. The negative regulation of Toll-like receptor and associated pathways. Immunol Cell Biol. 2007;85:425–434. doi: 10.1038/sj.icb.7100094. [DOI] [PubMed] [Google Scholar]
- Liew FY, Xu D, Brint EK, O’Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Maier MM, Gessler M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem Biophys Res Commun. 2000;275:652–660. doi: 10.1006/bbrc.2000.3354. [DOI] [PubMed] [Google Scholar]
- Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- Arias A. Martinez, Zecchini V, Brennan K. CSL-independent Notch signalling: a checkpoint in cell fate decisions during development? Curr Opin Genet Dev. 2002;12:524–533. doi: 10.1016/s0959-437x(02)00336-2. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Monsalve E, Perez MA, Rubio A, Ruiz-Hidalgo MJ, Baladron V, Garcia-Ramirez JJ, Gomez JC, Laborda J, Diaz-Guerra MJ. Notch-1 up-regulation and signaling following macrophage activation modulates gene expression patterns known to affect antigen-presenting capacity and cytotoxic activity. J Immunol. 2006;176:5362–5373. doi: 10.4049/jimmunol.176.9.5362. [DOI] [PubMed] [Google Scholar]
- Murray PJ. The primary mechanism of the IL-10-regulated antiinflammatory response is to selectively inhibit transcription. Proc Natl Acad Sci USA. 2005;102:8686–8691. doi: 10.1073/pnas.0500419102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa O, McFadden DG, Nakagawa M, Yanagisawa H, Hu T, Srivastava D, Olson EN. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc Natl Acad Sci USA. 2000;97:13655–13660. doi: 10.1073/pnas.250485597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi K, Varnum-Finney B, Serda RE, Anasetti C, Bernstein ID. The Notch ligand, Delta-1, inhibits the differentiation of monocytes into macrophages but permits their differentiation into dendritic cells. Blood. 2001;98:1402–1407. doi: 10.1182/blood.v98.5.1402. [DOI] [PubMed] [Google Scholar]
- Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak TW, Honjo T. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem. 2006;281:5106–5119. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- Osborne BA, Minter LM. Notch signalling during peripheral T-cell activation and differentiation. Nat Rev Immunol. 2007;7:64–75. doi: 10.1038/nri1998. [DOI] [PubMed] [Google Scholar]
- Ostroukhova M, Qi Z, Oriss TB, Dixon-McCarthy B, Ray P, Ray A. Treg-mediated immunosuppression involves activation of the Notch-HES1 axis by membrane-bound TGF-beta. J Clin Invest. 2006;116:996–1004. doi: 10.1172/JCI26490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaga T, Buranaruk C, Rengpipat S, Fauq AH, Golde TE, Kaufmann SH, Osborne BA. Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur J Immunol. 2008;38:174–183. doi: 10.1002/eji.200636999. [DOI] [PubMed] [Google Scholar]
- Plaisance S, Vanden Berghe W, Boone E, Fiers W, Haegeman G. Recombination signal sequence binding protein Jkappa is constitutively bound to the NF-kappaB site of the interleukin-6 promoter and acts as a negative regulatory factor. Mol Cell Biol. 1997;17:3733–3743. doi: 10.1128/mcb.17.7.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]
- Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- Skokos D, Nussenzweig MC. CD8- DCs induce IL-12-independent Th1 differentiation through Delta 4 Notch-like ligand in response to bacterial LPS. J Exp Med. 2007;204:1525–1531. doi: 10.1084/jem.20062305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, Kuroda K, Suzuki A, Nakano T, Honjo T. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8:451–456. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- Weijzen S, Velders MP, Elmishad AG, Bacon PE, Panella JR, Nickoloff BJ, Miele L, Kast WM. The Notch ligand Jagged-1 is able to induce maturation of monocyte-derived human dendritic cells. J Immunol. 2002;169:4273–4278. doi: 10.4049/jimmunol.169.8.4273. [DOI] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Yamazaki H, Yamane T, Yoshino M, Okuyama H, Tsuneto M, Kurino T, Hayashi S, Sakano S. Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells. Blood. 2003;101:2227–2234. doi: 10.1182/blood-2002-06-1740. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- Yoshiura S, Ohtsuka T, Takenaka Y, Nagahara H, Yoshikawa K, Kageyama R. Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc Natl Acad Sci USA. 2007;104:11292–11297. doi: 10.1073/pnas.0701837104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.