Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) is a member of the ATP-binding cassette (ABC) transporter superfamily, an ancient family of proteins found in all phyla. In nearly all cases, ABC proteins are transporters that couple the hydrolysis of ATP to the transmembrane movement of substrate via an alternating access mechanism. In contrast, CFTR is best known for its activity as an ATP-dependent chloride channel. We asked why CFTR, which shares the domain architecture of ABC proteins that function as transporters, exhibits functional divergence. We compared CFTR protein sequences to those of other ABC transporters, which identified the ABCC4 proteins as the closest mammalian paralogs, and used statistical analysis of the CFTR-ABCC4 multiple sequence alignment to identify the specific domains and residues most likely to be involved in the evolutionary transition from transporter to channel activity. Among the residues identified as being involved in CFTR functional divergence, by virtue of being both CFTR-specific and conserved among all CFTR orthologs, was R352 in the sixth transmembrane helix (TM6). Patch-clamp experiments show that R352 interacts with D993 in TM9 to stabilize the open-channel state; D993 is absolutely conserved between CFTRs and ABCC4s. These data suggest that CFTR channel activity evolved, at least in part, by converting the conformational changes associated with binding and hydrolysis of ATP, as are found in true ABC Transporters, into an open permeation pathway by means of intraprotein interactions that stabilize the open state. This analysis sets the stage for understanding the evolutionary and functional relationships that make CFTR a unique ABC transporter protein.
Keywords: ion channel, molecular evolution, CFTR, Type II divergence
Cystic fibrosis (CF) is the most common lethal, autosomal recessive disease affecting Caucasians in the United States. It arises from mutations in the gene encoding the CF transmembrane conductance regulator (CFTR), which is a member of the large superfamily of ATP-binding cassette (ABC) proteins (1, 2). The CFTR protein contains five functional domains: two transmembrane domains (TMD1 and TMD2), two cytoplasmic ATP-binding domains (ABC domains), and a cytoplasmic regulatory domain (R domain) [Fig. 1 and supporting information (SI) Fig. S1]. Activation of CFTR requires protein kinase A-mediated phosphorylation of the R domain followed by binding and hydrolysis of ATP at the ABC domains (2). CFTR is expressed in a variety of cells of epithelial origin; its loss or malfunction in epithelial cells of the small airways leads to alterations in the volume and composition of the airway surface liquid that are associated with chronic inflammation underlying CF lung disease (3).
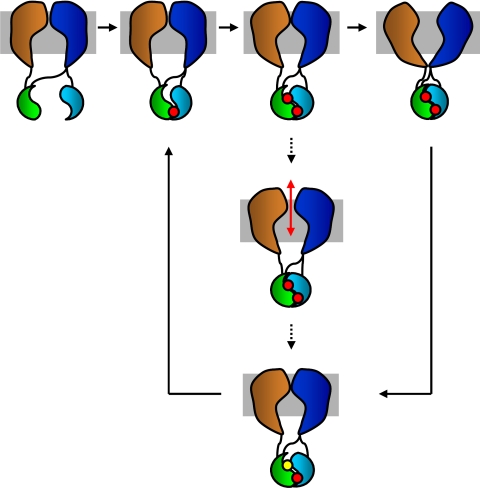
Fig. 1.
CFTR domain architecture, and schematic of transitions of ABC transporter gating. The solid arrows depict transporter activity during an alternating-access type mechanism whereas the dotted arrows show a theoretical cycle for CFTR activity during which the permeation pathway is open to both extracellular and intracellular solutions, allowing transport down an electrochemical gradient. Note that the final closing step is the same for both the alternating-access and channel gating mechanism. Gray, membrane; orange and dark blue, TMD1 and TMD2; green and light blue, ABC domains; red, ATP; yellow, ADP. The R-domain is omitted for clarity.
ABC transporter proteins are membrane-associated proteins that possess the characteristic, and highly conserved, ABC ATPase domain. Proteins of the ABC transporter superfamily are engaged in numerous cellular processes and their regulation via the import and export of a variety of substrates. Phylogenetic analysis of the ABC transporter superfamily revealed that CFTR proteins belong to class I, which includes most known exporters, and specifically within the OAD subclass that also includes the SUR proteins that regulate ion channel function (4). There are numerous ABC protein families (A–G) encoded in the human genome, and CFTR is a member of the ABCC family (1).
Although CFTR is a member of the ABC transporter superfamily, it clearly diverges from other ABC proteins in that it functions as an ion channel; no other member of the superfamily is known to bear channel activity. The ion channel function of CFTR has been directly demonstrated for three distantly related vertebrates: human, mouse, and Xenopus. For all three species, heterologous expression of a cDNA encoding the CFTR gene product has led to the appearance of CFTR-like channels (with characteristic chloride selectivity, activation by PKA, etc.) (5–7).
Channels and transporters are thought to operate by entirely divergent mechanisms. True transporters, such as the glucose transporter or excitatory amino acid transporters, move substrates across membranes down their electrochemical gradients by a mechanism commonly called the “alternating access” mechanism (8, 9). The transporter begins in a conformation permissive to binding substrate on one side of the membrane, undergoes a conformational change within the membrane domain, at least, and occupies a conformation permissive to release of substrate on the opposite side of the membrane; on release of substrate, the transporter returns to the initial conformation (see Fig. 1). At no time in this cycle is the substrate translocation pathway open at both ends. In secondary-active transporters, such as the Na+-coupled glucose cotransporter, and primary-active transporters, such as the ubiquitous Na+/K+-ATPase, the addition of chemical energy allows the transporter to also move substrate up its electrochemical gradient. Clearly, a leak in the substrate transport pathway, by way of being open simultaneously at both ends, would not support active transport.
In contrast, ion channels must occupy two major conformational states: one where the substrate transport pathway is blocked at one or multiple sites—the closed channel state—and one where the pathway is open throughout, thus allowing electrodiffusion—the open channel state. The membrane transport field has long considered transporters and channels to be fundamentally different, although there is some overlap in the absolute rates of substrate movement; some fast transporters move substrates at rates very similar to some very low-conductance ion channels. Indeed, the ClC family includes both transporters and channels that are structurally very similar (10, 11).
Because CFTR is related to a family of transport proteins, one may ask how this protein evolved to function as an ion channel. It seems likely that, in its evolutionary history, CFTR was responsible for accomplishing the movement of a substrate by a transporter mechanism; indeed, some evidence exists suggesting that CFTR may serve as the major pathway for transport of glutathione into the airway (12). At some point, CFTR diverged from its transporter relatives by adopting a conformation that allows diffusive movement of chloride, thus behaving as an ion channel (Fig. 1). Hence, the changes in conformation associated with the alternating access mechanism of true transporters may have been converted into the formation of a permeation pathway, open to both sides of the membrane; in CFTR, these changes in conformation would thus gate the channel between the closed and open states.
The evolutionary determinants of the functional differences between CFTR and closely related ABC transporters are poorly understood. We integrated computational approaches, including comparative sequence and structure analysis, and experimental approaches to determine the functional consequences of mutations at divergent residues to uncover the characteristic evolutionary differences in CFTR that are most likely to be relevant to its distinct function as an ion channel. The analysis identified a number of residues as likely to have been involved in the evolutionary transition from transporter to ion channel, including a pair of residues that we demonstrate to interact to stabilize the channel open state (13). Hence, this approach not only can help to make sense of existing functional data on CFTR, but can also guide further experimental analysis of the system by elucidating the residues most likely to be specifically involved in ion channel function.
Results
Domain Architecture Evolution of CFTR.
The CFTR protein consists of four distinct domains organized into two repeated units (Fig. 1 and Fig. S1). Each unit consists of a TMD, with six transmembrane helices (TMs), followed by the highly conserved ABC region, which is the ABC transporter superfamily characteristic domain. In CFTR, there is an additional R domain that lies between the repeated TMD-ABC units. CFTR and 47 paralogous human ABC sequences were characterized with respect to both their domain architectures and phylogenetic relationships (Fig. S2). Related members of the ABC transporter superfamily show different combinations and organizations of domains. In contrast to CFTR, some proteins, for example, members of the ABCB and ABCD families, are composed of a single TMD-ABC unit whereas others have a reversed ABC-TMD architecture (ABCG) or two ABC domains with no TMD (ABCE and ABCF) (Fig. S2).
Human ABC domain sequences from CFTR and related ABCs were isolated for phylogenetic analysis (Fig. S3A) to elucidate the dynamic process of gain, loss, and rearrangement that gave rise to the different domain architectures observed among these related proteins. Apparently, the ancestral state consisted of a single TMD-ABC domain arrangement, and repeated TMD-ABC units, such as seen for CFTR and all other closely related ABCC proteins, were generated by duplication independently on three occasions (Fig. S3B). In addition, the ABCE and ABCF families appear to have independently lost their TMD domains whereas the ABCA family gained a unique TMD that was subsequently duplicated along with its adjacent ABC domain. The ABCG family retained a single pair of domains state but rearranged their order to ABC-TMD.
Divergence Between CFTR and the ABCCs.
The CFTR protein is phylogenetically nested among a large group of related ABCC proteins that have demonstrated transporter activity (Fig. S2). Therefore, some specific sequence-structural features must have evolved along the CFTR lineage to enable its functional divergence into an ion channel. We used an analysis of position-specific amino acid sequence variation to search for the sites that distinguish CFTR proteins from their most closely related ABCC paralogs. ABCC4 was determined to be the closest paralog to CFTR by using a BLASTP search of the human proteome with CFTR as the query. The e-value for the similarity between CFTR and ABCC4 is 4e-155; the e-value for the next closest paralog, ABCC5, is 2e-80. CFTR and ABCC4 are also sister taxa in the phylogeny relating all human ABC proteins (Fig. S2).
Changes in selective constraints on protein sequences have been used to evaluate the functional divergence of gene families, and two types (I and II) of functional divergence have been described (Fig. S4A) (14, 15). Type I divergence occurs shortly after gene duplication because of changes in selective constraint at individual sites and is characterized by site-specific changes in evolutionary rates between paralogous groups. This results in amino acid sites that are highly conserved in one paralogous group of proteins and highly diverged in the other. Type II divergence occurs in the late phase after gene duplication when group specific functions undergo positive selection, and results in changes to paralogous group-specific amino acid properties at individual sites. Type II divergence is exemplified by alignment positions that: 1) are completely conserved (identical) within paralogous groups, and 2) have amino acids with radically different properties between paralogous groups, for example, positively versus negatively charged residues. Such sites are most likely to give rise to paralogous group-specific functional properties; accordingly, the approach we used to illuminate functionally relevant evolutionary changes between CFTR and other ABCCs relies on the identification of these so-called type II sites (16). It should be noted that this approach is fundamentally distinct from more straightforward techniques that use evolutionary conservation alone as a surrogate for functional relevance.
An alignment of 20 orthologous vertebrate CFTR proteins with 18 orthologous ABCC4 proteins (Fig. S4B) was broken down into the four domains: TMD1, ABC1, TMD2, and ABC2, and position-specific sequence differences within and between groups (CFTR vs. ABCC4) were evaluated for evidence of type I and type II divergence. All four domains show statistically significant type I divergence, and three of four domains show significant type II divergence (Table 1). Overall, the signal of type I divergence is stronger than that for type II divergence. This indicates that the so-called early phase of duplicate evolution continued to occur long after the initial duplication that gave rise to the two paralogous CFTR and ABCC4 groups, and suggests that site-specific evolutionary rate changes occur more frequently because of relaxation of selective constraint rather than by adaptive fixation of variants. Nevertheless, sites that have undergone type II divergence are more likely to encode group specific functional properties because they are clearly adaptive in one or both lineages.
Table 1.
Type I and type II sequence divergence between CFTR and ABCC4 domains
| Type I and type II sequences | |||||
|---|---|---|---|---|---|
| Vertebrates | |||||
| Type I divergence | |||||
| Domain | Residues | θ | SE | z | P |
| TMD1 | 281 | 0.70 | 0.07 | 9.65 | 5.1e-22 |
| ABC1 | 190 | 0.53 | 0.08 | 6.57 | 4.9e-11 |
| TMD2 | 300 | 0.62 | 0.06 | 10.98 | 4.7e-28 |
| ABC2 | 185 | 0.79 | 0.12 | 6.69 | 2.3e-11 |
| Type II divergence | |||||
| Domain | Residues | θ | SE | z | P |
| TMD1 | 281 | 0.49 | 0.08 | 6.42 | 1.3e-10 |
| ABC1 | 190 | 0.16 | 0.10 | 1.68 | 0.09 |
| TMD2 | 300 | 0.41 | 0.08 | 5.14 | 2.8e-7 |
| ABC2 | 185 | 0.60 | 0.08 | 7.25 | 4.1e-13 |
| Mammals | |||||
| Type I divergence | |||||
| Domain | Residues | θ | SE | z | P |
| TMD1 | 0.64 | 0.11 | 5.83 | 5.6e-9 | |
| ABC1 | 0.82 | 0.15 | 5.45 | 5.1e-8 | |
| TMD2 | 0.56 | 0.08 | 7.32 | 2.4e-13 | |
| ABC2 | 0.51 | 0.17 | 3.03 | 2.4e-3 | |
| Type II divergence | |||||
| Domain | Residues | θ | SE | z | P |
| TMD1 | 0.57 | 0.05 | 12.47 | 1.1e-35 | |
| ABC1 | 0.42 | 0.05 | 7.73 | 1.1e-14 | |
| TMD2 | 0.46 | 0.05 | 9.17 | 4.6e-20 | |
| ABC2 | 0.57 | 0.06 | 10.40 | 2.6e-25 | |
To further evaluate the role of adaptive divergence in the functional evolution of CFTR, we confined the type II divergence analysis to mammalian species only (Table 1). All four domains show statistically significant type II divergence in mammals, and the significance of type II divergence for mammals is greater than that for all vertebrates across all four domains. This is the opposite of what is seen for type I divergence, the significance of which decreases across all four domains when only mammals are considered. Furthermore, the number of individual type II sites increased substantially, for example, from 12 to 46 in TMD1, when mammals were considered alone. These data indicate that the CFTR and ABCC4 groups have continued to evolve functional refinements after mammals diverged from the other vertebrates. The increase of type II and decrease of type I divergence among mammals, compared with all vertebrates, is consistent with the more prominent role of adaptive evolution in the late phase after duplication, as proposed by Gu (15).
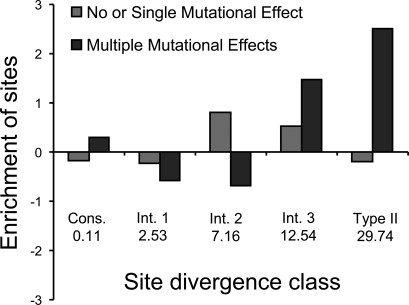
The high type II divergence value (θ) for TMD1 was particularly interesting in light of experimental results, reported here and elsewhere (see Table S1), pointing to the functional relevance of specific TMD1 residues for ion channel activity. Across all vertebrates, there are 12 individual type II sites in TMD1 that are completely conserved within groups (CFTR vs. ABCC4) and radically different between them; these are the positions that are likely to contribute most substantially to the functional differences between groups. The characteristic amino acid residues and biochemical changes for each of these positions are shown in Table S2. We explored the functional relevance of these type II sites in TMD1 by comparing their identity to a list of sites with known mutational effects that was compiled from the CFTR literature (Table S1). To do this, individual sites were broken down into four divergence site classes based on the posterior probability value that indicates the likelihood of a site to contribute to type II divergence between groups (Table S3). For each of the four divergence site classes in TMD1, across all vertebrates, we calculated the observed versus expected proportion of residues in that class that had 0 or 1 mutational effects and compared these with the class-specific proportions of residues that have multiple mutational effects (Fig. 2). Type II sites are the most substantially enriched class for residues that have multiple mutational effects, and intermediate class 3 sites, which are also highly conserved within and are radically different between groups (Table S3), are also highly enriched for residues that have multiple mutational effects. Residues with 0 or 1 mutational effects are most overrepresented in the intermediate class 2, which does not have a consistent pattern of within and between group variation. The enrichment distributions of the different mutational effect profiles are significantly different (χ2 = 15.5, df = 4, P = 3.8e-3). These patterns demonstrate the potential utility of type II sites for predicting functionally important CFTR sites. The predictive power of type II sites is underscored by the greater enrichment of sites with multiple mutational effects at type II versus absolutely conserved sites, which are typically used for functional predictions (Fig. 2). The functional relevance of type II sites is further supported by the finding that the number of known mutational effects per site is positively correlated with the average type II posterior probability, that is, the average type II divergence, per site (r = 0.16, t = 2.6, df = 279, P = 8.8e-3).
Fig. 2.
Type II sequence divergence patterns compared with known mutational effects for the first transmembrane domain (TMD1) of CFTR. Analysis of conservation within and between paralogous groups (CFTR vs. ABCC4) yields five divergence bins ranging from sites that are conserved within and between groups to the type II divergence sites that are maximally divergent between groups and conserved within groups (see Table S3). The 5 bins are: Conserved, Intermediate Class 1, Intermediate Class 2, Intermediate Class 3, and Type II Divergent. The figure shows the observed (O) versus expected (E) proportions [calculated as (O − E)/E] of sites in each divergence bin with 0–1 known mutational effects (light gray bars) versus those with 2 or more known mutational effects (dark gray bars). Values beneath the bars are the average type II posterior probability per bin.
Example of a Divergent Site with Physiological Relevance.
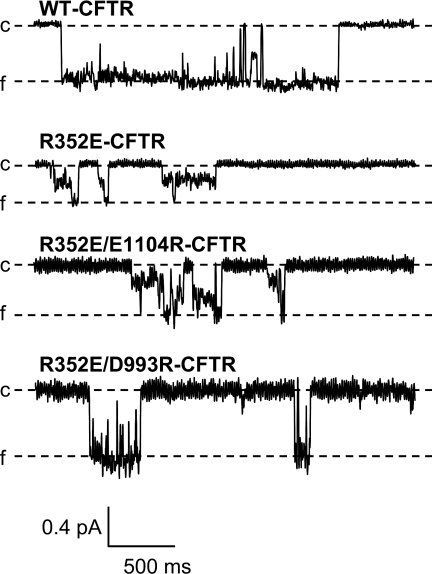
Within TMD1, one of the type II sites is R352 (L842 in ABCC4) (posterior probability = 29.74, Table S2). Our recent studies identified R352 as contributing to a pair of residues within the transmembrane domains of CFTR that interact, perhaps by forming a salt bridge (13). Here, we confirm that the interaction between R352 in TM6 and D993 in TM9 is critical to the maintenance of a stable open-channel conformation (Fig. 3). Mutations at R352 are also associated with CF. Disruption of this salt bridge, by charge-destroying mutations at either site in the interacting pair, altered several properties of open CFTR channels, including unitary conductance, ion selectivity, and susceptibility to pore blockade by organic drugs (13). In contrast, approximately wild-type channel behavior is retained in R352K-CFTR and the charge-swapping double mutant, R352E/D993R-CFTR (Fig. 3). R352 and D993 are conserved within CFTR across all species for which the polypeptide sequence is known. However, R352 is a type II site, divergent between CFTRs and ABCC4s, whereas D993 is conserved between CFTRs and ABCC4s (i.e., type II posterior probability = 0). These results suggest that the interaction between R352 and D993 may be critical to the evolution of channel activity in CFTR.
Fig. 3.
Charge-destroying mutations at R352 alter CFTR single channel behavior. Isolated bursts of channel activity from oocytes expressing WT-CFTR, R352E-CFTR, R352E/E1104R-CFTR, and R352E/D993R-CFTR. Channels were studied in excised, inside-out patches in the continuous presence of 1 mM ATP and 50 U/ml PKA. In each trace, the closed level (c) is marked by a dashed line, and downward deflections represent channel opening. The full conductance level (f) for each trace is indicated. Channels bearing the R352E mutation, or the double mutant R352E/E1104R, exhibited frequent transitions to subconductance levels. In contrast, WT-CFTR channels, and channels bearing the revertant mutation R352E/D993R, primarily exhibit transitions to the full conductance level.
Structural Analysis.
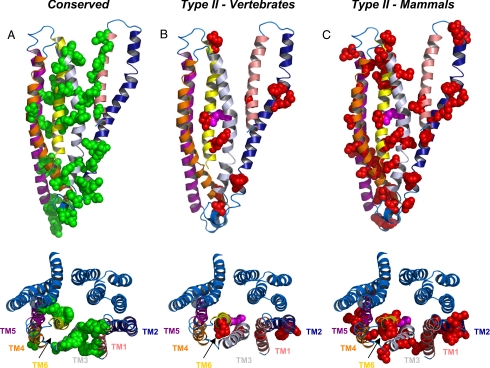
Sequence analysis of the CFTR and ABCC4 proteins revealed many residues that either are completely conserved between protein families, or show type II divergence within TMD1. To glean some insights into the roles that these residues may play, we mapped these sites onto TMD1 of the recently published CFTR homology model (Figs. 4 and S5) (17). All specific sites are shown in space-filling representation, with conserved sites in green and sites exhibiting type II divergence in red, R352 in magenta.
Fig. 4.
Structural environment of conserved and type II divergent CFTR residues. Conserved and type II residues are highlighted on a homology model of the CFTR protein. Helices are colored as follows: TM1, salmon; TM2, dark blue; TM3, silver; TM4, orange; TM5, purple; and TM6, yellow. TMD2 is shown as blue helices. (A) TMD1 of the CFTR homology model with residues conserved between CFTR and ABCC4 shown in green space-filling representation. (B) TMD1 of CFTR with type II divergent residues across all vertebrates shown in red space-filling representation. (C) TMD1 of CFTR with type II divergent residues in mammals only shown in red space-filling representation. Lower images are the view from the extracellular face. R352 is shown in magenta.
In TMD1, most of the conserved sites are located in TMs 1, 3, and 6, with a number of sites conserved in the linker regions between the ABC domains and the TMDs (Fig. 4A). The latter group is thought to comprise residues that are involved in the transmission of conformational changes from the ABC domains to the pore domain, which is likely to be a conserved mechanism between transporters and channels in the ABC superfamily, including the sites shown to interact with F508 (F495 in ABCC4), the highly conserved residue that is deleted in many CF patients (3). A top-down view of the protein shows that in TM6, with the exception of F337 (F352 in ABCC4), the conserved residues seem to be mostly involved in helix packing at the putative interfaces between TMs 1 and 3 and between TMs 5 and 6. Interestingly, the interaction interface between TMs 3 and 6 is entirely divergent. This suggests that this interface may be an important point of divergence between CFTR and ABCC4, and may be a critical region of the pore domain for CFTR channel function. Site R347 in TM6 was shown to form a salt-bridge with D924 in TM8 (18); both sites are conserved among CFTRs, suggesting that this interacting pair may contribute to structural stabilization within the channel cluster. However, although R347 is also conserved in ABCC4 (as R362), the position in ABCC4 equivalent to D924 (position 779) shows type I divergence.
Analysis of type II divergent sites in TMD1 also yields a number of insights. Unlike the conserved sites, the type II sites are mostly located within just two transmembrane helices, TMs 2 and 6 (Fig. 4 B and C). Unlike the conserved sites within TM2, which are mostly localized to the linker region, the type II sites in TM2 are spread throughout the helix, two are located within the linker region. Four of the 12 type II sites in TMD1, identified from analysis of vertebrate proteins, are located within TM6; these encompass residues that are known to be critical for CFTR channel function, including R334 (S349 in ABCC4) and R352 (L367) (Table S2). R334 is a critical basic residue generally thought to be involved in providing a net positive electrostatic potential at the outer mouth of CFTR, and would be involved in attraction of chloride to the channel pore (19, 20). In the model, this residue is located at the extracellular end of TM6 in such a position that it may play the aforementioned role. Toward the cytoplasmic end of the pore, R352 interacts functionally with D993, at least in the open channel state (13). In the current homology model, which is thought to represent the closed state of the channel, R352 points away from TM9, although R352 and D993 are predicted to be at similar positions within the plane of the membrane (Fig. 5). However, in the process of opening, it is likely that TM6 rotates, such that R352 may be brought in closer proximity to D993. This interaction may therefore be involved in stabilization of the pore structure during the open state and would promote ion conduction through the open channel. Such rotation of TM6 would also bring T338 and S341 closer to the pore axis; these residues are thought to make important contributions to open channel function (21–23). Interestingly, analysis of divergence of CFTR from ABCC4 within only mammalian species increases the number of type II sites to 46, which are spread throughout TMD1 (Fig. 4C). Additional sites within TM6, including T338 (V353 in ABCC4) and T339 (A354), have also been shown to be critical mediators of chloride conductance through CFTR. This analysis suggests that CFTR channel activity evolved, at least in part, because of the ability to lock the transport pathway into a channel-like conformation, open at both ends, via formation of the R352-D993 interaction; it then continued to evolve by selecting for residues that could optimize chloride channel function.
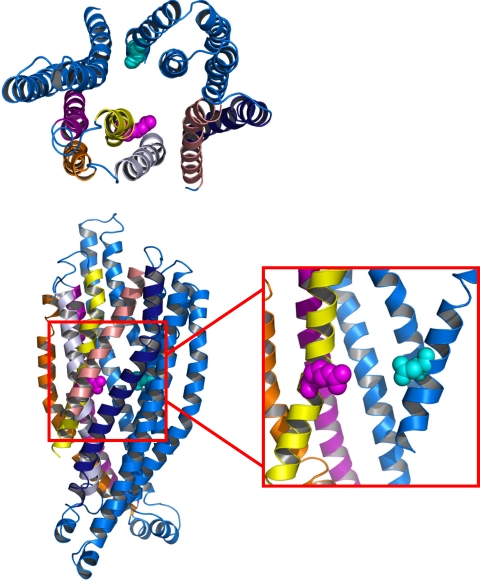
Fig. 5.
Structural environment of the R352-D993 salt bridge of CFTR shown in top-down (Upper) and side view (Lower). R352 (magenta) lies on the same plane as D993 (cyan), as can be seen in the blown up section contained within the red box, from which multiple helices were removed for clarity.
Discussion
In this study, we used comparative sequence, structure, and experimental analyses to identify sites of type II divergence between CFTR, an ion channel, and its closest relative, ABCC4, which functions as a transporter. It is precisely these type II sites, conserved within but divergent between these two groups, rather than the completely conserved sites, that we expect to be most relevant to ion channel specific function of CFTR. Type II sites and conserved sites are distributed throughout the protein (Movie S1 and Movie S2). Comparison of type II sites, identified by computational approaches, with specific experiments and structural features of CFTR further underscores their functional relevance; for example, mutation of R352, identified as a type II residue, led to destabilization of the open state critical to CFTR channel function. This approach may be useful in the design of experiments to identify the mechanisms by which CFTR, alone within the ABC superfamily, evolved to function as an ion channel.
Materials and Methods
CFTR Sequence Analysis.
The CFTR protein sequence (RefSeq accession NP 000483.3) was used as a query in BLASTP (24) searches against the GenBank database (25) to identify 47 human paralogous members of the ABC transporter superfamily, along with CFTR and ABCC4 orthologs from a variety of vertebrate species including primates, more distantly related mammals, amphibians, birds, and fish. Information on all sequences analyzed in this study can be found in Table S4. Sequences were aligned by using the T-COFFEE program (26) and phylogenetic analysis was conducted by using the program MEGA (27). Protein domain architectures for sequences were characterized by using the Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/) (28).
CFTR Functional Divergence.
The evolutionary determinants of the functional differences between CFTR and paralogous ABC transporter proteins were evaluated by using the program DIVERGE (16). DIVERGE was used to identify sites of type II functional divergence, which occurs via changes in the biochemical properties of amino acids at specific positions between groups of related proteins. The rationale behind this approach to identification of sites of divergence, and the kinds of sites it identifies, is illustrated in Fig. S5. Type II functional divergence was evaluated by comparing 20 orthologous CFTR proteins to 18 ABCC4 orthologs (Fig. S4B). The strength of type II functional divergence was measured by the values of the parameter θ, where a θ value significantly greater than zero indicates functional divergence. For CFTR domains showing a statistically significant value of θ, position-specific amino acid differences that underlie the functional divergence of CFTR from paralogous ABC transporter proteins were determined.
Structural analysis of conserved and type II divergent residues was performed on the recently published CFTR homology model (17) obtained from http://dokhlab.unc.edu/research/CFTR/. The CFTR model was visualized by using Pymol software (http://pymol.sourceforge.net/). Specific residues are shown in space filling representation whereas backbone atoms are shown in diagram representation. All images were rendered in Pymol and resized for publication by using Adobe Photoshop v 8.0. Movie S1 and Movie S2 were created by using VideoMach software.
Functional Analysis.
For single-channel recording, CFTR was expressed in Xenopus oocytes and studied in excised, inside-out patches at room temperature, as described (13, 29). Details are provided in the SI Text.
Supplementary Material
Acknowledgments.
We thank H.C. Hartzell for critically reading the manuscript. This work was supported by National Institutes of Health Grant DK-056481 (to N.A.M.), Cystic Fibrosis Foundation Grant MCCART07P0 (to N.A.M.), and the School of Biology, Georgia Institute of Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806306105/DCSupplemental.
References
- 1.Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- 2.Gadsby DC, Nairn AC. Regulation of CFTR channel gating. Trends Biochem Sci. 1994;19:513–518. doi: 10.1016/0968-0004(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 3.Guggino WB, Stanton BA. New insights into cystic fibrosis: Molecular switches that regulate CFTR. Nat Rev Mol Cell Biol. 2006;7:426–436. doi: 10.1038/nrm1949. [DOI] [PubMed] [Google Scholar]
- 4.Dassa E, Bouige P. The ABC of ABCS: A phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol. 2001;152:211–229. doi: 10.1016/s0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- 5.Price MP, Ishihara H, Sheppard DN, Welsh MJ. Function of Xenopus cystic fibrosis transmembrane conductance regulator (CFTR) Cl channels and use of human-Xenopus chimeras to investigate the pore properties of CFTR. J Biol Chem. 1996;271:25184–25191. doi: 10.1074/jbc.271.41.25184. [DOI] [PubMed] [Google Scholar]
- 6.Scott-Ward TS, et al. Chimeric constructs endow the human CFTR Cl- channel with the gating behavior of murine CFTR. Proc Natl Acad Sci USA. 2007;104:16365–16370. doi: 10.1073/pnas.0701562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang ZR, Cui G, Zeltwanger S, McCarty NA. Time-dependent interactions of glibenclamide with CFTR: Kinetically complex block of macroscopic currents. J Membr Biol. 2004;201:139–155. doi: 10.1007/s00232-004-0712-9. [DOI] [PubMed] [Google Scholar]
- 8.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell P. A general theory of membrane transport from studies of bacteria. Nature. 1957;180:134–136. doi: 10.1038/180134a0. [DOI] [PubMed] [Google Scholar]
- 10.Miller C. ClC chloride channels viewed through a transporter lens. Nature. 2006;440:484–489. doi: 10.1038/nature04713. [DOI] [PubMed] [Google Scholar]
- 11.Zifarelli G, Pusch M. CLC chloride channels and transporters: A biophysical and physiological perspective. Rev Physiol Biochem Pharmacol. 2007;158:23–76. doi: 10.1007/112_2006_0605. [DOI] [PubMed] [Google Scholar]
- 12.Gao L, Kim KJ, Yankaskas JR, Forman HJ. Abnormal glutathione transport in cystic fibrosis airway epithelia. Am J Physiol. 1999;277:L113–118. doi: 10.1152/ajplung.1999.277.1.L113. [DOI] [PubMed] [Google Scholar]
- 13.Cui G, Zhang ZR, O'Brien AR, Song B, McCarty NA. Mutations at Arginine 352 Alter the Pore Architecture of CFTR. J Membr Biol. 2008;222:91–106. doi: 10.1007/s00232-008-9105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu X. Statistical methods for testing functional divergence after gene duplication. Mol Biol Evol. 1999;16:1664–1674. doi: 10.1093/oxfordjournals.molbev.a026080. [DOI] [PubMed] [Google Scholar]
- 15.Gu X. Maximum-likelihood approach for gene family evolution under functional divergence. Mol Biol Evol. 2001;18:453–464. doi: 10.1093/oxfordjournals.molbev.a003824. [DOI] [PubMed] [Google Scholar]
- 16.Gu X. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol Biol Evol. 2006;23:1937–1945. doi: 10.1093/molbev/msl056. [DOI] [PubMed] [Google Scholar]
- 17.Serohijos AW, et al. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci USA. 2008;105:3256–3261. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotten JF, Welsh MJ. Cystic fibrosis-associated mutations at arginine 347 alter the pore architecture of CFTR. Evidence for disruption of a salt bridge. J Biol Chem. 1999;274:5429–5435. doi: 10.1074/jbc.274.9.5429. [DOI] [PubMed] [Google Scholar]
- 19.Dawson DC, Liu X, Zhang Z-R, McCarty NA. In: The CFTR Chloride Channel. Kirk K, Dawson DC, editors. Georgetown: Landes Publishing; 2003. pp. 1–34. [Google Scholar]
- 20.Smith SS, et al. CFTR: Covalent and noncovalent modification suggests a role for fixed charges in anion conduction. J Gen Physiol. 2001;118:407–431. doi: 10.1085/jgp.118.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Zhang ZR, Fuller MD, Billingsley J, McCarty NA, Dawson DC. CFTR: A cysteine at position 338 in TM6 senses a positive electrostatic potential in the pore. Biophys J. 2004;87:3826–3841. doi: 10.1529/biophysj.104.050534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarty NA, Zhang ZR. Identification of a region of strong discrimination in the pore of CFTR. Am J Physiol Lung Cell Mol Physiol. 2001;281:L852–867. doi: 10.1152/ajplung.2001.281.4.L852. [DOI] [PubMed] [Google Scholar]
- 23.McDonough S, Davidson N, Lester HA, McCarty NA. Novel pore-lining residues in CFTR that govern permeation and open-channel block. Neuron. 1994;13:623–634. doi: 10.1016/0896-6273(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank Nucleic Acids Res. 2008;36:D25–30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 28.Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuller MD, et al. State-dependent inhibition of cystic fibrosis transmembrane conductance regulator chloride channels by a novel peptide toxin. J Biol Chem. 2007;282:37545–37555. doi: 10.1074/jbc.M708079200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.